Cardiac mitochondrial function depends on BUD23 mediated ribosome programming
Figures
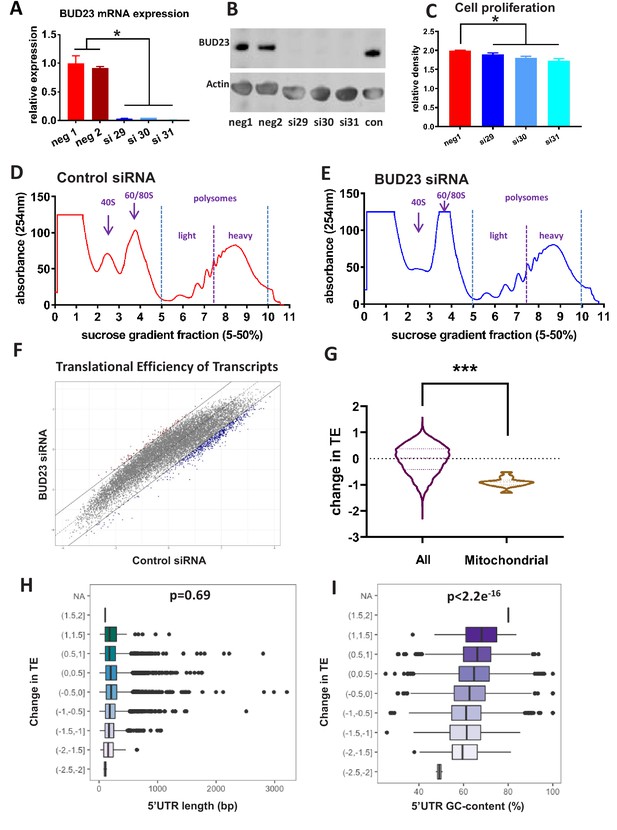
Analysis of BUD23-dependent ribosome function.
(A) Relative mRNA expression levels of BUD23. A549 cells were transfected for 48 hr with either BUD23 specific siRNA (si29, 30, 31) or non-targeting control (neg1, 2). Total RNA was extracted before reverse transcription to cDNA and qPCR (TaqMan). (B) Protein expression was determined by western blotting using antibody GTX105840. Actin was used as a loading control. (C) siRNA treated cells were plated into cell culture wells and allowed to grow in DMEM supplemented with 10% FBS for 48 hr. Total cell volume was then measured using SRB assay to indicate amount of proliferation. Polysome profiles were obtained from A549 cell lysates treated with (D) Control siRNA or (E) BUD23 specific siRNA (si30) for 48 hr (n = 3). Changes in the ratio of the 40S peak to the 60/80S peak were observed between the two conditions. Polysome fractions were divided into heavy (more than three ribosomes) or light (three or less ribosomes) before RNA sequencing.. A translational efficiency (TE) score was derived by dividing the relative proportion of transcript abundance in the heavy fraction by that in the light fraction. (F) The TE scores in the two conditions derived from the samples in A and B are plotted against each other. (G) Violin plot showing the change in translational efficiency upon BUD23 siRNA treatment (‘change in TE score’) for all detected transcripts (All) versus mitochondrially-encoded transcripts (Mitochondrial). (H) 5’UTR length of the transcripts was plotted against change in translational efficiency (TE, as defined in Figure 1G). The data are plotted in bins with ‘change in TE’ (BUD23 TE/Control TE) windows of 0.5. (I) 5’UTR GC content was of the same transcripts was plotted against change in TE. The data are plotted in bins with TE windows of 0.5.
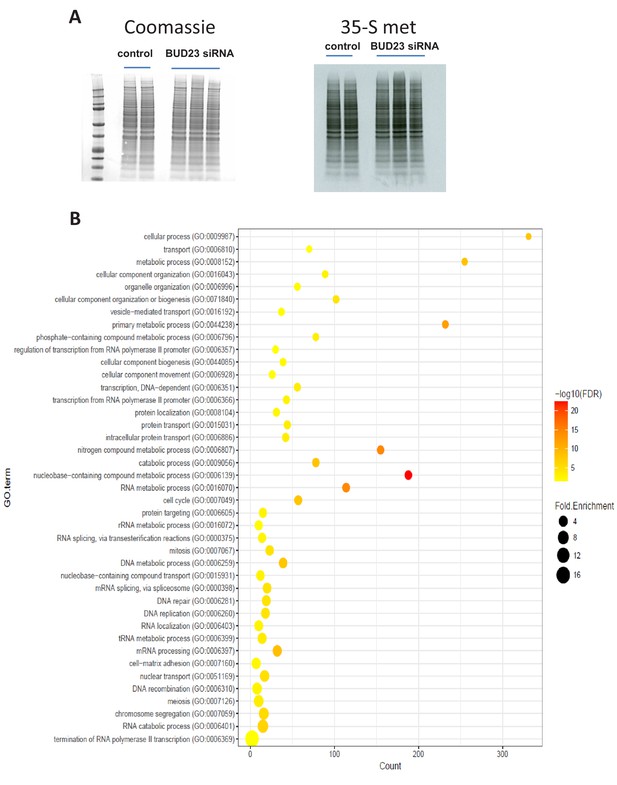
Further analysis of BUD23-dependent ribosome function in A549 cells.
(A) Global translation rates in A549 cells after siRNA treatment were measured by 35-S-methionine incorporation assay. Equal protein loading was confirmed by Coomassie. (B) Data-set enrichment analysis using the PANTHER DB GO-Slim Biological processes database, of transcripts with >2 fold decrease in TE in the BUD23-depleted condition relative to control. These transcripts form a highly inter-connected network with a PPI enrichment value <1e−16.
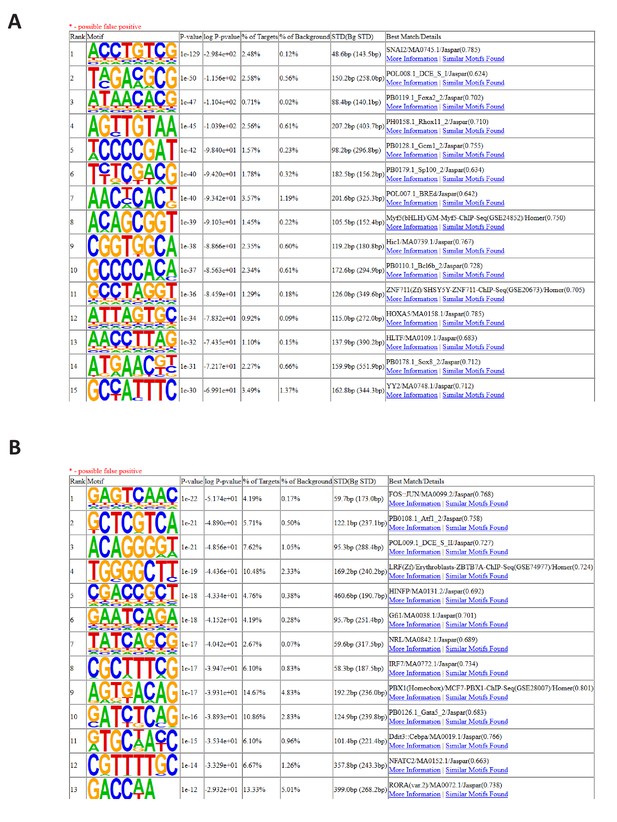
5'UTR motif analysis.
(A and B) De-novo motif analysis of 5’UTRs from the highly down-regulated (A) and up-regulated (B) transcripts identified by comparing TE from control and BUD23 knockdown A549 cells.
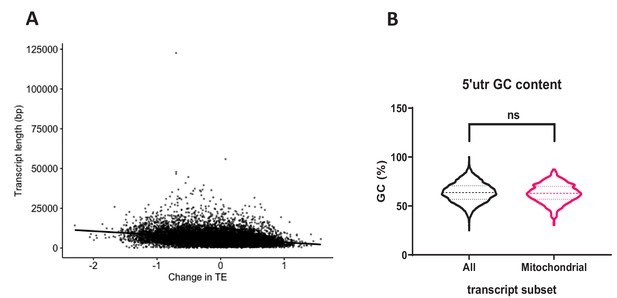
Further analysis of polysome profile dataset.
(A) Change in translational efficiency (TE) after BUD23 siRNA treatment is plotted against transcript length. Transcript length was retrieved using the EDAseq package in R. (B) Distribution of 5’UTR GC content for ‘All’ transcripts analysed in the polysome profiling dataset (Figure 1F), compared to the transcripts annotated to the ‘mitochondrial’ compartment. No significant difference was detected using the Mann-Whitney statistical test.
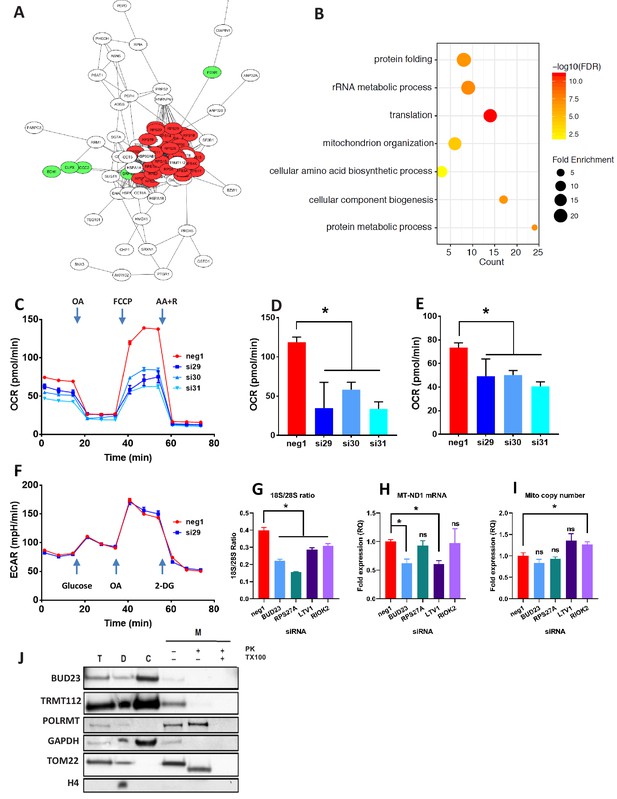
Proteomic analysis of BUD23 deficiency reveals mitochondrial phenotype Proteomics was performed on A549 cell lysates 48 hr after transfection with either BUD23-targeting siRNA or control siRNA.
Significantly down-regulated and up-regulated proteins were defined using Welch’s T-test with a permutation-based FDR < 0.05. (A) Significantly down-regulated proteins were visualised as a network using String 10 online software. Red nodes are 40S ribosomal proteins, Green nodes are mitochondrial proteins (B) Over-representation analysis of the significantly down-regulated protein dataset was performed using the PANTHER GO-Slim Biological Processes, and significantly over-represented terms were visualise using ggplot2. (C) Mitochondrial stress test was conducted on A549 cells treated with BUD23-targeting or non-targeting control siRNA (n = 3). Oxygen consumption rate (OCR) was measured over time to obtain mitochondrial respiration rates. Values for ETS (D) and OXPHOS (E) were derived from the OCR measurements after the addition of oligomycin A (OA), FCCP and antimycin A + rotenone (AA+R) in succession. (F) Glycolytic capacity was tested using a Seahorse glycolytic stress test and measuring extracellular acidification rate (ECAR). (G) A549 cells were treated with control siRNA (neg1) or siRNA specifically targeting BUD23 (si30), RPS27A, LTV1 or RIOK2. 18S/28S ratio was measured using Tapestation to confirm ribosome subunit imbalance. (H) MT-ND1 mRNA was measured by qPCR as an indicator of mitochondrial transcript abundance (relative to control gene HPRT). (I) Mitochondrial copy number was measured relative to genomic copy number by qPCR. (J) Cellular fractionation into total ‘T’, cytosolic ‘C’, cellular debris ‘D’ and mitochondrial ‘M’ fractions. Protein was isolated from these fractions before SDS-PAGE and immuno-blotting for BUD23 and TRMT112. POLRMT was blotted as a control for mitochondrial proteins, TOM22 as a control for a mitochondrial membrane protein and GAPDH and Histone H4 as control non-mitochondrial proteins.
-
Figure 2—source data 1
Proteomics data table.
Proteomics was performed on A549 cell lysates 48 hr after transfection with either BUD23-targeting siRNA or control siRNA. Significantly down-regulated and up-regulated proteins were defined using Welch’s T-test with a permutation-based FDR < 0.05.
- https://cdn.elifesciences.org/articles/50705/elife-50705-fig2-data1-v2.xlsx
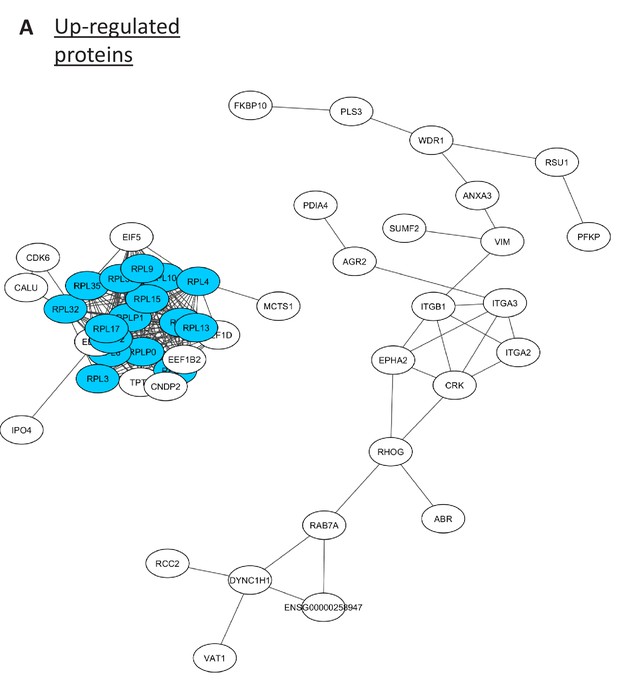
Network analysis of up-regulated proteins.
(A) The up-regulated proteins from proteomic analysis of BUD23-deficient A549 cells are visualised as a STRING10 network.
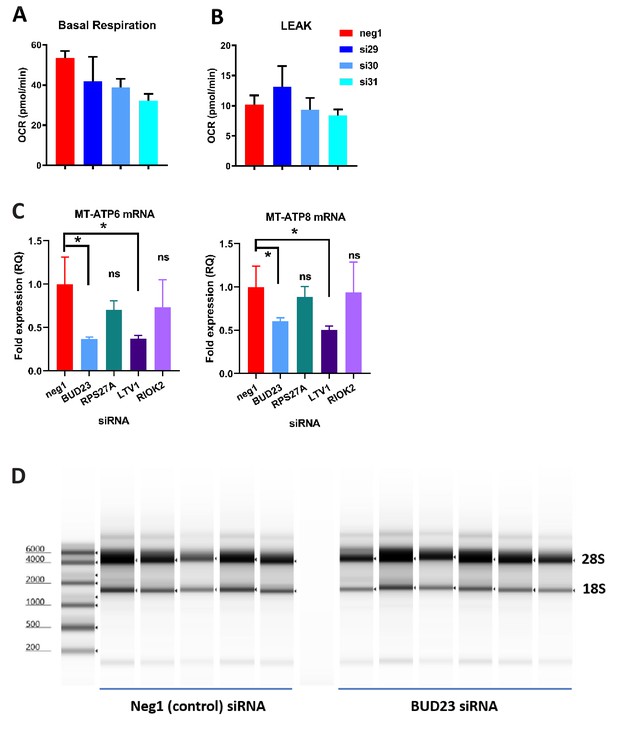
Further analysis of ribosomal deficiency in A549 cells.
(A and B) Further analysis of the data in Figure 2C: Basal respiration was determined from average OCR measurements of A549 cells in an unstimulated state. LEAK respiration was determined by subtracting the average OCR after addition of OA from the average OCR after addition of AA+R. (C) qPCR analysis of mitochondrial gene expression (MT-ATP6 and MT-ATP8) in A549 cells treated with control siRNA (neg1) or siRNA specifically targeting BUD23, RPS27A, LTV1 or RIOK2. (D) Example RNA screentape gel showing 28S and 18S bands for A549 cells treated with either control siRNA or BUD23 specific siRNA.
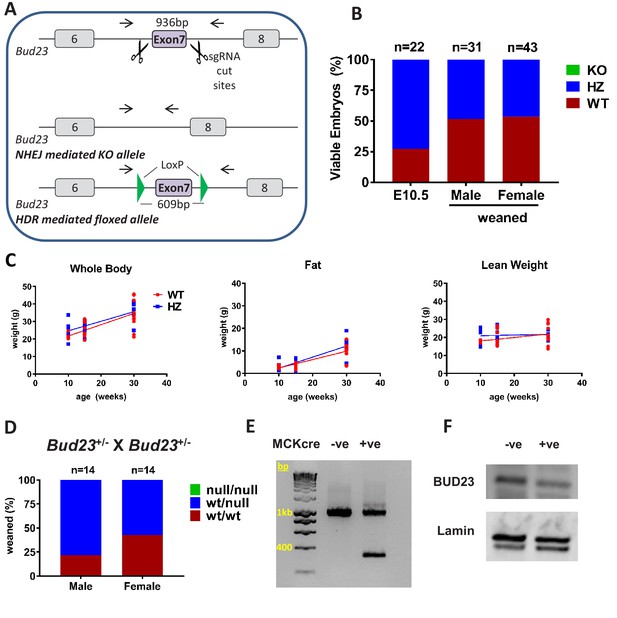
Generation and characterisation of murine Bud23 null and floxed alleles.
(A) Schematic of the targeting strategy used to create a murine Bud23 knockout allele and a floxed allele were generated by CRISPR-Cas9. (B) Genotype analysis of day E10.5 embryos and the offspring from heterozygous breeding pairs. (C) Bud23 Heterozygous (HZ) and wildtype (WT) mice (10–30 weeks old) were analysed over time for body composition by ECHO-MRI. (D) Genotype analysis of offspring from heterozygous recombined Bud23 (Bud23rec/wt) allele crosses. (E) PCR amplification of the Bud23 genomic locus in Bud23fl/fl mice positive or negative for MCKcre expression in heart tissue, shows successful recombination of the mutant Bud23 allele (bp = base pairs). (F) Disruption of BUD23 protein in the cre positive animals was confirmed by Western blot.
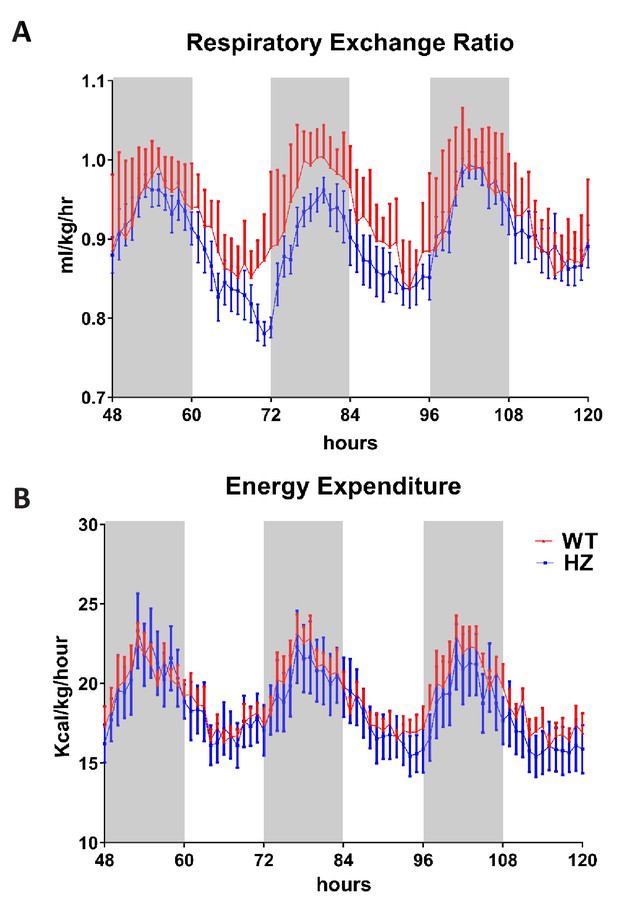
Metabolic analysis of BUD23 heterozygous mice.
(A and B) Metabolic function of adult HZ and WT mice was analysed over 3 days by CLAMS cage. (A) respiratory exchange ratio was calculated; (B) energy expenditure was calculated. Grey shading represents subjective night time.
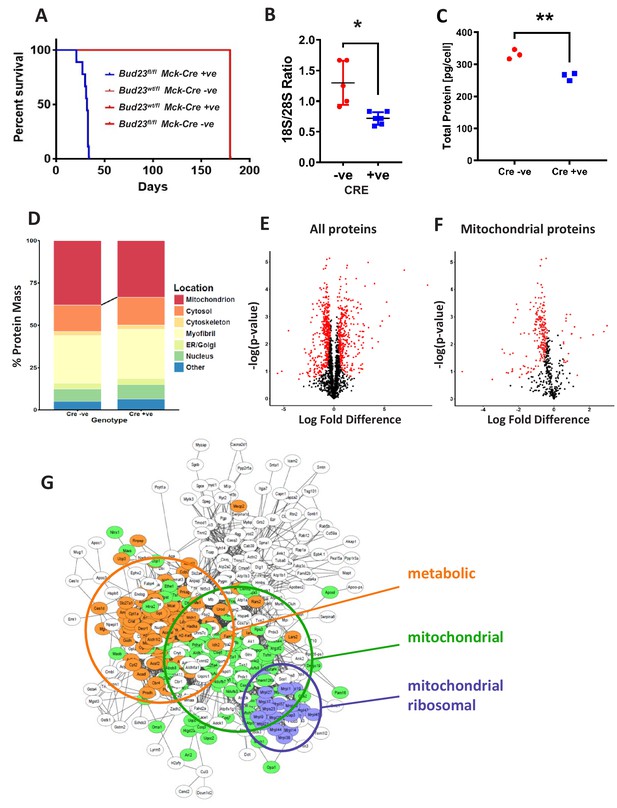
Proteomic analysis of BUD23 deficient heart tissue shows selective mitochondrial phenotype A murine Bud23 knockout allele and a floxed allele were generated by CRISPR-Cas9.
(A) Survival curve for Bud23fl/fl Mck-Cre+/- (Bud23 knock-out targeted to heart and muscle) shown in blue, compared to littermate controls (Bud23wt/fl Mck-Cre-/-, Bud23wt/fl Mck-Cre+/-, Bud23fl/fl Mck-Cre-/-), all shown in red. For subsequent analyses, hearts were collected from 26 day old Bud23fl/fl Mck-Cre+/-, and Bud23fl/fl Mck-Cre-/- littermate controls. (B) 18S/28S ratio analysis of RNA extracted from BUD23 positive or negative mouse hearts using TapeStation. (C) Proteomics was performed on heart tissue from Cre +ve and Cre -ve littermates (n = 3). Total protein concentration per cell is shown, as determined via the proteomic ruler approach. (D) Relative contribution levels of each organelle to total protein mass based on ontology. The decrease in mitochondrial mass observed in Cre positive hearts is indicated. (E and F) Volcano plots of either total protein or mitochondrial protein fold change differences in Cre positive hearts relative to Cre negative hearts. Significantly up- and down-regulated proteins are indicated in red (FDR < 0.05, s0 = 0.2). (G) Functional enrichment network of total down-regulated proteins. The network was built using String 10 online software prior to export into Cytoscape.
-
Figure 4—source data 1
Proteomics data table.
Proteomics was performed on heart tissue from BUD23 deficient mice and littermate control mice (n = 3). Excel workbook tabs: ‘All detected’ are all proteins detected in the analysis; ‘mitochondrial-only’ are proteins which have a mitochondrial subcellular location listed; ‘UpreginKO’ are proteins which were defined as significantly increased in the BUD23-deficient condition (FDR < 0.05); ‘DownregInKO’ are proteins defined as significantly decreased in the BUD-23 deficient condition (FDR < 0.05); ‘ProteomicRuler_SubcellLocation’ shows the calculation of subcellular location for detected proteins; ‘MassCalculations’ shows the final calculation used to produce Figure 4D.
- https://cdn.elifesciences.org/articles/50705/elife-50705-fig4-data1-v2.xlsx
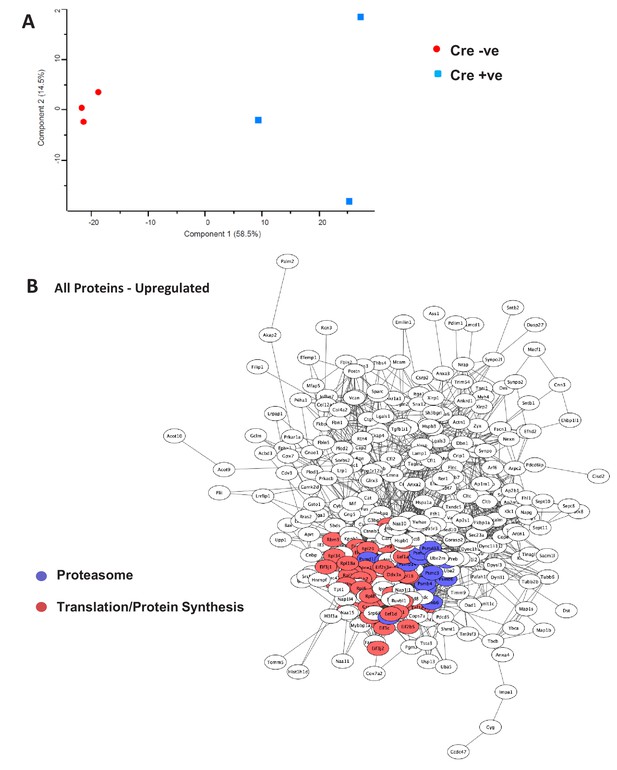
Proteomic analysis of BUD23 deficient mouse hearts.
(A) PCA analysis of proteomic data from BUD23 deficient cardiac homogenates and littermate controls. (B) Network analysis of all significantly upregulated proteins. The cluster of proteins involved in translation/protein synthesis are coloured red; the cluster of proteins associated with the proteasome are highlighted in purple.
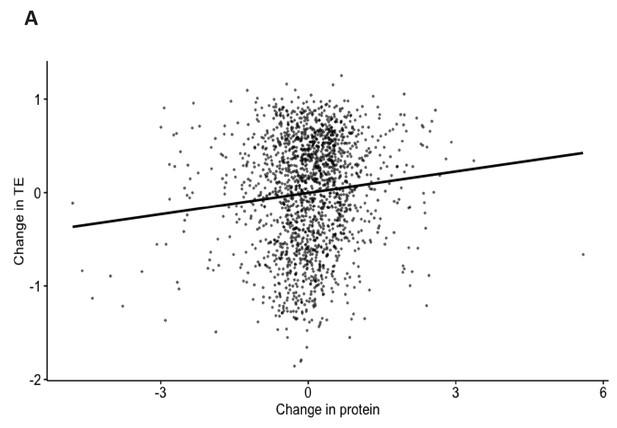
Change in TE correlates poorly with change in protein abundance.
(A) Correlation analysis of change in TE after BUD23 knockdown (x-axis, change in TE) plotted against change in protein abundance in BUD23-deficient murine hearts (y-axis, change in protein). Spearman’s rank correlation coefficient: R = 0.11, p=4e^−6.
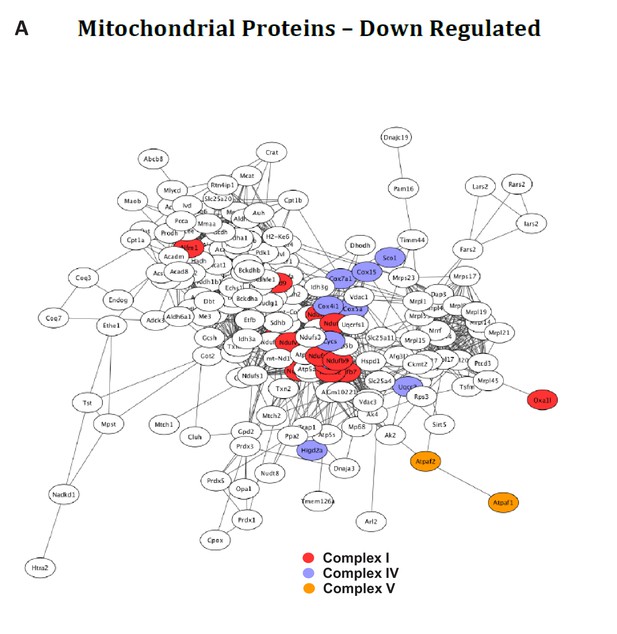
Network analysis of down-regulated proteins in BUD23 deficient mouse hearts.
(A) Network analysis of all downregulated mitochondrial proteins. Electron transport chain complexes I, IV and V are highlighted in red purple and orange respectively.
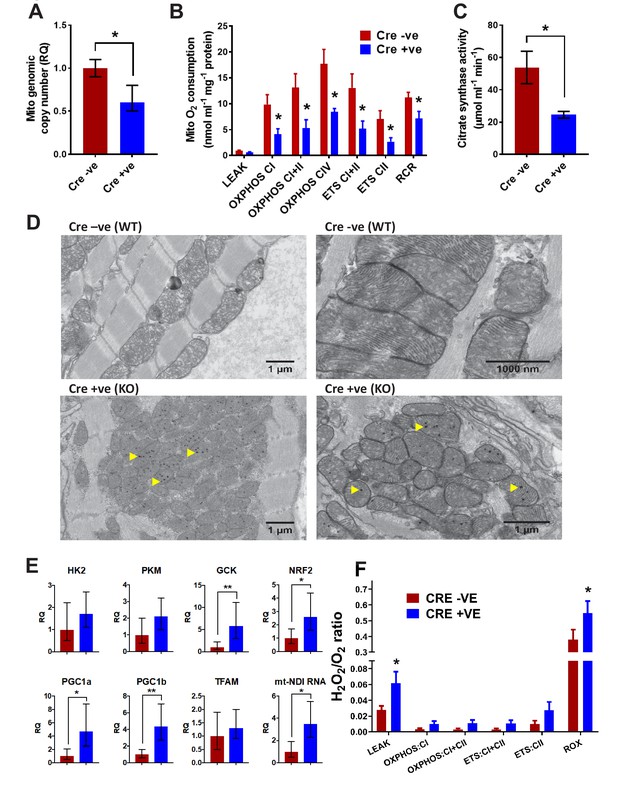
Analysis of mitochondrial structure and function in BUD23 depleted heart.
Mouse tissues were collected at 26 days old. (A) Mitochondrial genome copy number measured in the heart tissue by qPCR, relative to autosomal genome copy number (n = 6). (B) Mitochondrial respiration was measured in heart homogenates (n = 5–6). Oxygen consumption was measured under various mitochondrial states and normalised to protein content: LEAK conditions (malate, pyruvate and glutamate as substrates); OXPHOS (routine respiration rate) through Complex I, I+II or IV; ETC (maximal respiration rate) through Complex I+II and II; Respiratory Control Ratio (RCR, measure of mitochondrial efficiency). (C) Citrate Synthase activity was measured in the heart homogenates as a marker of mitochondrial content. (D) Electron microscopy images of mitochondria in cardiomyocytes from either Bud23fl/fl (WT) hearts, or Bud23fl/fl Mck-Cre +ve (KO) hearts. Accumulation of electron dense granules is visible in Bud23fl/fl Mck-Cre +ve (KO) hearts, denoted by arrows. (E) These hearts were further analysed for expression of mitochondrial biogenesis genes, markers of glycolysis, mitochondrial biogenesis, oxidative stress and indicators of mitochondrial transcription using qPCR. (F) ROS was measured simultaneously with O2 consumption (see Figures 1–3) under various mitochondrial states and normalised to citrate synthase activity: LEAK conditions (malate, pyruvate and glutamate as substrates); OXPHOS (routine respiration rate) through Complex I or I+II; ETS (maximal respiration rate) through Complex I+II and II, and in the presence of antimycin-A (residual oxygen consumption, ROX)(n = 5–6).
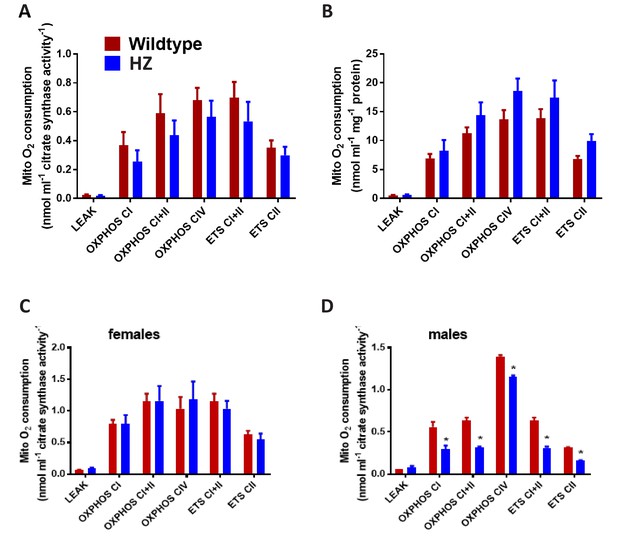
Oroboros analysis of mitochondrial function in cardiac homogenates from HZ mice and WT littermate controls.
Analysed to citrate synthase activity and protein content respectively in (A) and (B). (C and D) The data in Figure 4B separated into males and females respectively (normalised to citrate synthase activity).
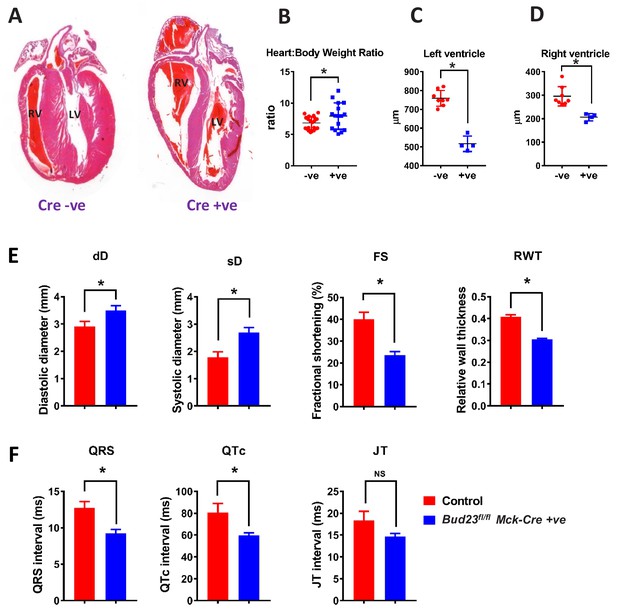
Targeted disruption of Bud23 in cardiac tissue results in dilated cardiomyopathy.
(A) Heart sections cut in the coronal plane and stained with haematoxylin and eosin from day 26 mice. Representative images are shown. (B) Heart weight measured as a ratio to total bodyweight. (C and D) Analysis of heart ventricle wall thickness in H and E stained heart sections. (E) Echocardiography of Bud23fl/flMck-Cre+/- mice and littermate controls at 26–28 days of age (n = 9–10). Mice were lightly anesthetized maintaining the heart rate at approximately 450 beats/min. Data for diastolic diameter (dD), systolic diameter (sD), fractional shortening (FS) and relative wall thickness (RWT) are shown; full parameter measurements are shown in supplementary. (F) Electrocardiography of Bud23fl/flMck-Cre+/- mice and littermate controls at 26–28 days of age (n = 9–10). Data for QRS, QTc and JT intervals are shown. Full parameter measurements are shown in supplementary. * denotes statistical significance (p<0.05) using Student’s T-test.
-
Figure 6—source data 1
Data table of echocardiogram analysis results and electrocardiogram analysis results.
LW/BW, lung weight/body weight; IVSD, interventricular septum thickness in diastole; IVSS, interventricular septum thickness in systole; LVPWD, posterior wall thickness in diastole; LVPWS, posterior wall thickness in systole; BPM, beat/per minute; RWT, relative wall thickness. Statistical p-values were defined using Student’s T-test.
- https://cdn.elifesciences.org/articles/50705/elife-50705-fig6-data1-v2.docx
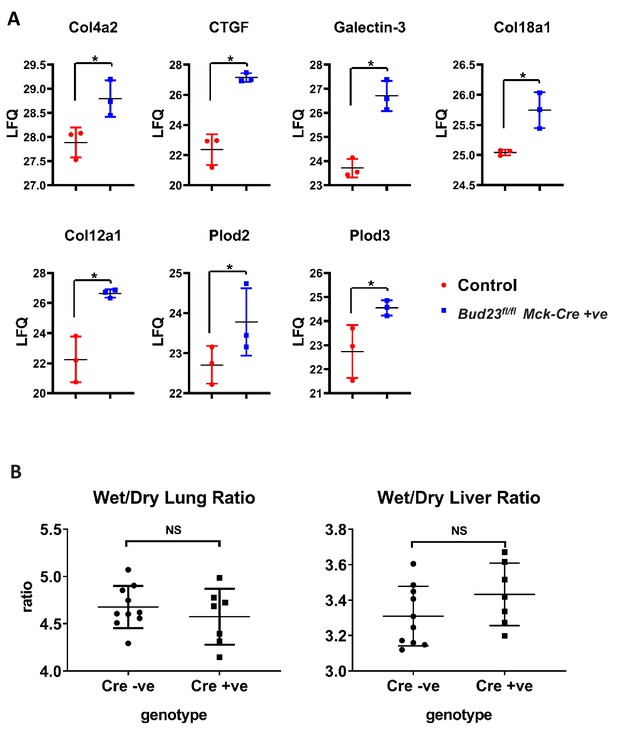
Fibrosis markers are up-regulated in BUD23 deficient mouse hearts.
(A) Fibrosis related proteins identified as significantly up-regulated in heart tissue from Bud23fl/flMck-Cre +/- mice and littermate controls Data are shown as Log2(LFQ) values. (B) Lungs and livers were removed en bloc from mice deficient for cardiac BUD23 or littermate controls, then weighed before and after drying to give a wet/dry ratio.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | BUD23 | Gene ID: 114049 | Aliases: WBSCR22, MERM1 | |
| Genetic reagent (Mus musculus) | Bud23null | This paper | Bud23null | Mutant allele: deleted exon 7 of BUD23 gene |
| Genetic reagent (Mus musculus) | BUD23fl/fl | This paper | BUD23fl/fl | Mutant allele: floxed exon 7 of BUD23 gene |
| Genetic reagent (Mus musculus) | Mck-Cre | Whitnall et al., 2008 | B6.FVB(129S4)-Tg(Ckmm-cre)5Khn/J | Heart and muscle specific Cre-driver |
| Cell line (Homo-sapiens) | A549 | ATCC | CCL-185 | Airway epithelial adenocarcinoma |
| Antibody | anti-BUD23 (Rabbit polyclonal) | GeneTex | Cat# GTX105840 | WB (1:500) |
| Sequence-based reagent | BUD23_F | This paper | Genotyping primers | 5’-CCTGGCTGATGTGTTGCTTT-3’ Annealing temperature: 62°C |
| Sequenced-based reagent | BUD23_R | This paper | Genotyping primers | 5’-GCTGCACATCTCTCCTCACT-3’ Annealing temperature: 62°C |
| Sequence-based reagent | MT-ND1_F | This paper | qPCR primers | 5’-GAAGTCACCCTAGCCATCATTC-3’ |
| Sequenced-based reagent | MT-ND1_R | This paper | qPCR primers | 5’GCAGGAGTAATCAGAGGTGTTC-3’ |
| Sequenced-based reagent | siRNA: nontargeting control 1 | Thermo Fisher | AM4611 | Silencer Select |
| Sequenced-based reagent | siRNA: nontargeting control 2 | Thermo Fisher | 4390847 | Silencer Select |
| Sequenced-based reagent | siRNA: BUD23 | Thermo Fisher | 41529 | Silencer Select |
| Sequenced-based reagent | siRNA: BUD23 | Thermo Fisher | 41530 | Silencer Select |
| Sequenced-based reagent | siRNA: BUD23 | Thermo Fisher | 41531 | Silencer Select |
| Software, algorithm | R | www.r-project.org | Key packages: -EDGER -BIOCONDUCTOR -EDASEQ | |
| Commercial assay or kit | Seahorse XF Mito stress test | Agilent | 103015–100 | |
| Commercial assay or kit | Seahorse XF Glyco stress test | Agilent | 103020–100 |
Additional files
-
Supplementary file 1
Primer sequences.
- https://cdn.elifesciences.org/articles/50705/elife-50705-supp1-v2.xlsx





