Roles of C/EBP class bZip proteins in the growth and cell competition of Rp (‘Minute’) mutants in Drosophila
Figures
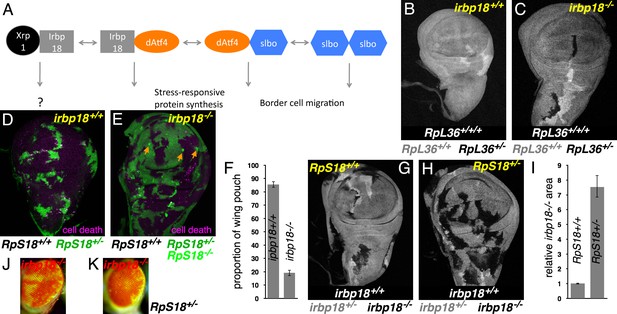
Xrp1 and related bZip proteins in cell competition.
(A) Known dimers of Drosophila C/EBP –class bZip proteins and their potential functions. (B,C) Mitotic recombination in RpL36+/+ wing discs (grey) generates clones of RpL36+/+/+ cells (light grey) and reciprocal clones of RpL36+/- cells (black, lacking beta-Gal labeling). RpL36+/- clones that did not survive in the irbp18+/+ background (B) always survived in the irbp18-/- background (C). (D,E) Mitotic recombination in RpS18+/- wing discs (green) generates clones of RpS18+/+ cells (black,)lacking GFP expression. In the irbp18+/+ background (D) these have a growth and competitive advantage and come to dominate wing disc territory, eliminating remaining RpS18+/- cells by cell death (anti-active caspase DCP1 labeling in magenta). RpS18+/+ cells had less advantage in the irbp18-/- background (E). In addition, reciprocally recombinant RpS18-/- cells survived as small clones (bright green, eg arrows in E). (F) Quantitative comparison of RpS18+/+ clone growth in irbp18+/+ and irbp18-/- backgrounds. Wing pouch areas were the same in irbp18+/+ and irbp18-/- backgrounds (p=0.191, two-tailed t-test), but the RpS18+/+ fractions were not (p<0.0001, two-tailed t-test). Data derived from measurements of 4 irbp18+/+ discs and five irbp18-/- discs. (G,H) Mitotic recombination of the irbp18 locus in the RpS18+/+ and RpS18+/- backgrounds. Reciprocal clones of RpS18+/+ irbp18+/+ and RpS18+/+ irbp18-/- cells grew comparably (G) whereas clones of RpS18+/-irbp18-/- cells expanded at the expense of RpS18+/-irbp18+/- cells and RpS18+/-irbp18+/+ cells (H). (I) Quantification of growth in RpS18+/- and RpS18+/+ wing discs (eg panels G,H). irbp18-/- clone area was compared to reciprocal irbp18+/+ controls. Probability that clone areas are the same in the RpS18+/+ and RpS18+/- backgrounds = 1.4 × 10−9 (2-tailed t-test with unequal variances). Data derived from measurements of 7 RpS18+/+ discs and 9 RpS18+/- discs. (J) irbp18-/- clones (pigmented) contributing to the adult eye. (K) irbp18-/- clones (pigmented) occupy nearly all the RpS18+/- eye. Genotypes B) hsFlp/M(1)Bld; P[RpL36+ w+] arm-LacZ FRT80B/FRT80B. C) hsFlp/M(1)Bld; P[RpL36+ w+] arm-LacZ FRT80B/irbp18f05006 FRT80B. D) y w hs-FLP; FRT42D ubi-GFP M( f05006)56F/FRT42D. E) y w hs-FLP; FRT42D ubi-GFP M(2)56F/FRT42D; irbp18f05006/irbp18f05006. G) y w hs-FLP; +/+; irbp18f05006 FRT80B /arm LacZ FRT80B. H) y w hs-FLP; FRT42D ubi-GFP M(2)56F*/+; irbp18f05006 FRT80B/arm LacZ FRT80B. J) y w ey-FLP/Y; FRT42D ubi-GFP/+; irbp18f05006 FRT80B/FRT80B K) y w ey-FLP/Y; FRT42D ubi-GFP RpS18/+; irbp18f05006 FRT80B/FRT80B. Note that the irbp18f05006 allele includes a w+ element that is responsible for most of the eye pigmentation.
-
Figure 1—source data 1
Measurements of clone and wing pouch sizes corresponding to Figure 1F.
- https://cdn.elifesciences.org/articles/50535/elife-50535-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Clone size data corresponding to Figure 1I.
- https://cdn.elifesciences.org/articles/50535/elife-50535-fig1-data2-v1.xlsx
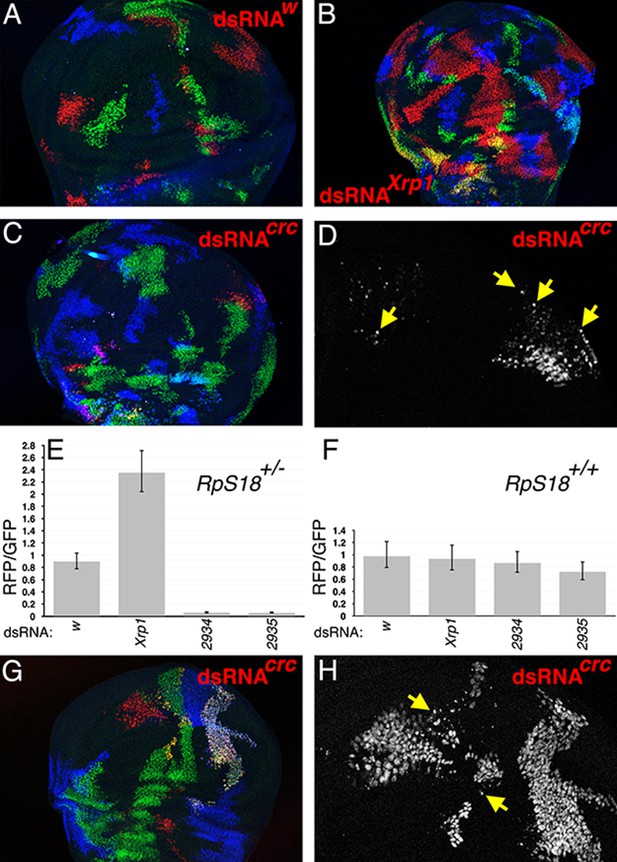
dATF4/crc requirement in wild type and Rp+/- wing discs.
(A) In the TIE-DYE method, independent recombination events generate parallel clones expressing GFP (green), Gal4 (detected through UAS-RFP expression in red), and b-Gal (blue). These parallel clones grow equivalently in RpS18+/- wing discs. In this control, dsRNA targeting transcripts from the white gene was co-expressed with RFP under Gal4 control. (B) Co-expression of dsRNA targeting Xrp1 increased the contribution of RFP-positive clones in RpS18+/- wing discs. (C) Co-expression of dsRNA targeting dATF4/crc decreased the contribution of RFP-positive clones in RpS18+/- wing discs. (D) Enlarged portion of the wing disc from panel C to show the fragmentation of RFP-positive, dATF4/crc knock-down RpS18+/- cells (eg arrows). (E) Quantification of growth for various dsRNA-expressing RFP-positive clones in RpS18+/- wing discs. Shown is the mean ratio of RFP-positive to GFP-positive areas for each wing disc. Error bars represent ± 1 SEM. Probabilities that clone sizes are the same as for the w control (2-tailed t-tests): Xrp1 - 3.4 × 10−5; 2934–3.1 × 10−12; 2935–2.2 × 10−11. VDRC2934 and VDRC2935 encode independent, previously-validated dsRNAs targeting dATF4/Crc (Kang et al., 2017). Number of wing discs analyzed = 25 (w), 23 (Xrp1), 17 (2934), 26 (2935). (F) Quantification of growth for various dsRNA-expressing RFP-positive clones in RpS18+/+ wing discs. Probabilities that clone sizes are the same as for the w control (2-tailed t-tests): Xrp1 – 0.88; 2934–1.0; 2935–0.68. Number of wing discs analyzed = 17 (w), 26 (Xrp1), 16 (2934), 19 (2935). (G) An example of dATF4/Crc knock-down in RFP-positive clones in a RpS18+/+ wing disc. (H) Enlarged portion of the wing disc shown in G illustrating that, although substantial RFP-positive territories are present, some of these cells are fragmented (eg arrows). Genotypes A) y w hs-FLP; Act < stop < lacZ-NLS Ubi < stop < eGFP-NLS M(2)56 F/ +; Act < stop < GAL4 UAS-His2A::mRFP/P{TRiP.HMS00017}attP2. B) y w hs-FLP; Act < stop < lacZ-NLS Ubi < stop < eGFP-NLS M(2)56 F/ +; Act < stop < GAL4 UAS-His2A::mRFP/P{TRiP.HMS00053}attP2. C,D) y w hs-FLP; Act < stop < lacZ-NLS Ubi < stop < eGFP-NLS M(2)56 F/ +; Act < stop < GAL4 UAS-His2A::mRFP/UAS-dsRNA(Crc)VDRC2935. G,H) y w hs-FLP; Act < stop < lacZ-NLS Ubi < stop < eGFP-NLS / +; Act < stop < GAL4 UAS-His2A::mRFP/UAS-dsRNA(Crc)VDRC2935.
-
Figure 2—source data 1
Source data for the graph shown in Figure 2E.
- https://cdn.elifesciences.org/articles/50535/elife-50535-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Source data for the graph shown in Figure 2F.
- https://cdn.elifesciences.org/articles/50535/elife-50535-fig2-data2-v1.xlsx
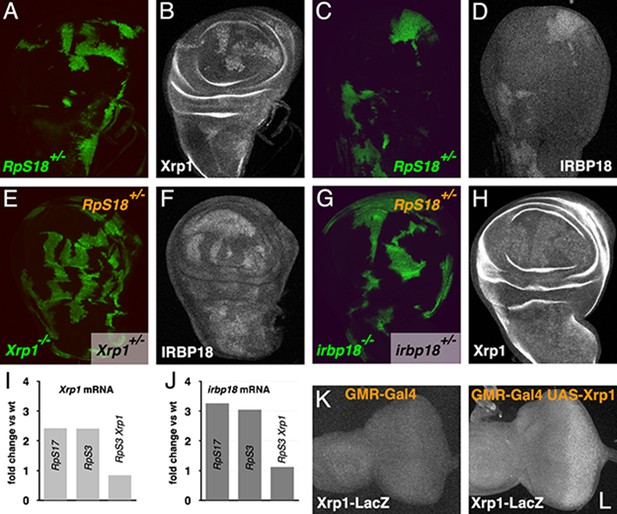
Cross-regulation of Xrp1 and IRBP18 expression.
(A,B) Xrp1 protein (labeled in B) is elevated in RpS18+/- cells (green in A) compared to RpS18+/+ cells (unlabelled in A). (C,D) Irbp18 protein (labeled in D) is elevated in RpS18+/- cells (green in C) compared to RpS18+/+ cells (unlabelled in C). (E,F) Irbp18 protein (labeled in F) is elevated in RpS18+/-Xrp1+/- cells or RpS18+/-Xrp1+/+ cells (unlabelled in E) compared to RpS18+/-Xrp1-/- cells (green in E), showing that Irbp18 protein expression is Xrp1-dependent. (G,H) Xrp1 protein (labeled in H) is higher in RpS18+/-irbp18+/- and RpS18+/-irbp18+/+ cells (both unlabeled in G) than in RpS18+/-irbp18-/- cells (green in G), showing that Xrp1 protein up-regulation is Irbp18-dependent. (I) mRNA fold-change in wing imaginal discs of various genotypes in comparison to wild type controls. Both Xrp1 and irbp18 mRNA levels were elevated in Rp+/- wing discs in an Xrp1-dependent manner. Fold changes compared to wild type were determined using DESEQ2 from three biological replicates (Lee et al., 2018). The adjusted probabilities that expression levels differed from the wild type control were 7.95 × 10−153 (Xrp1 in RpS17+/-), 1.21 × 10−146 (Xrp1 in RpS3+/-), 0.0149 (Xrp1 in RpS3+/-Xrp1+/-), 6.75 × 10−32 (irbp18 in RpS17+/-), 9.72 × 10−34 (irbp18 in RpS3+/-), 0.714 (irbp18 in RpS13+/-RpS3+/-Xrp1+/-). (K,L) Over-expression of Xrp1 in the posterior eye disc under control of GMR-Gal4 (L) upregulates the Xrp1-LacZ enhancer trap compared to the GMR-Gal4 control (K), confirming transcriptional auto-regulation. Genotypes A-D) y w hs-FLP; FRT42D ubi-GFP M(2)56F/FRT42. E,F) y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/FRT42D ubi-GFP M(2)56F; FRT82B Xrp1M2-73/FRT82B tubP-GAL80. G,H) y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/FRT42D ubi-GFP M(2)56 f*; irbp18f05006 FRT80B/tubP-GAL80 FRT80B. K) GMR-Gal4/+; Xrp102515/+. L) GMR-Gal4/UAS-Xrp1; Xrp102515/+.
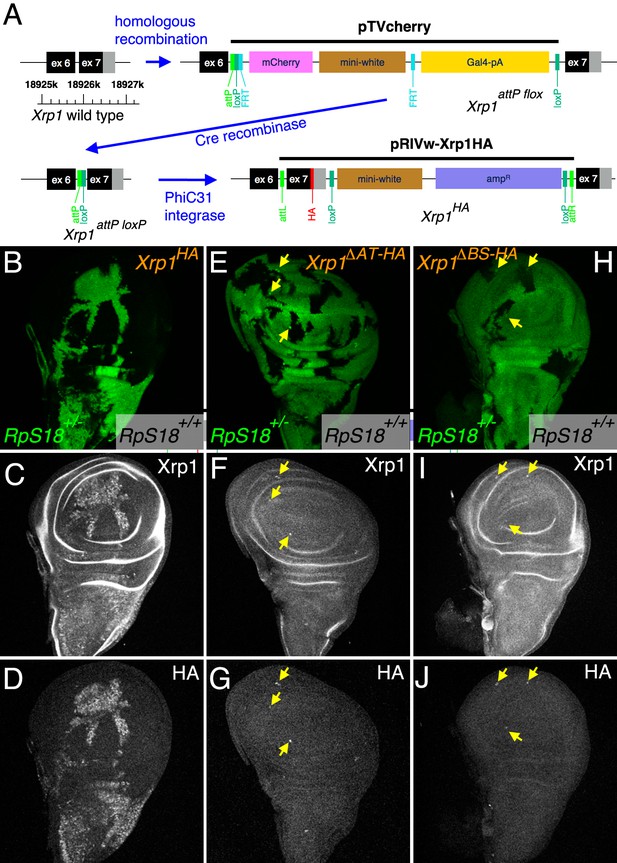
Modifications of the endogenous Xrp1 locus and their consequences for cell competition.
(A) Sequential modifications of the Xrp1 locus in the 18925–18926 kb region of chromosome arm 3R by homologous recombination, Cre recombination and PhiC31 integration introduced modified and HA-tagged exon seven sequences into intron 6. (B–D) Cell competition in the homozygous Xrp1HA background. RpS18+/+ Xrp1HA/HA clones (unlabeled in B) grow to occupy most of the wing disc at the expense of RpS18+/-Xrp1 HA/HA cells (green in B), similar to what is seen in the Xrp1+/+ background (see Figure 3A and C). (C) Xrp1 protein is elevated in RpS18+/-Xrp1 HA/HA cells, as was seen for RpS18+/-Xrp1+/+ cells (see Figure 3B). (D) HA-tagged protein is detected only in the RpS18+/-Xrp1 HA/HA cells. (E–G) Cell competition in the homozygous Xrp1ΔΑT-ΗΑ background. RpS18+/+ Xrp1ΔΑT-ΗΑ/ΔΑT-ΗΑ clones (unlabeled in E) occupy less of the wing disc than RpS18+/-Xrp1ΔΑT-ΗΑ/ΔΑT-ΗΑ cells (green in E), similar to what is seen in the Xrp1-/- or irbp18-/- backgrounds (see Figure 1E). Note the survival of small clones of RpS18-/-Xrp1ΔΑT-ΗΑ/ΔΑT-ΗΑ (brighter green in E, eg arrows). Survival of Rp-/- genotypes is a feature of Xrp1 mutants. (F) Xrp1 protein is not elevated in RpS18+/-Xrp1ΔΑT-ΗΑ/ΔΑT-ΗΑ cells, but is in RpS18-/-Xrp1ΔΑT-ΗΑ/ΔΑT-ΗΑ (eg arrows). (G) As expected, the Xrp1 protein is HA-tagged (eg arrows). (H–J) Cell competition in the homozygous Xrp1ΔBR-HA background. RpS18+/-Xrp1ΔBR-HA/ΔBR-HA cells (green in H) are not out-grown by RpS18+/+ Xrp1ΔBR-HA/ΔBR-HA cells. Note the survival of small clones of RpS18-/-Xrp1ΔBR-HA/ΔBR-HA (brighter green in H, eg arrows), a feature of Xrp1 mutant genotypes. These RpS18-/-Xrp1ΔBR-HA/ΔBR-HA clones are the only cells where Xrp1 protein is elevated (eg arrows I) or HA-tag detected (eg arrows J). Genotypes B-D) w hs-FLP; FRT42D ubi-GFP M(2)56F/FRT42; Xrp1HA/Xrp1HA. E–G). w hs-FLP; FRT42D ubi-GFP M(2)56F/FRT42; Xrp1ΔAT-HA/Xrp1ΔAT-HA. H–J) w hs-FLP; FRT42D ubi-GFP M(2)56F/FRT42; Xrp1ΔBR-HA/Xrp1ΔBR-HA..
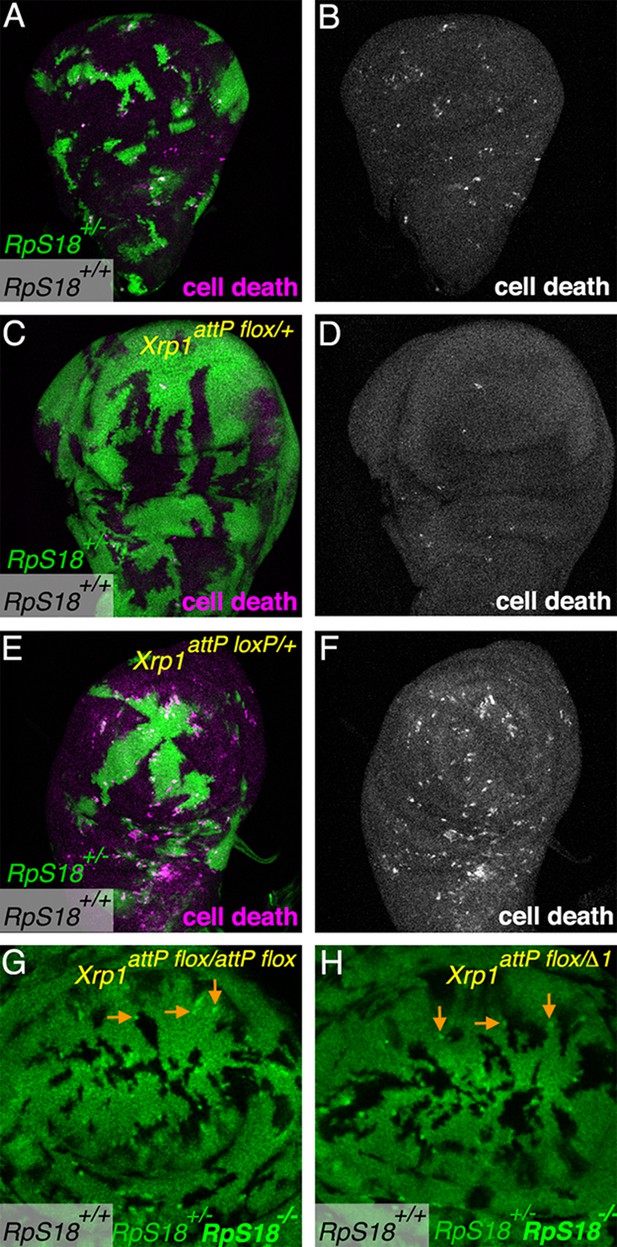
Xrp1attP flox is a loss-of-function allele.
(A) As clones of RpS18+/+ cells (unlabelled) expand to occupy most of a RpS18+/- wing imaginal disc (green, GFP labeling), apoptosis is elevated especially at the boundaries between the cells (magenta labeling for activated effector caspase DCP1). (B) anti-DCP1 labeling from panel A). (C). RpS18+/+ clones occupy less of an RpS18+/-Xrp1 attP flox/+ wing imaginal disc, and cell death is rarely detected, as in other Xrp1 mutant genotypes (eg see Figure 1D). (D) anti-DCP1 labeling from panel C). (E). In the Xrp1attP loxP/+ background, RpS18+/+ cells expand to occupy most of a RpS18+/- wing imaginal disc, inducing apoptosis at the boundaries, as typical of Xrp1+/+ genotypes. (F) Anti-DCP1 labeling form panel E). (G). Small RpS18-/- clones persist in the Xrp1attP flox/attp flox background (eg arrows), as in other Xrp1-/- backgrounds (eg see Figure 1E). (H) Small RpS18-/- clones persist in the Xrp1attP flox/Δ1background (eg arrows), as in other Xrp1-/- backgrounds (eg see Figure 1E). Xrp1Δ1 is a deletion encompassing the Xrp1 locus (Lee et al., 2018).
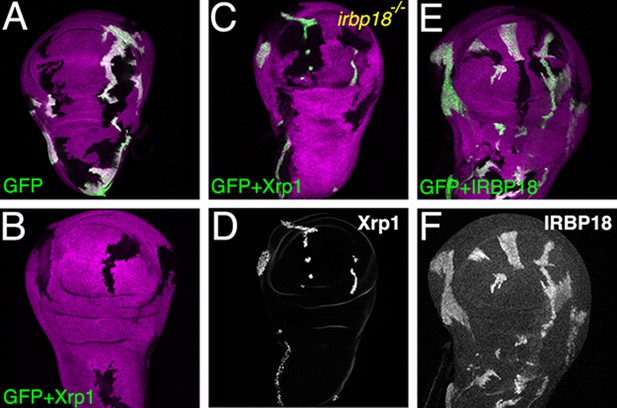
Ectopic Xrp1 requires IRBP18 to affect clone survival.
(A) Wing imaginal discs containing clones of control cells lacking b-galactosidase (magenta) and reciprocal clones expressing GFP (green). In panels B-D) Xrp1 is co-expressed with GFP. Xrp1 led to complete elimination of GFP-positive lineages in the irbp18+/+ background (B), but not when irbp18 was mutated (C). Panel D shows that the GFP-marked clones nevertheless highly over-expressed Xrp1 protein in the irbp18-/- background. (E,F) co-expression of Irbp18 with GFP had no effect on clone survival, even though Irbp18 protein was highly over-expressed (F). Genotypes A) y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/+; FRT82B arm-LacZ/FRT82B tubP-GAL80. B) y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/UAS-Xrp1; FRT82B arm-LacZ/FRT82B tubP-GAL80. C–D). y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/UAS-Xrp1; irbp18f05006 arm-LacZ FRT80/tubP-GAL80 FRT80. E,F). y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/UAS-irbp18; arm-LacZ FRT80/tubP-GAL80 FRT80.
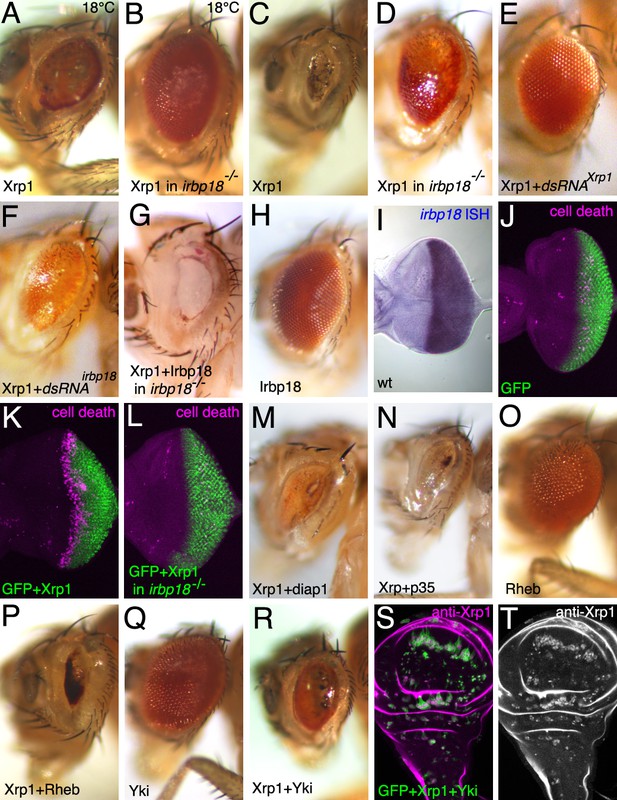
Xrp1 over-expression affects the eye in an irbp18-dependent manner.
Most panels show adult eyes where GMR-Gal4 has been used to express the indicated proteins and dsRNAs posterior to the morphogenetic furrow. Temperature was 25°C except where indicated otherwise. Panels (J-L) show eye imaginal discs immunolabeled for mGFP (green) and for cleaved caspase DCP1 (magenta) to reveal apoptosis. Panels S,T show a wing imaginal disc containing clones over-expressing GFP, Xrp1 and Yki. Clones of these cells survived, unlike clones expressing GFP and Xrp1 alone (compare Figure 5B), despite expressing highly elevated Xrp1 (panel T). Genotypes A,C,K) GMR-Gal4/UAS-Xrp1; B,D,L) GMR-Gal4/UAS-Xrp1; irbp18f05006/irbp18f05006. E) GMR-Gal4/UAS-Xrp1; UAS-Xrp1RNAi /+. F). GMR-Gal4/UAS-Xrp1; UAS-irbp18RNAi /+. G) GMR-Gal4/UAS-Xrp1 UAS-irbp18; irbp18f05006/f05006. H,I) GMR-Gal4/UAS-irbp18. J) GMR-Gal4/+. M) GMR-Gal4/UAS-Xrp1; UAS-diap1/+. N) GMR-Gal4/UAS-Xrp1; UAS-p35/+. O) GMR-Gal4/+; UAS-Rheb/+. P) GMR-Gal4/UAS-Xrp1; UAS-Rheb/+. Q) GMR-Gal4/+; UAS-yki/+. R) GMR-Gal4/UAS-Xrp1; UAS-yki/+. S,T) y w hs-FLP; tubP-GAL4 UAS-mCD8::GFP/UAS-Xrp1 UAS-yki; FRT82B arm-LacZ/FRT82B tubP-GAL80..
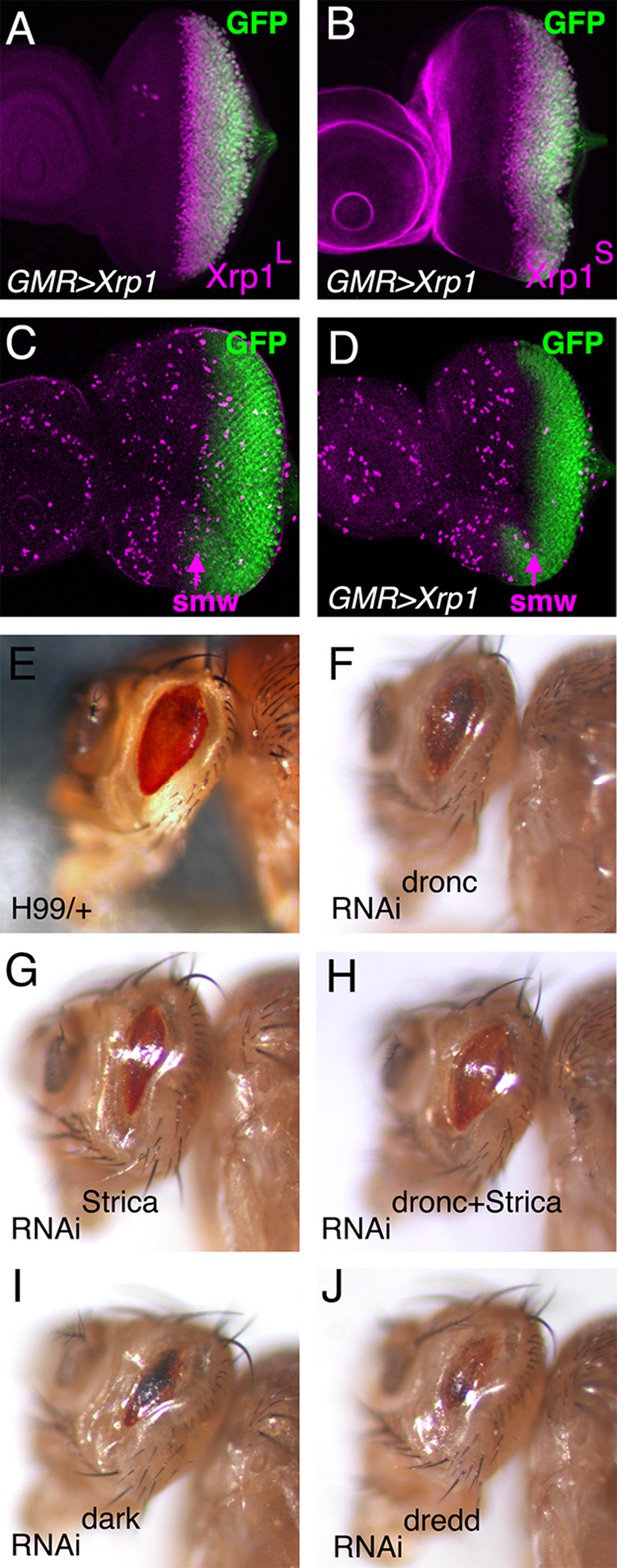
Cell cycle and genetic interactions in the GMR>Xrp1 genotype.
(A) GMR-Gal4 drives UAS-GFP expression (green) posterior to the morphogenetic furrow in the eye imaginal disc. An antibody specific for the Xrp1 long isoform detects co-expressed Xrp1 protein slighty earlier (magenta). (B) An antibody raised against the Xrp1 short isoform detects all Xrp1 proteins expressed posterior to the morphogenetic furrow (magenta). (C) Mitotic figures detected with anti-pH3 (magenta) are concentrated anterior to the morphogenetic furrow and in the Second Mitotic Wave (SMW) that immediately follows the morphogenetic furrow. (D) SMW mitoses are greatly reduced in GMR-Gal4 UAS-Xrp1 eye discs. (E) Heterozygosity for Df(3L)H99, encoding the proapoptotic genes rpr, grim, hid and skl, has little effect on the GMR-Gal4 UAS-Xrp1 eye phenotype (compare Figure 6C). (F–J) RNAi knockdown of various caspases and pro-apoptotic genes has little effect on the GMR-Gal4 UAS-Xrp1 eye phenotype.
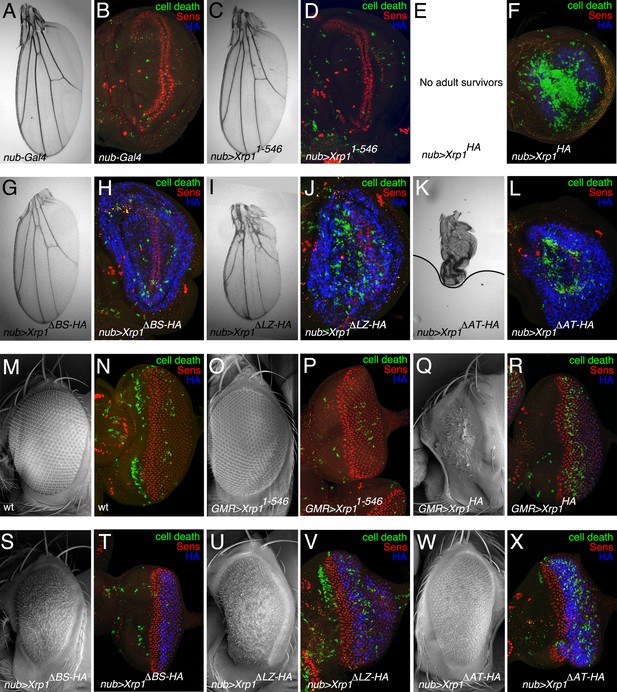
Ectopic expression of mutated Xrp1 proteins.
(A–L) Over-expression of Xrp1 proteins during wing development using nub-Gal4 at 18°C. A,C,E,G,I,K show wings from males misexpressing the indicated proteins. Results were similar from females although the male wings were affected more. (B,D, F, H, J, L) show third instar wing imaginal discs labeled for Senseless (red) to reveal the neural differentiation pattern along the future wing margin, anti-active caspase Dcp1 (green) to reveal cell death, and anti-HA (blue) to detect expression of mutated Xrp1 proteins. The Xrp11-546 protein, which had little or no effect on wing development, was not detected by anti-HA because it was not tagged (D) Notably, each of the other deletion proteins was expressed more highly than the wild type (compare blue signal in panels H,J,L with F) although it can’t be excluded this reflects apoptosis of many cells expressing wild type Xrp1 (F) No adults survived expression of full-length Xrp1 in wings (E). (M–X) Over-expression of Xrp1 proteins during eye development using GMR-Gal4 at 18°C. M,O,Q,S,U,W show eyes from males misexpressing the indicated proteins. Results were similar from females. N,P, R, T, V, X show third instar eye imaginal discs labeled for Senseless (red) to reveal the retinal differentiation pattern posterior to the morphogenetic furrow, anti-active caspase Dcp1 (green) to reveal cell death, and anti-HA (blue) to detect expression of mutated Xrp1 proteins. Genotypes A,B) nub-Gal4/+. C,D) nub-Gal4/+; UAS-Xrp11-546. E,F) nub-Gal4/+; UAS-Xrp1HA. G,H) nub-Gal4/+; UAS-Xrp1ΔBR-HA. I,J) nub-Gal4/+; UAS-Xrp1ΔLZ-HA. K,L) nub-Gal4/+; UAS-Xrp1ΔAT-HA. (M,N) w11-18. (O-P) GMR-Gal4/+; UAS-Xrp11-546. (Q-R) GMR-Gal4/+; UAS-Xrp1HA. (S-T) GMR-Gal4/+; UAS-Xrp1ΔBR-HA. (U-V) GMR-Gal4/+; UAS-Xrp1ΔLZ-HA. (W-X) GMR-Gal4/+; UAS-Xrp1ΔAT-HA..
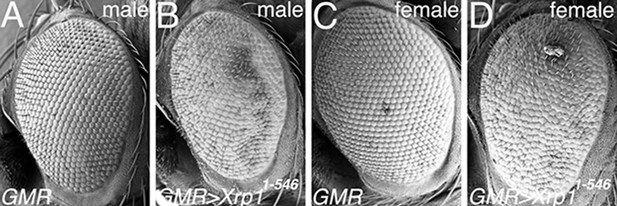
Xrp11-546 expression at higher temperature.
(A) Scanning Electron Micrograph (SEM) of male GMR-Gal4/+ eye, raised at 25°C. Morphology is almost wild type (compare Figure 7M). (B) At 25°C, expression of the Xrp11-546 protein that lacks the C-terminus results in mild eye abnormalities, including irregular arrangement of facets and of interommatidial bristles. (C) Female GMR-Gal4/+ eye resembles the male (see panel A). (D) Female GMR-Gal4/UAS-Xrp11-546 eye resembles the male (see panel B).
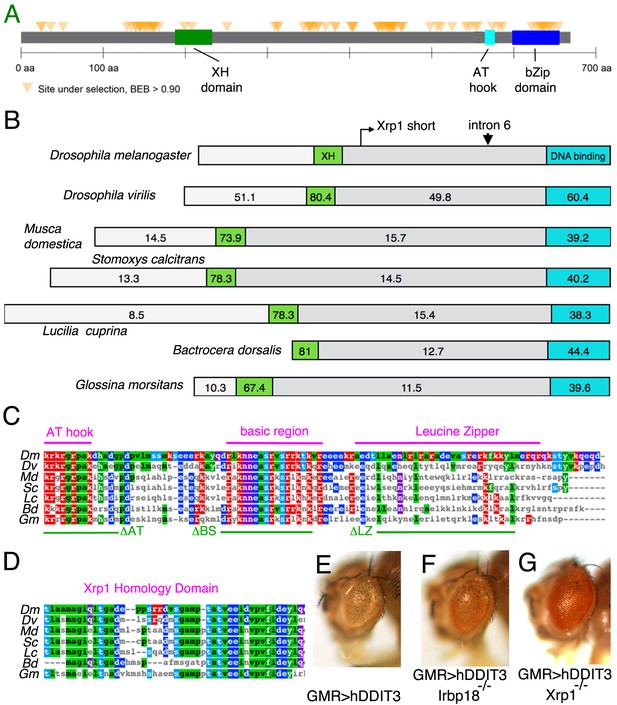
Xrp1 gene conservation.
(A) PAML results for 12 Drosophila species showing the location of sites under strong positive selection in the Xrp1 protein. (B) The conservation (% amino-acid identity to Xrp1 from Drosophila melanogaster) is plotted for Drosophila virilis and for predicted proteins from five other Dipterans. The C-terminal DNA-binding domain region (corresponding to amino acids 565–668 from the long form of D. melanogaster Xrp1) is highlighted in Cyan, and the more amino-terminal conserved sequence (corresponding to amino acids 189–234 from the long form of D. melanogaster) in green. The locations of the alternative amino-terminus of Xrp1 short isoforms in D. melanogaster, and the position of intron six whose modification was described in Figure 6A are indicated. (C) Clustal Omega alignment of the C-terminal DNA-binding domain region of insect Xrp1 sequences. The core consensus sequences defining the AT-hook domain, basic region of bZip domains, and the Leucine Zipper, are overlined in magenta. The deletions made in this study for structure-function analysis of D. melanogaster Xrp1 are underlined in green. (D) Clustal Omega alignment of the Xrp1 Homology domain in the amino-terminal portion of the insect Xrp1 sequences. The Bactrocera dorsalis sequence is perhaps not optimally aligned by this program. (E) Expression of hDDIT3 under GMR-Gal4 control (at 25°C) reduces eye size (compare the effects of Xrp1 overexpression, Figure 6A,C). (F) Eye size reduction by hDDIT3 depended in part on the Drosophila irbp18 gene. (G) Eye size reduction by hDDIT3 depended in part on the Drosophila Xrp1 gene.
Additional files
-
Supplementary file 1
Key Resources.
- https://cdn.elifesciences.org/articles/50535/elife-50535-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50535/elife-50535-transrepform-v1.pdf






