Microglial depletion disrupts normal functional development of adult-born neurons in the olfactory bulb
Figures
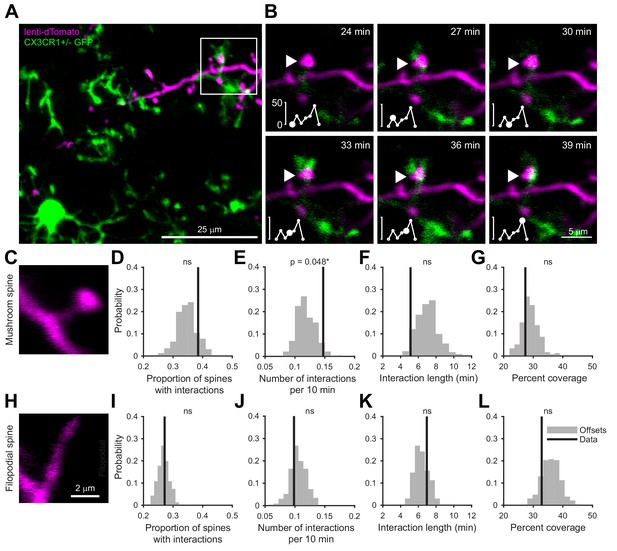
Microglia preferentially interact with mushroom spines on developing abGCs.
(A) Maximum intensity projection (10 µm volume at the first imaging timepoint) showing dTomato-labeled abGCs in a CX3CR1-GFP heterozygous mouse imaged 4 weeks after lentivirus injection. Brightness and contrast adjusted for display only. (B) Single plane time series showing the region marked in (A). Inset shows the calculated percent microglial coverage for the spine marked with the arrowhead (images shown for six frames, 7th frame not shown but the value is plotted, showing the end of the interaction) with the larger circle marking the value for the corresponding frame. Brightness and contrast adjusted with the same parameters for each timepoint for display only. (C) Single plane image showing an example of a spine classified as a mushroom spine because the spine head is wider and brighter than the spine neck. (D) Probability distribution showing the proportion of mushroom spines with at least one microglial interaction for mushroom spines in the real data (black line, ‘Data’) compared to values calculated from iteratively translating the microglial channel relative to the dendritic imaging channel (gray histogram, ‘Offsets’). The proportion of spines with interactions was not significantly higher than chance (one-tailed permutation test, p=0.13). (E) Probability distribution showing the number of interactions (normalized to 10 min) for mushroom spines. The mean number of interactions (value for each dendrite is the mean number across all mushroom spines on that dendrite) in the real data was significantly higher than chance (one-tailed permutation test, p=0.048). (F) Probability distribution showing interaction length for mushroom spines (for spines that had at least one frame that met the criteria for an interaction, see Materials and methods). The mean interaction length across all dendrites (value for each dendrite is the mean interaction length across all interactions for all mushroom spines) was not higher than chance (one-tailed permutation test, p=0.96). (G) Probability distribution showing maximum percent coverage (mean across all interactions for a given spine for spines that had at least one frame that met the criteria for an interaction). The mean maximum percent coverage across all dendrites (value for each dendrite is the mean interaction length across all interactions for all mushroom spines) was not higher in the real data (one-tailed permutation test, p=0.72). (H) Single plane image of a spine classified as a filopodial spine because it has no clear spine head. (I) Probability distribution showing the proportion of spines with at least one microglial interaction for filopodial spines. The proportion of filopodial spines with interactions was not significantly higher than chance (one-tailed permutation test, p=0.39). (J) Probability distribution showing the number of interactions (normalized to 10 min) for filopodial spines. The mean number of interactions (value for each dendrite is the mean number across all filopodial spines on that dendrite) in the real data was not significantly higher than chance (one-tailed permutation test, p=0.72). (K) Probability distribution showing interaction length for filopodial spines (for spines that had at least one frame that met the criteria for an interaction, see Materials and methods). The mean interaction length across all dendrites (value for each dendrite is the mean interaction length across all interactions for all filopodial spines) was not significantly higher than chance (one-tailed permutation test, p=0.23). (L) Probability distribution showing maximum percent coverage (mean across all interactions for a given spine for spines that had at least one frame that met the criteria for an interaction). The mean maximum percent coverage across all dendrites (value for each dendrite is the mean interaction length across all interactions for all filopodial spines) was not higher in the real data (one-tailed permutation test, p=0.84). n = 726 spines (271 mushroom spines and 455 filopodial spines) from 48 dendrites combined at 1, 2, 3, and 4 weeks post injection in three mice. ns, not significant; *p<0.05.
-
Figure 1—source data 1
This spreadsheet contains the values used to create the histograms in Figure 1D–G and I–L.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig1-data1-v1.xlsx
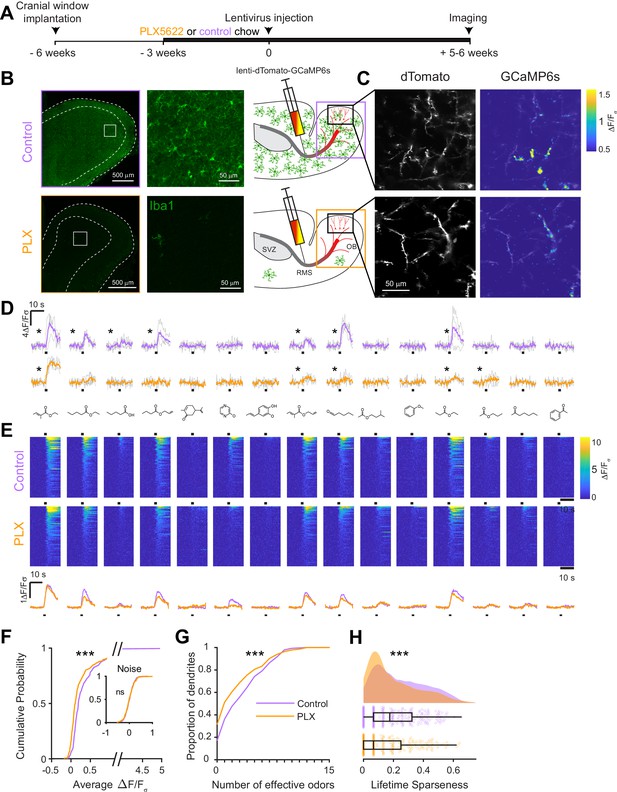
Microglial depletion during development reduces odor-evoked responses of abGCs in anesthetized mice.
(A) Experimental timeline for microglial depletion during development of abGCs. A cranial window was implanted and 3 weeks later mice were given control or PLX5622-containing chow for the remainder of the experiment. After 3 weeks of chow consumption, a lentivirus was injected into the RMS to label abGCs, which were imaged 5–6 weeks later. (B) Left, images of Iba1 staining in the olfactory bulb of control (top) and PLX-treated (bottom) mice. White squares show the locations of the enlarged insets. Dotted lines mark the upper edge of the glomerular and granule cell layers. Right, schematic showing injection of a lentivirus encoding dTomato and GCaMP6s and microglial depletion. (C) Example fields of view showing an average intensity projection of dTomato structural images of abGC dendrites (left) and overlaid heatmaps of GCaMP6s-recorded activity (right) in response to ethyl valerate in control (top) and PLX5622-treated (bottom) mice. (D) GCaMP6s traces showing odor responses of example ROIs from control (top) and PLX-treated (bottom) mice (chosen to have the same ranked response to the first odor). Gray traces represent responses on individual trials and colored trace is the mean across trials. Individual trial traces were median filtered over three frames before averaging for presentation. *, odor responses for which the mean response was above threshold (E) Heatmap traces from the 100 ROIs with the largest odor-evoked Ca2+ signals across all mice ranked separately for each of 15 odors (molecular structures shown above). Black bar denotes odor time. Bottom, mean response time course for each odor across all ROIs. (F) Cumulative distribution showing that the distribution of responses (averaged across odors for each dendrite) is shifted to the left in PLX-treated mice (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.25, p=2.56e-08) while the noise distributions constructed from blank trials are not different (D = 0.042, p=0.96). (G) Cumulative distribution showing the number of effective odors (odors that evoked responses above the ROC threshold 0.39, which was calculated across all dendrites from both groups). The median number of effective odors was significantly lower in the PLX-treated group (Wilcoxon rank sum test, z = 3.86, p=1.15e-04). (H) Raincloud plot showing the distribution of lifetime sparseness across all dendrites. Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per dendrite). Median lifetime sparseness was significantly lower in the PLX-treated group (Wilcoxon rank sum test, z = 3.53, p=4.18e-04). n = 287 dendrites from five control mice and 277 dendrites from 7 PLX-treated mice. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 2—source data 1
This spreadsheet contains values from each dendrite from each mouse for Figure 2F,G and H.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig2-data1-v1.xlsx
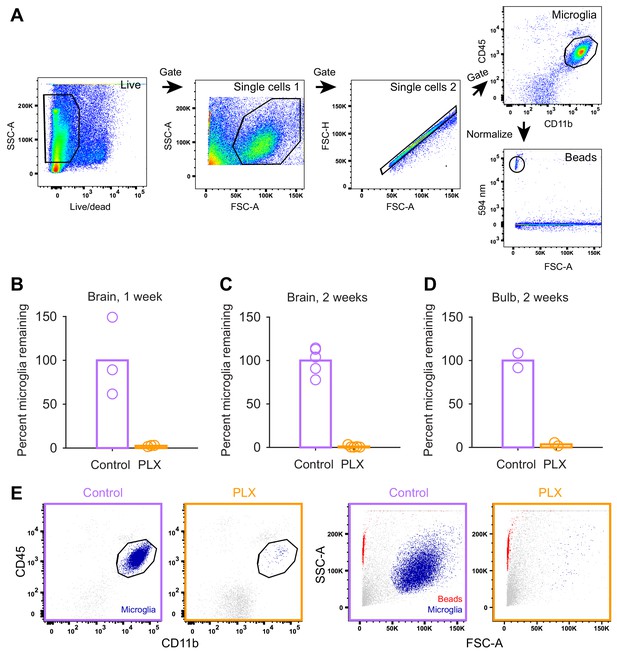
Quantification of microglial depletion with flow cytometry.
(A) Microglial cells were gated on CD45intermediate and CD11bhigh using the following gating strategy: Debris and dead cells were excluded based on the fluorescent intensity of a dead cell stain. Next, single cells where determined based on of their light scattering properties (first by side over forward scatter, SSC-A/FSC-A, and second by forward scatter height over area, FSC-H/FSC-A). The counting beads were identified based on their fluorescent emission at 594 nm. (B) Plot showing the percent of live microglia remaining (normalized to the mean of the controls) in mice treated with PLX compared to controls (number of microglia in each sample normalized to counting beads) in whole brain samples after 1 week of control or PLX diet demonstrating 97.4% ablation. (C) Plot showing the percent of live microglia remaining in mice treated with PLX (normalized to the mean of the controls) compared to controls (number of microglia in each sample normalized to counting beads) in whole brain samples after 1 week of control or PLX diet, demonstrating 99.0% ablation. (D) Plot showing the percent of live microglia remaining in mice treated with PLX (normalized to the mean of the controls) compared to controls measured via flow cytometry in olfactory bulb samples after 2 weeks of control or PLX diet, demonstrating 96.4% ablation. (E) Interrogation of light scattering properties (SSC-A/FSC-A) independent of surface marker labeling revealed that hardly any cells remained in the microglia enriched samples after treatment with PLX compared to control samples. This becomes visually more apparent when microglia cells (CD45intermediate, CD11bhigh, control and PLX-treated plots on left) are backgated to SSC-A/FSC-A plots on right (in dark blue). Note that counting beads can be observed in red. Bars represent the mean across mice (circles). Whole brain: n = 5 control and 5 PLX mice. OB: n = 2 control and 2 PLX samples (each sample contained both OBs from two littermates which were combined after dissection).
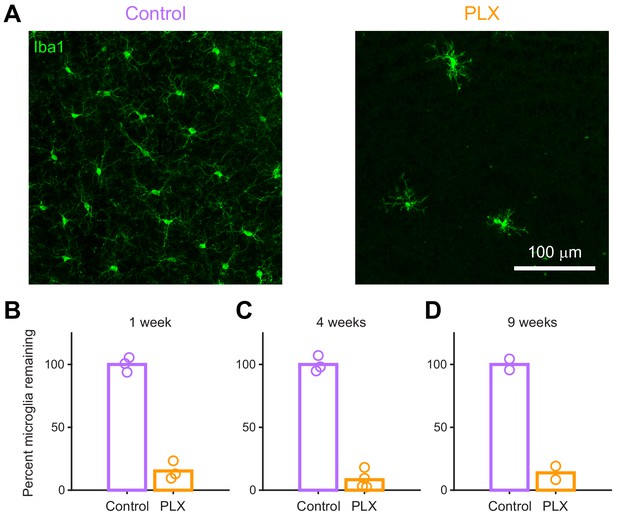
Quantification of microglial depletion with immunohistochemistry.
(A) Maximum intensity projection showing microglia stained with anti-Iba1 in the granule cell layer of the olfactory bulb in control (left) and PLX-treated (right) mice after 4 weeks of treatment. (B) Plot showing the percent of microglia remaining in mice treated with PLX compared to control littermates based on counting of cell bodies stained with Iba1 after 1 week of treatment, demonstrating 84.7% ablation. (C) Percent microglia remaining after 4 weeks of treatment (same mice used for spine quantification in Figure 5), demonstrating 91.7% ablation. (D) Percent microglia remaining after 9 weeks of treatment (PLX mice are the same mice used for imaging at the 9 weeks timepoint in Figure 4—figure supplement 1 and control mice are age-matched controls), demonstrating 86.2% ablation. Bars represent the mean across mice (circles). n = 3 mice (1 week), four mice (4 weeks), and two mice (9 weeks).
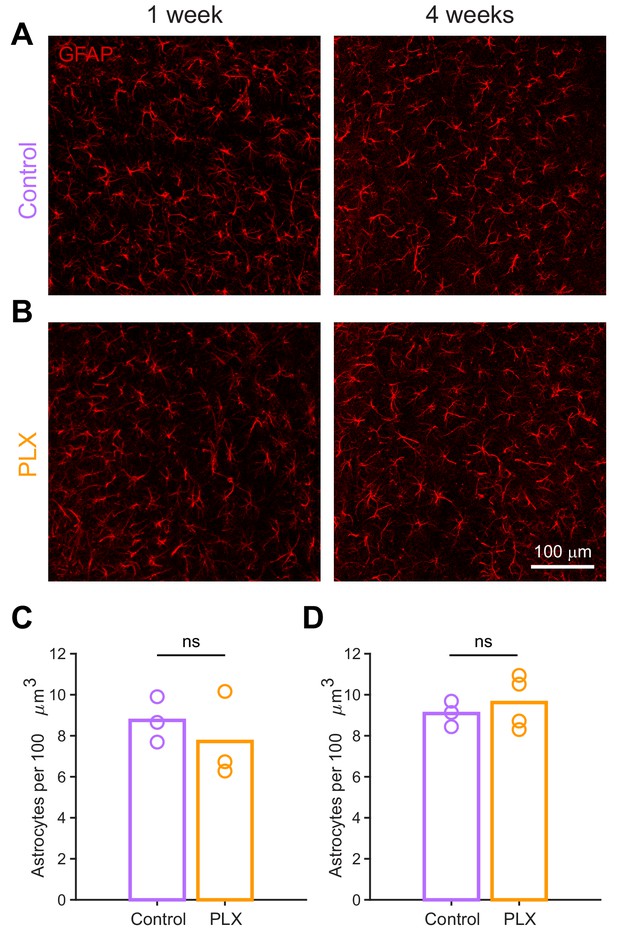
Astrocytic response to microglial depletion.
(A) Maximum intensity projection showing astrocytes stained with anti-GFAP in the granule cell layer of the olfactory bulb in control mice 1 week (left) and 4 weeks (right) after beginning treatment. (B) Maximum intensity projection showing astrocytes stained with anti-GFAP in the granule cell layer of the olfactory bulb in PLX-treated mice 1 week (left) and 4 weeks (right) after beginning treatment. (C) Plot showing the number of astrocytes counted in control versus PLX-treated mice 1 week after the start of treatment. The median numbers were not significantly different between groups (Wilcoxon rank sum test, rank sum = 12, p=0.70). (D) Plot showing the number of astrocytes counted in control versus PLX-treated mice 4 weeks after the start of treatment. The median numbers were not significantly different between groups (Wilcoxon rank sum test, rank sum = 11, p=0.86). Bars represent the median across mice (circles). n = 3 control versus 3 PLX-treated mice (1 week) and 3 control versus 4 PLX-treated mice (4 weeks). ns, not significant.
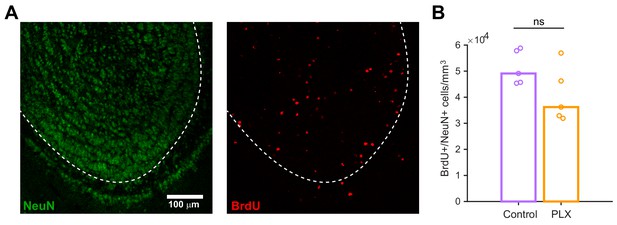
Microglial depletion does not affect the number of maturing abGCs.
(A) Maximum intensity projection showing NeuN staining (left) and BrdU staining (right) in the olfactory bulb. Dotted line indicates boundary of the granule cell layer, where positive cells were quantified. (B) Plot showing the density of BrdU+/NeuN+ cells per mm3 in the granule cell layer of the olfactory bulb in control mice and mice treated with PLX for 4 weeks, beginning 3 days after BrdU injection. The density was not different between groups (Wilcoxon rank sum test, rank sum = 35, p=0.15). Bars represent the median across mice (circles). n = 5 control and 5 PLX-treated mice. ns, not significant.
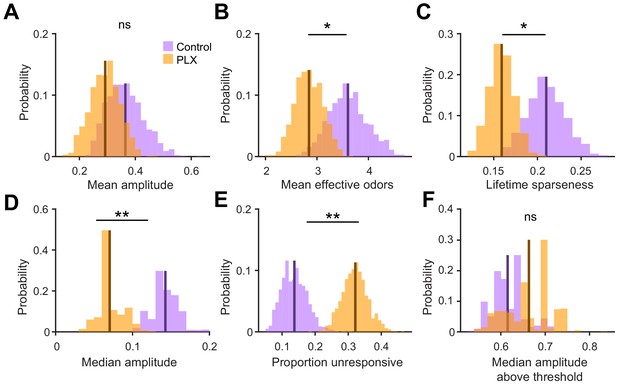
Further analysis of odor responses in abGCs in control versus PLX-treated mice.
(A) Sampling distributions of the mean response amplitude obtained with the hierarchical bootstrap. The mean amplitude (estimated by the mean of the sampling distribution, dark line) were not significantly different between dendrites in control versus PLX-treated mice (hierarchical bootstrap, p=0.21). (B) Sampling distributions of the mean number of effective odors (of the 15-odor panel) evoking a significant response. The mean number of effective odors was significantly greater in control than PLX-treated mice (hierarchical bootstrap, p=0.050). (C) Sampling distributions of the mean lifetime sparseness. The mean lifetime sparseness was significantly greater in control than PLX-treated mice (hierarchical bootstrap, p=0.023). (D) Sampling distributions of the median response amplitude. The median amplitude was significantly higher in control than PLX-treated mice (hierarchical bootstrap, p=0.0011). (E) Sampling distributions of the proportion of dendrites that did not respond to any of the odors. The mean proportion of unresponsive dendrites was significantly greater in PLX-treated mice (hierarchical bootstrap, p=0.0013). (F) Sampling distributions of the median response amplitude, only considering responses that were above the threshold. The median response amplitude above threshold was not different between groups (hierarchical bootstrap, p=0.77). The means of the sampling distributions are shown as dark lines and were used to estimate the value of the parameter (x-axis label) for each group. n = 287 dendrites from 5 control mice and 277 dendrites from 7 PLX-treated mice. ns, not significant; *p<0.05, **p<0.01.
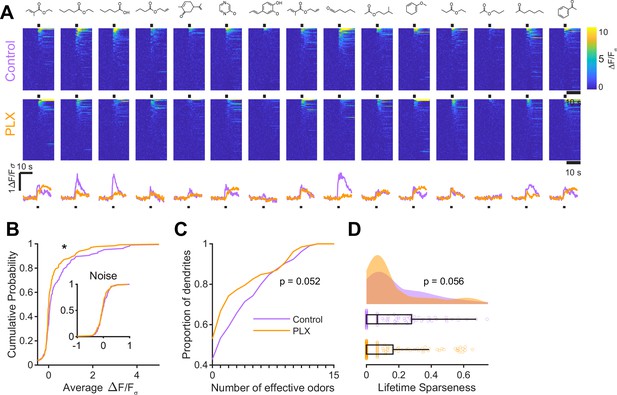
Microglial depletion during development reduces odor-evoked responses of abGCs in awake mice.
(A) Above, Heatmap traces from the 100 ROIs with the largest odor-evoked Ca2+ signals across all mice ranked for each of 15 odors (molecular structures shown above). Below, Mean response time course for each odor across all ROIs. Black bar denotes odor time. (B) Cumulative distribution showing that the distribution of responses (averaged across odors for each dendrite) is shifted to the left in PLX-treated mice (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.18, p=0.037) while the noise distributions constructed from blank trials are not different (D = 0.11, p=0.45). (C) Cumulative distribution showing the number of effective odors (odors that evoked responses above the ROC threshold 0.52, which was calculated across all dendrites from both groups). There was a trend toward a lower median number of effective odors in the PLX-treated group (Wilcoxon rank sum test, z = 1.95, p=0.052). (D) Raincloud plot showing the distribution of lifetime sparseness across all dendrite. Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per dendrite). There was a trend toward lower median lifetime sparseness in the PLX-treated group (Wilcoxon rank sum test, z = 1.91, p=0.056). n = 105 dendrites from three control mice and 132 dendrites from 4 PLX-treated mice. *p<0.05.
-
Figure 3—source data 1
This spreadsheet contains values from each dendrite for Figure 3B,C and D.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig3-data1-v1.xlsx
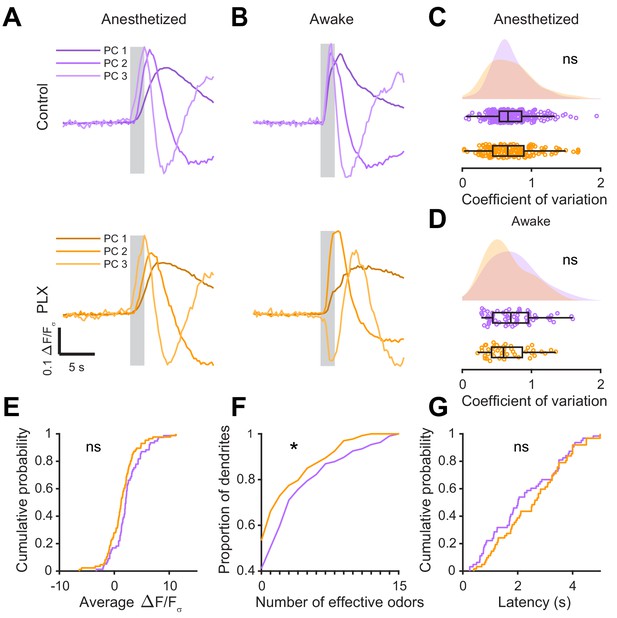
Further analysis of abGC odor responses in awake versus anesthetized mice.
(A) Top, the first three principal components (PCs) obtained from PCA of the temporal profiles of responses of all abGC dendrites (ΔF/Fσ traces, GCaMP6s) in anesthetized control mice to all odors. The x-axis represents time, and the y-axis represents the magnitude of ΔF/Fσ for each PC. Percent variance explained: 36.3%, 4.19%, 1.85% for the first three PCs, respectively. Bottom, The first three PCs for odor responses in anesthetized PLX-treated mice. Percent variance explained: 33.7%, 3.82%, 1.96%. Shading indicates the odor presentation period (2 s). The time courses of responses for the odor analysis period (5 s after odor onset) for the first three PCs was not different between control and PLX-treated mice (permutation test, p=0.41). (B) Top, the first three PCs for odor responses in awake control mice. Percent variance explained: 43.7%, 12.6%, 3.36%. Bottom, the first three PCs for odor responses in awake PLX-treated mice. Percent variance explained: 44.9%, 10.2%, 2.86%. The time courses of responses for the odor analysis period for the first three PCs were significantly different between control and PLX-treated mice (permutation test, p=0.011). (C) Raincloud plot showing the distribution of coefficient of variation values for all dendrites in anesthetized mice averaged across all odors for which the dendrite had a mean response above threshold (ROC thresholds calculated separately for control (threshold = 0.37) and PLX (threshold = 0.5) to ensure the same proportion of true responses). Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per cell, unless the cell had no responses above threshold in which case it is not included). The median CVs were not significantly different (Control: 0.59, PLX: 0.62, Wilcoxon rank sum test, z = 0.58, p=0.56). (D) Raincloud plot showing the distribution of coefficient of variation values for all dendrites in awake mice averaged across all odors for which the dendrite had a response above threshold (ROC thresholds calculated separately for control (threshold = 0.46) and PLX (threshold = 0.85) to ensure the same proportion of true responses). The median CVs were not significantly different (Control: 0.55, PLX: 0.48, Wilcoxon rank sum test, z = 1.47, p=0.14). (E) Cumulative distribution (responses averaged across odors for each dendrite) showing that the distribution of responses detected with event analysis (ROC threshold calculated across both groups combined, event detected during the odor analysis period if six or more frames were at least 2.6 standard deviations above baseline) was not significantly different in PLX-treated mice (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.18, p=0.23). (F) Cumulative distribution showing the number of effective odors (odors for which an event was detected in at least one repetition). The median number of effective odors was lower in the PLX-treated group (Wilcoxon rank sum test, z = 2.09, p=0.037). (G) Cumulative distribution showing that the latency from odor onset to event detection (averaged across odors for each dendrite) was not significantly different in PLX-treated mice (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.17, p=0.31). ns, not significant; *p<0.05.
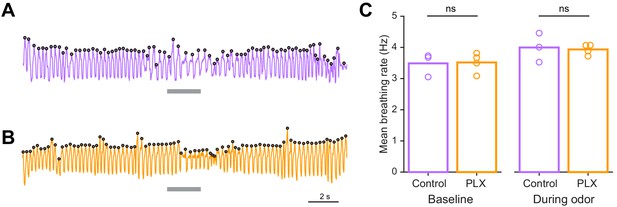
Sniffing rates are not different in control versus PLX-treated mice.
(A) Example trace showing breathing recordings during one trial from a control mouse. The gray bar indicates the time the odor is on and the black circles indicate sniffs that were detected. (B) Example trace showing breathing recordings during one trial from a PLX-treated mouse. (C) Mean breathing rate during baseline periods (10 s before the odor comes on) and during the odor period (2 s) calculated from all sniffs recorded in all trials from each mouse. The breathing rates in the two groups were not different during the baseline period (Two sample t test, t = −0.11, p=0.91) or the odor period (Two sample t test, t = 0.25, p=0.81). Bars represent the mean across mice (circles). n = 3 control mice (Baseline: 15,086 detected sniffs, Odor: 2895 detected sniffs) and 4 PLX-treated mice (Baseline: 19,248 detected sniffs, Odor: 3468 detected sniffs) ns, not significant.
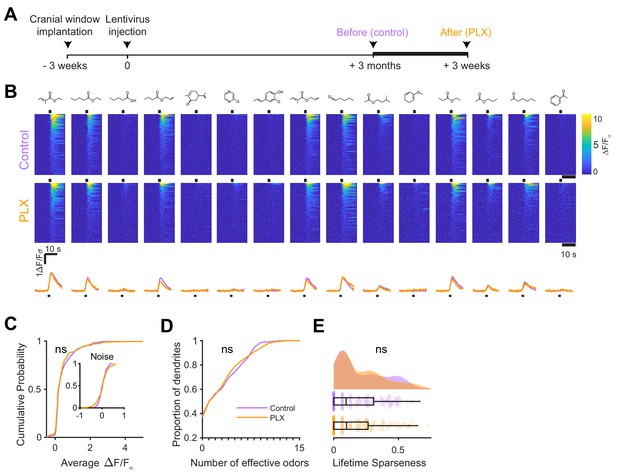
Microglial depletion after development has no effect on odor-evoked responses of abGCs.
(A) Experimental timeline for microglial depletion after development of abGCs. AbGCs were labeled via lentivirus injection and allowed to mature for 3 months. A control imaging session was performed immediately before administration of PLX chow and the second imaging session occurred 3 weeks later. (B) Heatmap traces from the 100 ROIs with the largest odor-evoked Ca2+ signals across all mice ranked for each of 15 odors (molecular structures shown above). Black bar denotes odor time. Bottom, mean response time course for each odor across all ROIs. (C) Cumulative distribution showing that the distribution of responses (averaged across odors for each dendrite) is not different before and after PLX diet administration (Two sample Kolmogorov–Smirnov test, D = 0.087, p=0.45) and the noise distributions constructed from blank trials are also not different (D = 0.11, p=0.16). (D) Cumulative distribution showing the number of effective odors (odors that evoked responses above the ROC threshold 0.53, which was calculated across all dendrites from both groups). The median number of effective odors was not different between groups (Wilcoxon rank sum test, z = 0.0038, p=1.00). (E) Raincloud plot showing the distribution of lifetime sparseness across all dendrite. Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per dendrite). Median lifetime sparseness was not different between groups (Wilcoxon rank sum test, z = 0.18, p=0.87). n = 198 dendrites before and 185 dendrites after 3 weeks of PLX administration from three mice. ns, not significant.
-
Figure 4—source data 1
This spreadsheet contains values from each dendrite for Figure 4C,D and E.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig4-data1-v1.xlsx
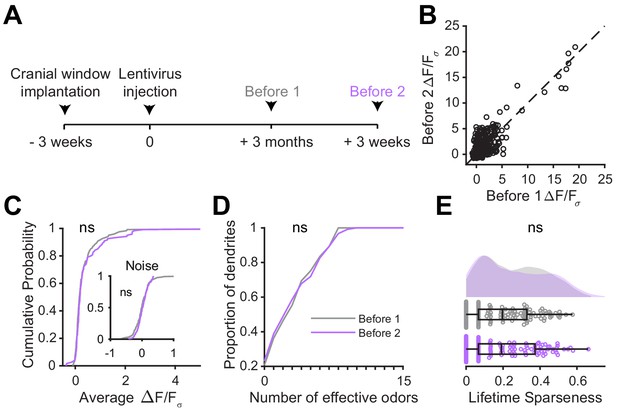
Odor responses are stable in mature abGCs.
(A) Experimental timeline for microglial depletion after development of abGCs. AbGCs were labeled via lentivirus injection and allowed to mature for 3 months. One imaging session was performed and a second was performed 3 weeks later in which the same dendrites were imaged again (the second is the same ‘Control’ imaging session from the main figure). (B) Scatterplot showing the responses of all dendrite-odor pairs for the first and second imaging sessions. Dotted line is unity. Linear correlation coefficient R2 = 0.73, p=0. (C) Cumulative distribution showing that the distribution of responses (averaged across odors for each dendrite) is not different between the two imaging sessions (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.097, p=0.61) and the noise distributions constructed from blank trials are also not different (D = 0.11, p=0.48). (D) Cumulative distribution showing the number of effective odors (odors that evoked responses above the ROC threshold 0.39, which was calculated across all dendrites from both groups). The median number of effective odors was not different between groups (Wilcoxon rank sum test, z = 0.20, p=0.84). (E) Raincloud plot showing the distribution of lifetime sparseness for all dendrites. Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per dendrite). The median lifetime sparseness values were not significantly different (Wilcoxon rank sum test, z = 0.30, p=0.76). n = 121 dendrites from three mice (same mice from the main figure). ns, not significant.
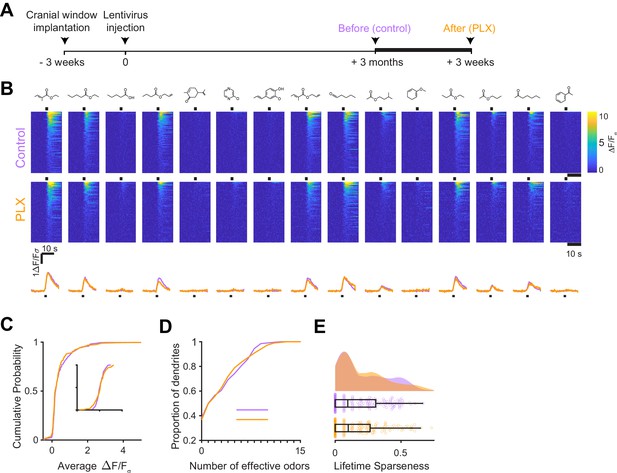
Microglial depletion for a prolonged period after abGC development has no effect on odor-evoked responses of abGCs.
(A) Experimental timeline for microglial depletion after development of abGCs. AbGCs were labeled via lentivirus injection and allowed to mature for 3 months. A control imaging session was performed immediately before administration of PLX chow and the second imaging session occurred 9 weeks later. (B) Heatmap traces from the 100 ROIs with the largest odor-evoked Ca2+ signals across all mice ranked for each of 15 odors (molecular structures shown above). Black bar denotes odor time. Bottom, mean response time course for each odor across all ROIs. (C) Cumulative distribution showing that the distribution of responses (averaged across odors for each dendrite) is not different before and after PLX diet administration (Two sample Kolmogorov–Smirnov test for probability distributions, D = 0.089, p=0.61) and the noise distributions constructed from blank trials are also not different (D = 0.14, p=0.095). (D) Cumulative distribution showing the number of effective odors (odors that evoked responses above the ROC threshold 0.78, which was calculated across all dendrites from both groups). The median number of effective odors was not different between groups (Wilcoxon rank sum test, z = −0.17, p=0.87). (E) Raincloud plot showing the distribution of lifetime sparseness for all dendrites in control and PLX-treated mice. Above, kernel density estimate. Below, boxplot showing the median, interquartile range (box), and 1.5 times the interquartile range (whiskers) superimposed on a dot plot of all the data (one dot per dendrite). Median lifetime sparseness was not different between groups (Wilcoxon rank sum test z = −0.26, p=0.79). n = 168 dendrites before and 122 dendrites after 9 weeks of PLX administration from three mice (one mouse was also included in the 3 weeks group that is shown in the main figure). ns, not significant.
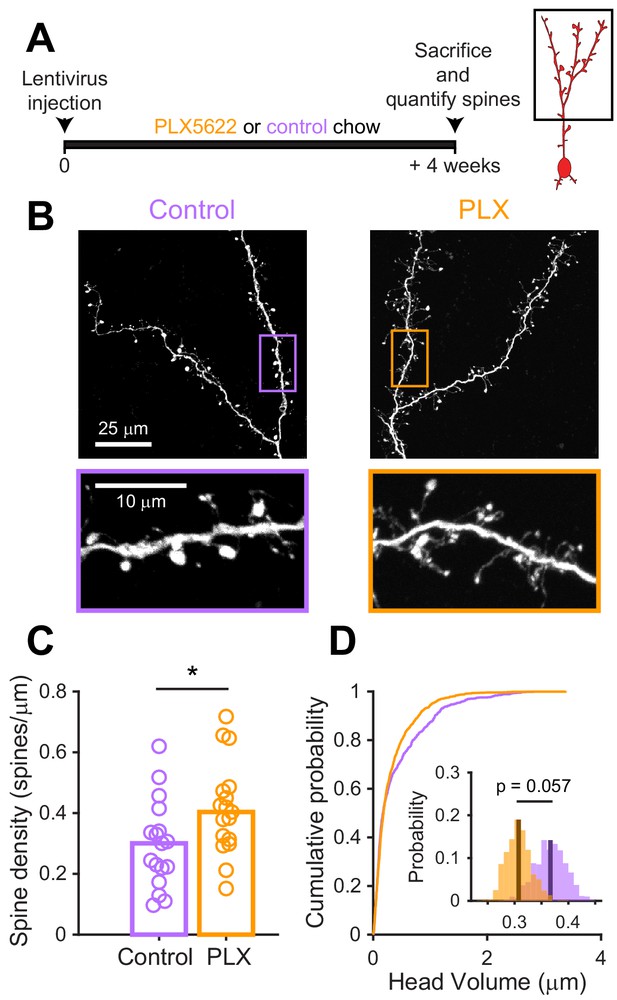
Microglial depletion during development reduces spine head volume in abGCs.
(A) Experimental timeline for microglial depletion during development of abGCs. Mice were given control or PLX5622-containing chow on the same day that a lentivirus was injected into the RMS to label abGCs. Spine numbers and morphology were quantified 4 weeks later. (B) Above, sample images showing two apical dendrites from one cell that were analyzed in a control mouse (left) and PLX-treated mouse (right). Below, insets from the images shown above, showing spine morphology in more detail. (C) Spine density averaged across 1–5 apical dendrites from each abGC. Spine density was higher in PLX-treated mice (Wilcoxon rank sum test, z = −2.07, p=0.039). Bars indicate medians across cells (circles). (D) Cumulative distribution showing the volume of all spines analyzed. Inset, sampling distributions of the mean head volume obtained with the hierarchical bootstrap. The mean head volume (dark lines) was greater in control versus PLX-treated mice (hierarchical bootstrap, p=0.057). n = 810 spines from 17 abGCs from three control mice and 1551 spines from 17 abGCs from 4 PLX mice. *p<0.05.
-
Figure 5—source data 1
This spreadsheet contains spine density values for each dendrite for Figure 5C and values for the head volume of each spine from each dendrite for Figure 5D.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig5-data1-v1.xlsx
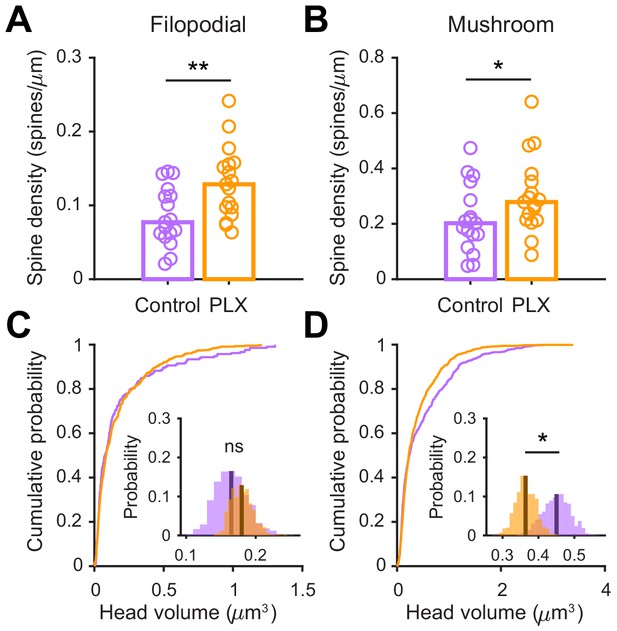
Further analysis of spine density and volume in control versus PLX-treated mice.
(A) Spine density for filopodial spines (defined as spines with spine head width less than 1.5 times the mean width of the spine neck). Spine density was higher in PLX-treated mice (Wilcoxon rank sum test, z = −2.69, p=0.0072). Bars indicate medians across cells (circles). (B) Spine density for mushroom spines (defined as spines with the width of the spine head greater than or equal to 1.5 times greater than the mean width of the spine neck). Spine density was higher in PLX-treated mice (Wilcoxon rank sum test, z = −1.96, p=0.050). Bars indicate medians across cells (circles). (C) Cumulative distribution showing filopodial spine volume. Inset, sampling distributions of the mean filopodial head volume obtained with the hierarchical bootstrap. The mean head volume (dark lines) was not different between groups (hierarchical bootstrap, p=0.72) (D) Cumulative distribution showing mushroom spine volume. Inset, sampling distributions of the mean mushroom head volume obtained with the hierarchical bootstrap. The mean head volume (dark lines) was significantly larger in control mice (hierarchical bootstrap, p=0.036). n = 810 spines from 17 abGCs from three control mice and 1551 spines from 17 abGCs from 4 PLX mice. ns, not significant; *p<0.05; **p<0.01.
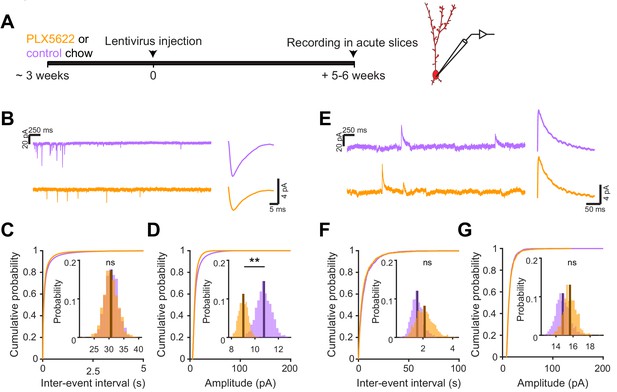
Microglial depletion during abGC development reduces the amplitude of excitatory synaptic currents but does not affect inhibitory synaptic currents.
(A) Experimental timeline for electrophysiological recording in abGCs (B) Left, sample sections from raw traces recorded from abGCs in control (top) and PLX-treated (bottom) mice. Right, median EPSCs across all EPSCs detected from all mice. (C) Cumulative distribution showing the inter-event intervals from all recorded EPSCs. Inset, sampling distributions of the mean frequency obtained with the hierarchical bootstrap. The median frequencies (dark lines) were not significantly different (hierarchical bootstrap, p=0.48). (D) Cumulative distribution showing the amplitudes from all recorded EPSCs. Inset, sampling distributions of the mean amplitude obtained with the hierarchical bootstrap. The mean amplitude (dark line) was significantly higher in control cells (hierarchical bootstrap, p=0.0068). (E) Left, Sample sections from raw traces recorded from abGCs in control (top) and PLX-treated (bottom) mice. Right, median IPSCs across all IPSCs detected from all mice. (F) Cumulative distribution showing the inter-event intervals from all recorded IPSCs. Inset, sampling distributions of the median frequency obtained with the hierarchical bootstrap. The mean frequencies (dark lines) were not significantly different (hierarchical bootstrap, p=0.77). (G) Cumulative distribution showing the amplitudes from all recorded IPSCs. Inset, sampling distributions of the mean amplitude obtained with the hierarchical bootstrap. The mean amplitudes (dark lines) were not significantly different (hierarchical bootstrap, p=0.79). For EPSCs: n = 30 abGCs from four control mice and 33 abGCs from 4 PLX mice. For IPSCs: n = 29 abGCs from four control mice and 30 abGCs from 4 PLX mice (same mice in both cases and cells used for both EPSCs and IPSCs if the recordings met criteria stated in Materials and methods). ns, not significant; **p<0.01.
-
Figure 6—source data 1
This spreadsheet contains the frequency and amplitude values for all detected events from each dendrite for Figure 6C,D,F and G.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig6-data1-v1.xlsx
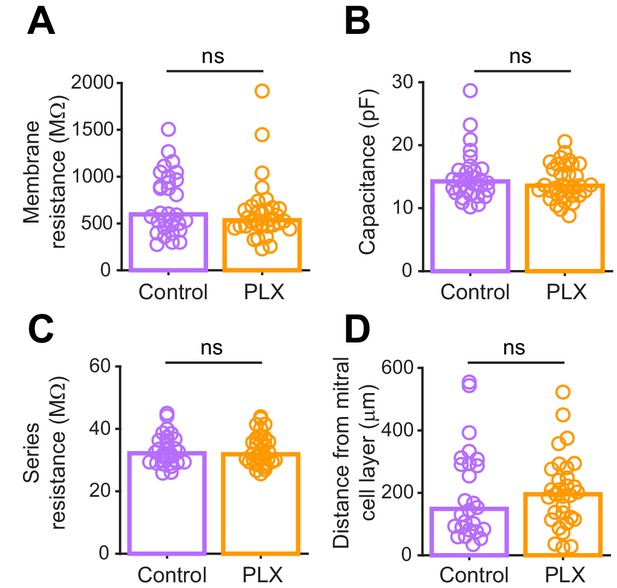
Passive electrophysiological properties and recording conditions are similar for abGCs in control versus PLX-treated mice.
(A) Membrane resistance (mean of measurements taken before and after EPSC recordings for each cell) in abGCs. There was no difference between the two groups (Wilcoxon rank sum test, z = 1.01, p=0.31). (B) Membrane capacitance (mean of measurements taken before and after EPSC recordings for each cell) in abGCs. There was no difference between the two groups (Wilcoxon rank sum test, z = 0.52, p=0.61). (C) Series resistance (mean of measurements taken before and after EPSC recordings) during abGC recordings. There was no difference between the two groups (Wilcoxon rank sum test, z = 0.048, p=0.96). (D) Distance of the cell body from the mitral cell layer (measured in 2D, not taking into account depth from the surface of the slice) for all cells recorded. Both superficial and deep abGCs were included in the dataset and there was no difference in the mean distance to the mitral cell layer between the two groups (Wilcoxon rank sum test, z = −0.52, p=0.60). Bars indicate medians across cells (circles). n = 30 abGCs from four control mice and 33 abGCs from 4 PLX mice. ns, not significant.
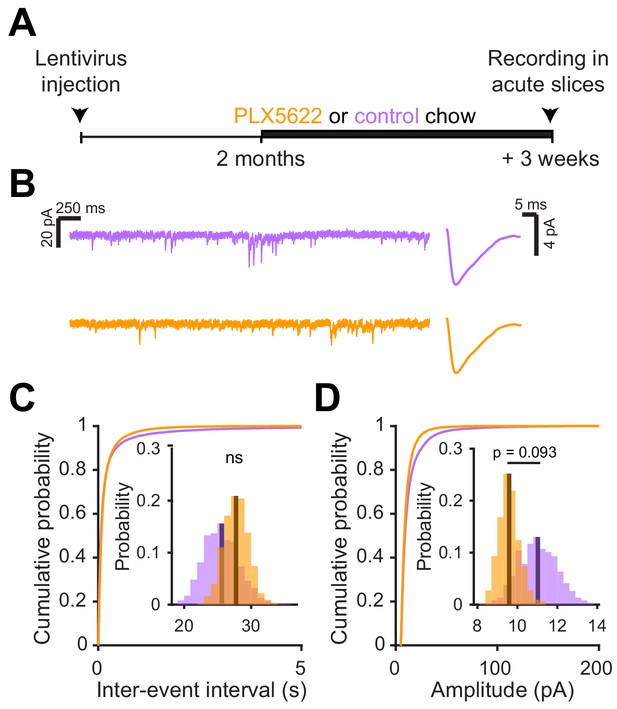
Microglial depletion after abGC development has no effect on excitatory synaptic currents.
(A) Experimental timeline for electrophysiological recordings in abGCs after their development (B) Left, Sample sections from raw traces recorded from abGCs in control (top) and PLX-treated (bottom) mice. Right, median EPSCs detected from all mice. (C) Cumulative distribution showing the inter-event intervals from all recorded EPSCs. Inset, sampling distributions of the mean frequency obtained with the hierarchical bootstrap. The mean frequencies (dark lines) were not significantly different (hierarchical bootstrap, p=0.76). (D) Cumulative distribution showing the amplitude from all recorded EPSCs. Inset, sampling distributions of the mean frequency obtained with the hierarchical bootstrap. The mean amplitudes (dark lines) were not significantly different (hierarchical bootstrap, p=0.093). n = 23 abGCs from three control mice and 27 abGCs from 3 PLX mice. ns, not significant.
-
Figure 7—source data 1
This spreadsheet contains the frequency and amplitude values for all sEPSCs from each dendrite for Figure 7C and D.
- https://cdn.elifesciences.org/articles/50531/elife-50531-fig7-data1-v1.xlsx
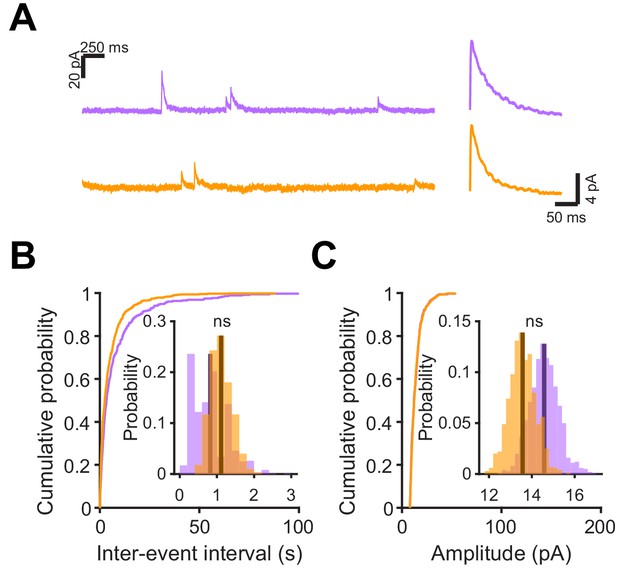
Microglial depletion after abGC development has no effect on inhibitory synaptic currents.
(A) Left, Sample sections from raw traces recorded from abGCs in control (top) and PLX-treated (bottom) mice. Right, median IPSCs calculated across all IPSCs detected from all mice. (B) Cumulative distribution showing the inter-event intervals from all recorded IPSCs. Inset, sampling distributions of the mean frequency obtained with the hierarchical bootstrap. The mean frequencies (dark lines) were not significantly different (hierarchical bootstrap, p=0.73). (C) Cumulative distribution showing the amplitudes from all recorded IPSCs. Inset, sampling distributions of the mean amplitude obtained with the hierarchical bootstrap. The mean amplitudes (dark lines) were not significantly different (hierarchical bootstrap, p=0.14). n = 11 abGCs from three control mice and 13 abGCs from 3 PLX mice (subset of the cells from the main figure that met criteria stated in Materials and methods). ns, not significant.
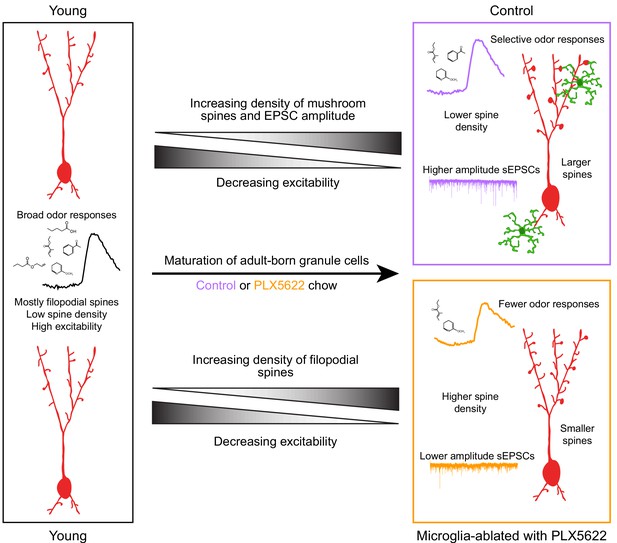
Summary of results and model for the role of microglia in abGC maturation.
Young abGCs enter existing circuits and begin to make synapses, a process that involves the extension of filopodial spines that sample potential synaptic partners (Breton-Provencher et al., 2014). In control mice, abGCs undergo a coordinated process of synaptic formation and elimination (Mizrahi, 2007; Sailor et al., 2016), which leads to an increasing number of mushroom spines with mature synapses and higher frequency and amplitude excitatory synaptic currents as they mature (Whitman and Greer, 2007; Kelsch et al., 2008; Breton-Provencher et al., 2014). Maturation likely also involves a decrease in dendritic excitability (Carleton et al., 2003; Nissant et al., 2009; Wallace et al., 2017), such that stronger, coordinated synaptic inputs with particular odor tuning are necessary to activate mature abGCs, leading to more selective odor responses. In mice treated with PLX5622 to ablate microglia throughout the time course of abGC development, we hypothesize that abGCs undergo some parts of the maturation process, but not others. We show that aspects of excitability, such as input resistance, mature normally, and overall spine density increases to levels even above controls. However, synapses fail to mature, leading to overall smaller spines and lower amplitude sEPSCs. This may contribute to lower odor responsiveness in abGCs that have matured in the absence of microglia.
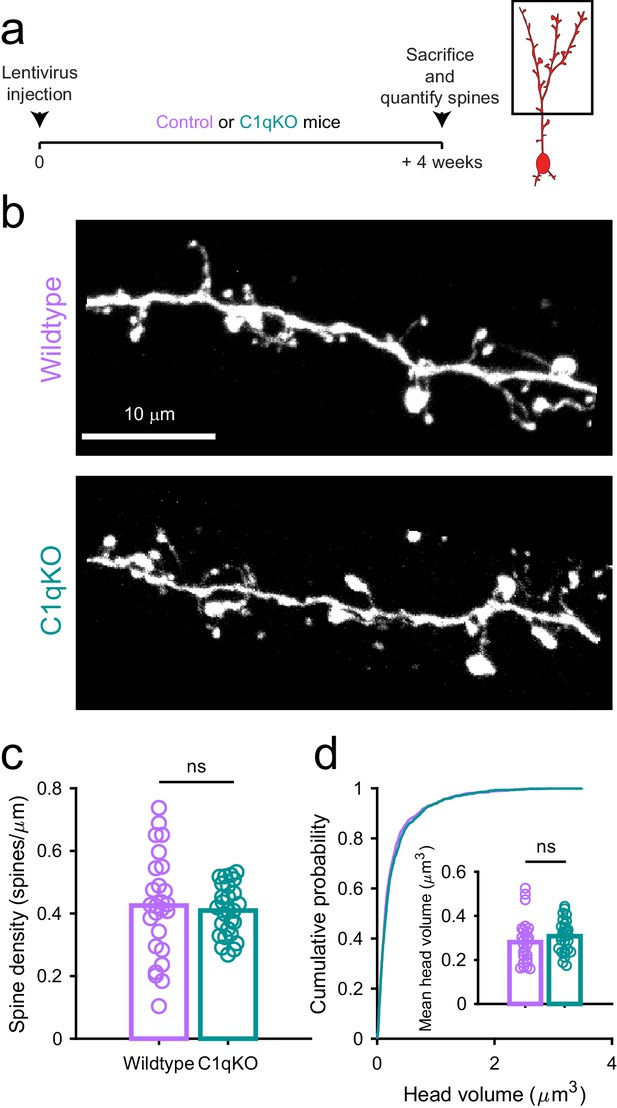
Spine density and volume are unaffected in C1qKO mice.
(a) Experimental timeline for spine quantification in wild type versus C1qKO mice. Lentivirus was injected into the RMS to label abGCs. Spine numbers and morphology were quantified 4 weeks later. (b) Sample image showing sections of apical dendrites from an abGC in a control mouse (top) and a C1qKO mouse (below). (c) Spine density averaged in apical dendrites of abGCs. (d) Cumulative distribution showing the volume of all spines analyzed. Inset, head volume averaged across all spines in each cell. Bars indicate medians across cells (circles). n = 1215 spines from 26 abGCs from 3 wild type mice and n = 1262 spines from 27 abGCs from 3 C1qKO mice.
Videos
Z stack showing microglia and abGC dendrites.
Time series showing interactions between microglia and dendritic spines of abGCs.
The movie shows a single plane taken from the z stack in Video 1 across 48 min of imaging (images taken 3 min apart). The time course of the interaction between a microglial process and the mushroom spine shown in the example in Figure 1 can be observed. The analysis in Figure 1 was performed on individual planes from such time series.
Tables
Common names and chemical structures for monomolecular odor stimuli.
| Ethyl tiglate | 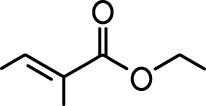 | Valeraldehyde | 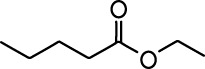 |
| Ethyl valerate | 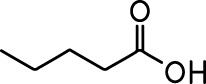 | Isoamyl acetate | 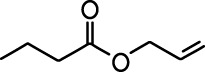 |
| Valeric acid | 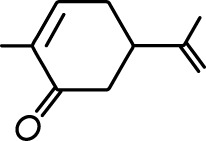 | Anisole | 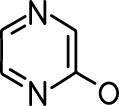 |
| Allyl butyrate | 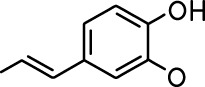 | Ethyl propionate | 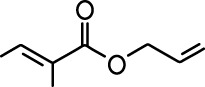 |
| Carvone | 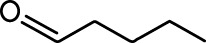 | Propyl acetate | 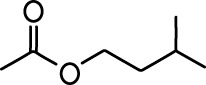 |
| 2-methoxypyrazine | 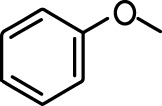 | 2-heptanone |  |
| Isoeugenol |  | Acetophenone |  |
| Allyl tiglate |  |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | CX3CR1-GFP heterozygote | Jung et al., 2000 | Cat# Jax: 008451 | |
| Genetic reagent (lentivirus) | pLenti-TRE-t2A-dTomato-GCaMP5 | Wallace et al., 2017 | Produced in house (see Materials and methods, 'Viral Vectors') | |
| Genetic reagent (lentivirus) | pLenti-TRE-t2A-dTomato-GCaMP6s | Wallace et al., 2017 | Produced in house (see Materials and methods, 'Viral Vectors') | |
| Genetic reagent (lentivirus) | pLenti-TRE-t2A-dTomato | Wallace et al., 2017 | Boston Children’s Hospital Viral Core | |
| Genetic reagent (lentivirus) | pLenti-hSynapsin - tTad | Hioki et al., 2009 | Produced in house or by Boston Children’s Hospital Viral Core | |
| Antibody | Anti-Iba1 (Rabbit polyclonal) | Wako | Cat # 019–19741 RRID:AB_839504 | 1:500 |
| Antibody | Anti-GFAP (Rabbit polyclonal) | Dako | Cat # Z0334 RRID:AB_10013382 | 1:1000 |
| Antibody | Anti-BrdU (Rat monoclonal) | Abcam | Cat # ab6326 | 1:200 |
| Antibody | Anti-NeuN (Mouse monoclonal) | Millipore | MAB377 | 1:200 |
| Antibody | Anti-CD45 (Rat monoclonal) | Biolegend | 103116 | 1:100 (final concentration of 2 µg/mL) |
| Antibody | Anti-CD11b (Rat monoclonal) | Biolegend | 101217 | 1:200 (final concentration of 2.5 µg/mL) |
| Antibody | Anti CD16/CD32 (Rat) | BD Bioscience | 553141 | 1:50 (final concentration of 10 µg/mL) |
| Chemical compound, drug | PLX5622 | Plexxikon | PLX5622 | 1200 mg/kg |
| Chemical compound, drug | BrdU | Sigma Aldrich | Cat # 19–160 | 100 mg/kg |
| Software, algorithm | FlowJo | FlowJo | Version 10.2 | https://www.flowjo.com/solutions/flowjo/downloads/previous-versions |
| Software, algorithm | Imaris | Oxford Instruments | Versions 8 and 9 | https://imaris.oxinst.com/ |
| Software, algorithm | pClamp | Molecular Devices | Version 10.3 | |
| Software, algorithm | Matlab | Mathworks | Version 2019b | |
| Software, algorithm | MiniAnalysis | Synaptosoft | Version 6.0.3 | http://www.synaptosoft.com/MiniAnalysis/ |
| Other | Blue Dead Cell Stain | ThermoFisher | L34961 | |
| Other | Counting Beads (CountBright) | ThermoFisher | C36950 |





