Essential role for InSyn1 in dystroglycan complex integrity and cognitive behaviors in mice
Figures
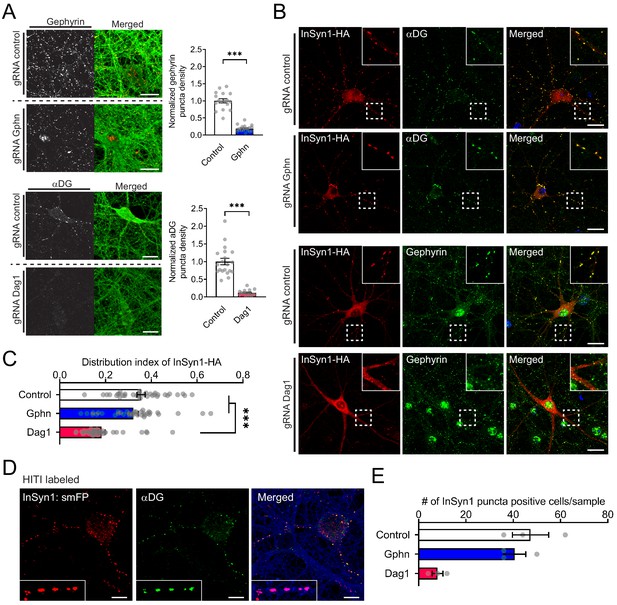
InSyn1 localization to the iPSD is DGC dependent.
(A) Depletion of Gephyrin or αDG by CRISPR in neurons. Cas9 knock-in hippocampal neurons were transduced with AAV:Cre/(control)gRNA [control], AAV:Cre/(Gphn)gRNA [gephyrin] or AAV:Cre/(Dag1)gRNA [αDG] at DIV1 and stained with gephyrin or αDG at DIV13 (left panel). GFP fluorescence of the Cas9-2A-GFP (right panel). Graphs to the right show the normalized puncta density. Gphn vs control (two-tailed t-test, gephyrin n = 19, control n = 17, p<0.0001), Dag1 vs control (two-tailed t-test, αDG n = 16, control n = 20, p<0.0001). (B, C) InSyn1-HA localization after αDG or gephyrin CRISPR depletion. Neurons were depleted of αDG or gephyrin, followed by AAV:InSyn1-HA transduction 3 days before fixation. Exogenously expressed InSyn1-HA is shown in red. Endogenous gephyrin or αDG are shown in green. Bar graph showing the distribution index of InSyn1-HA as arbitrary units. Control (n = 36), Gphn (n = 36), Dag1 (n = 36). One-way ANOVA followed by Tukey's multiple comparisons test, F (2, 105)=25.49, ***p<0.001. Scale bars, 20 µm. (D). HITI labeling of endogenous InSyn1 with smFP-HA in Cas9 KI neurons. InSyn1 is shown in red and αDG is shown in green. Of note, InSyn1:smFP showed clear puncta staining colocalized with αDG. (E) InSyn1 puncta-positive cells were quantified in either Control, Gphn, or Dag1 depleted neurons (n = 3).
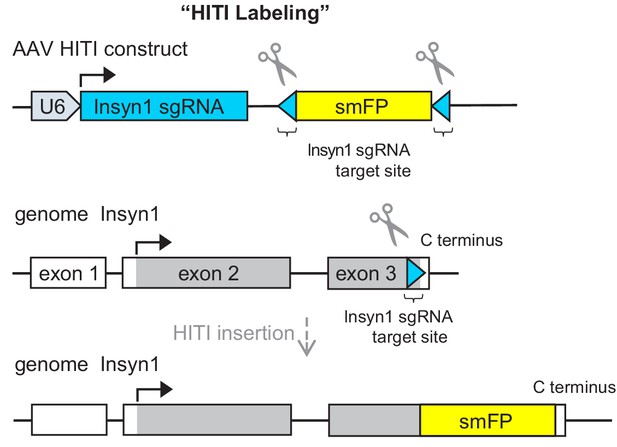
Schematic illustration of HITI labeling.
To epitope tag the C-terminus of InSyn1, a guide targeting the end of the coding region was chosen. After CRISPR-dependent cutting, smFP-HA that contains 12 HA epitopes was directly inserted in-frame to label endogenous InSyn1 in neurons.
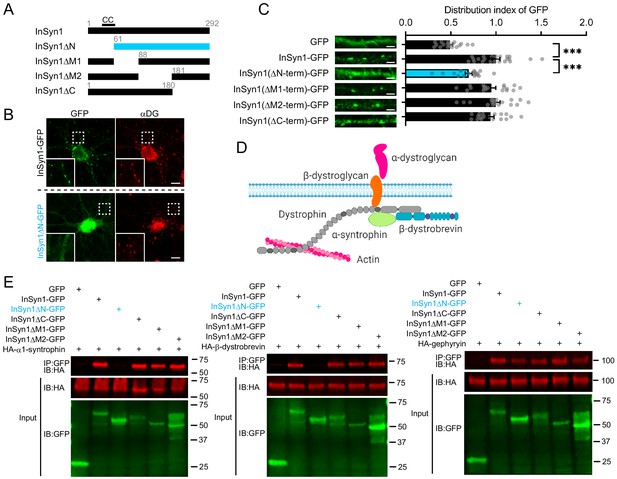
The N-terminus of InSyn1 is essential for its localization and interaction with DGC components.
(A) Schematic of InSyn1 deletion mutants used for co-immunoprecipitation or localization in neurons. CC: putative coiled-coil domain. (B) Representative images of full-length or ΔN InSyn1-GFP expression (green) in neurons immunostained with antibodies to αDG (red). Scale bar; 5 µm. (C) Left; representative images of dendrites expressing each construct. Right, graphs depicting the distribution index of InSyn1 versus negative control (GFP) or InSyn1 mutants. GFP (n = 18), InSyn1-GFP (n = 22), InSyn1ΔN-GFP (n = 18), InSyn1ΔM1-GFP (n = 18), InSyn1ΔM2-GFP (n = 18), InSyn1ΔC-GFP (n = 18). One-way ANOVA followed by Tukey’s post-hoc tests, F (5, 106)=23.99, ***p<0.001. Scale bars, 2 µm. (D) Schematic of the dystrophin/dystroglycan complex (DGC) in neurons. Created with BioRender.com (E) Representative immunoblots following co-immunoprecipitation of GFP (negative control), InSyn1-GFP, or InSyn1-GFP mutants with HA-α1-syntrophin, HA-β-dystrobrevin, or HA-gephyrin. Protein constructs were expressed in HEK293T cells and precipitated by GFP-Trap. IP: immunoprecipitated, IB: immunoblot.
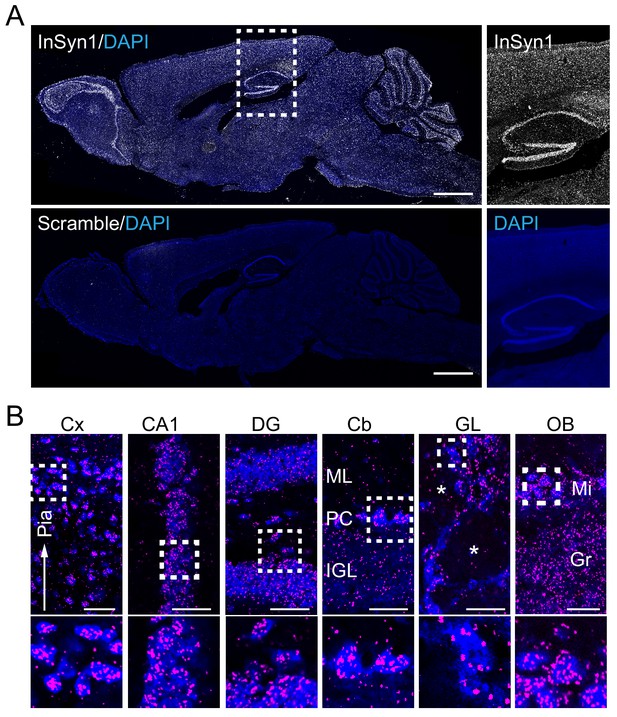
InSyn1 expression distribution in the mouse brain.
(A) InSyn1 mRNA (white) was detected throughout the adult mouse brain with strong signals in the hippocampus, cerebellum and olfactory bulb. Nissle stain (blue). Magnified images of the hippocampus and the cortex are shown. (B) Numerous clusters were found in cells in different layers of the cortex (Cx), pyramidal cell layers (CA1) and dentate gyrus granule cells (DG) in the hippocampus, Purkinje cells in the cerebellum (Cb), cells surrounding the glomerulus (GL) and in the mitral cell layer of the olfactory bulb (OB). Cx; cerebrum cortex, CA1; hippocampus CA1, DG; dentate gyrus, Cb; cerebellum, OB; olfactory bulb, GL; glomerular layer, Mi; mitral cell layer, Gr; granular cell layer, ML; molecular cell layer, PCL; Purkinje cell layer, IGL; internal granule layer. The asterisk represents the glomerulus. Scale bars, 1 mm (A), 50 um (B).
-
Figure 3—source data 1
Hybridization probes.
- https://cdn.elifesciences.org/articles/50712/elife-50712-fig3-data1-v2.xlsx
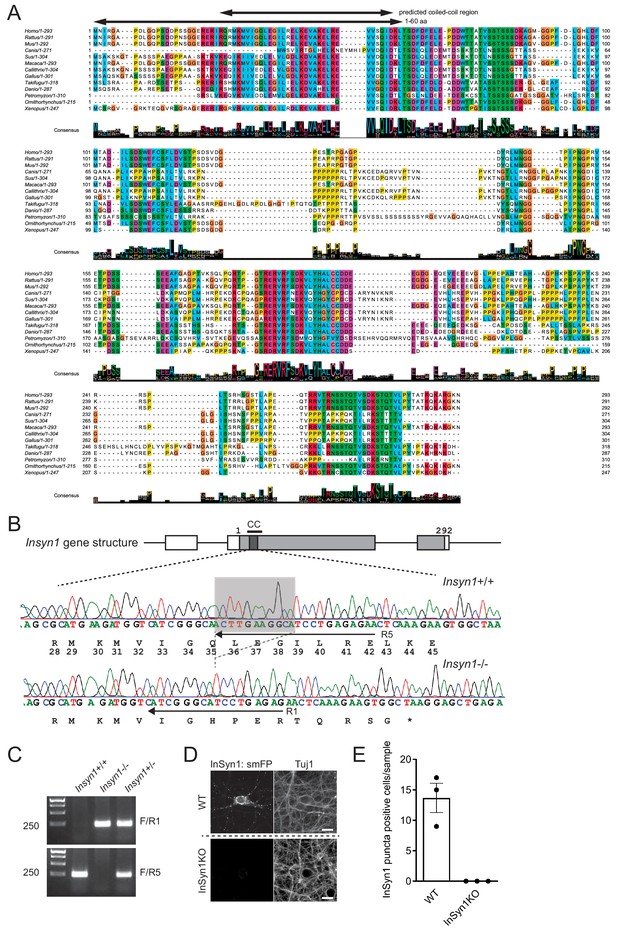
Generation of InSyn1 KO mice.
(A) Multiple protein sequence alignment of InSyn1 from human (Homo sapiens), rat (Rattus norvegicus), mouse (Mus musculus), Dog (Canis lupus), Wild pig (Sus scrofa), monkey (Macaca fascicularis), marmoset (Callithrix jacchus), chicken (Gallus gallus), Lamprey (Petromyzon marinus), platypus (Ornithorhynchus anatinus), xenopus (Xenopus laevis), and two fish species (Takifugu rubripes), (Danio rerio), The set of sequences were chosen from the Gene Tree of human InSyn1 (ENSGT00910000144204). The sequences were aligned by multiple sequence alignment algorithm Kalign2 and manually curated by JalView (Lassmann et al., 2009). The consensus sequence with sequence logos is depicted below. The predicted coiled-coil region and the N terminus 60 amino acid-deleted InSyn1 mutant (InSyn1ΔN) are indicated above. Note the sequence similarities of InSyn1 between different species. (B) Schematic of InSyn1 gene structure (top, gray box regions representing coding sequence) and the alignment of DNA sequencing from WT (top trace) and InSyn1 (bottom trace) KO mice showing an 11bp-deletion in exon two in InSyn1 KO mice. (C) Representative images of PCR-based genotyping. F; a common forward primer. R1 and R5; reverse primers depicted in B. (D and E). HITI labeling of endogenous InSyn1 in WT and KO hippocampal neurons. (D) WT neurons exhibit clear puncta staining along the neurite while InSyn1 null neurons lost puncta staining. scale bars, 10 µm. (E) InSyn1 puncta-positive cells were quantified from three different samples. Of note, no InSyn1-positive cells were detected from null hippocampal neurons.
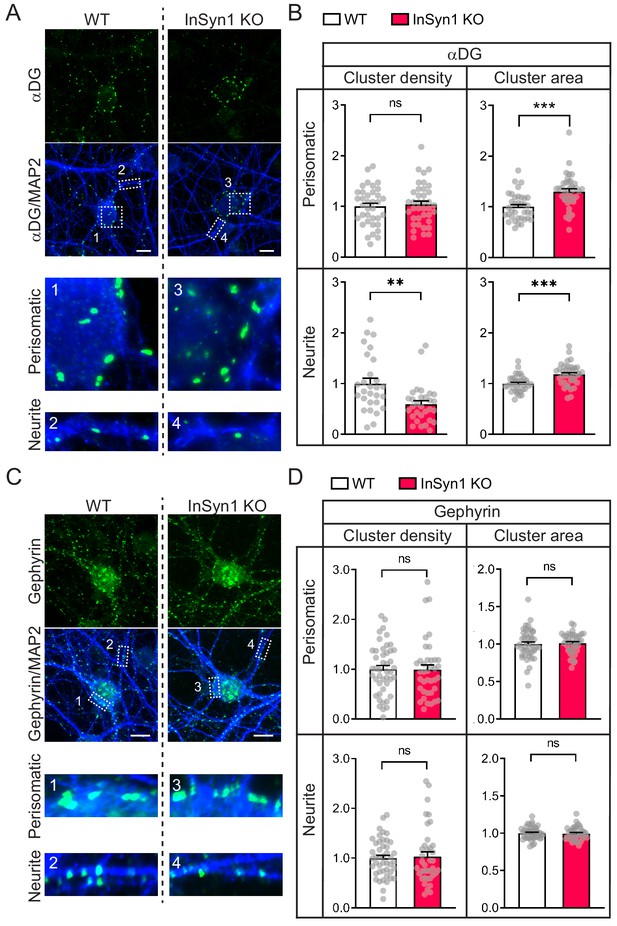
InSyn1 modulates the distribution of the DGC but not gephyrin in neurons.
(A) Representative images of WT and InSyn1 KO hippocampal neurons at DIV13, labeled with antibodies to αDG (green) and MAP2 (blue). Scale bars, 10 um. (B) Bar graphs showing normalized αDG cluster density or area size either at perisomatic regions or at neurite regions. Perisomatic cluster density (WT n = 36, KO n = 38, two-tailed t-test, p=0.6491), neurite cluster density (WT n = 28, KO n = 30, two-tailed t-test, p=0.0010), perisomatic cluster area (WT n = 36, KO n = 38, two-tailed t-test, p<0.0001), and neurite cluster area (WT n = 36, KO n = 38, two-tailed t-test, p<0.0001). *p<0.05, **p<0.001, ***p<0.0001. (C) Representative images of WT and InSyn1 KO hippocampal neurons labeled with antibodies to gephyrin (green) and MAP2 (blue). Scale bars, 10 µm. (D) Bar graphs showing normalized gephyrin cluster density or area size. Perisomatic cluster density (WT n = 45, KO n = 38, two-tailed t-test, p=0.5963), neurite cluster density (WT n = 45, KO n = 38, two-tailed t-test, p=0.6331), perisomatic cluster area (WT n = 45, KO n = 39, two-tailed t-test, p=0.7923), and neurite cluster area (WT n = 36, KO n = 38, two-tailed t-test, p=0.8098).
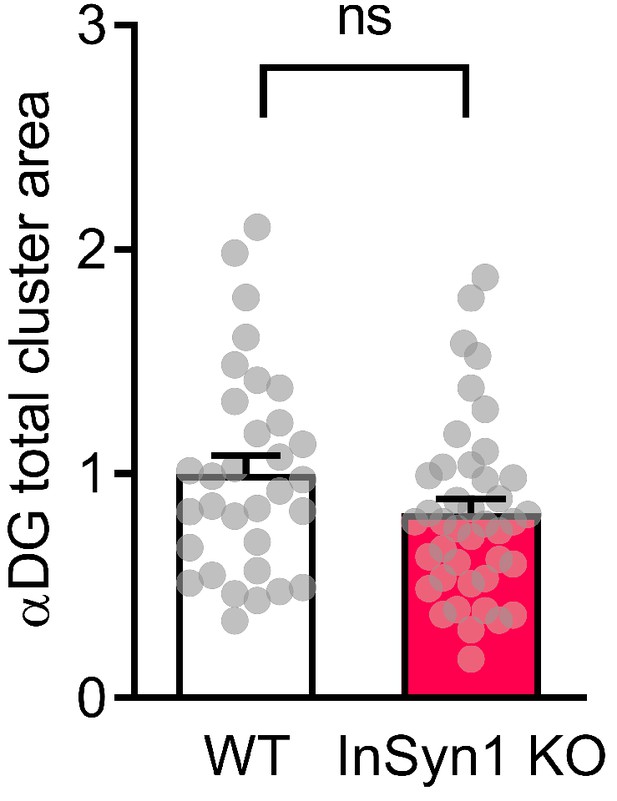
Quantification of αDG area in hippocampal neurons.
Bar graphs showing normalized total αDG cluster area size (WT n = 32, KO n = 41, two-tailed t-test, p=0.0839).
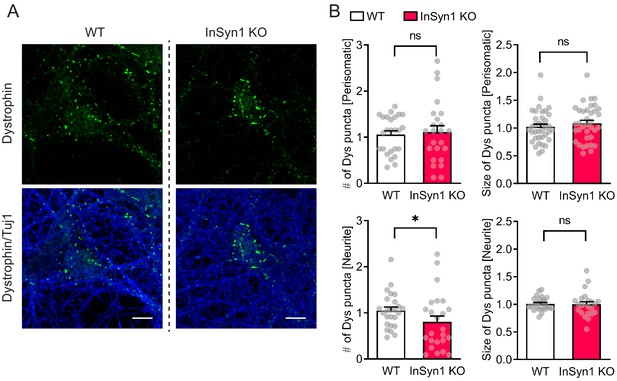
InSyn1 modulates the distribution of dystrophin in neurons.
(A) Representative images of WT and InSyn1 KO hippocampal neurons at DIV13, labeled with antibodies to dystrophin (green) and Tuj-1 (blue). Scale bars, 10 um. (B) Bar graphs showing normalized dystrophin cluster density or area size either at perisomatic regions or at neurite regions. Perisomatic cluster density (WT n = 25, KO n = 24, two-tailed t-test, p=0.9249), neurite cluster density (WT n = 25, KO n = 24, two-tailed t-test, p=0.0371), perisomatic cluster area (WT n = 25, KO n = 24, two-tailed t-test, p<0.6096), and neurite cluster area (WT n = 25, KO n = 23, two-tailed t-test, p<0.6234). *p<0.05.
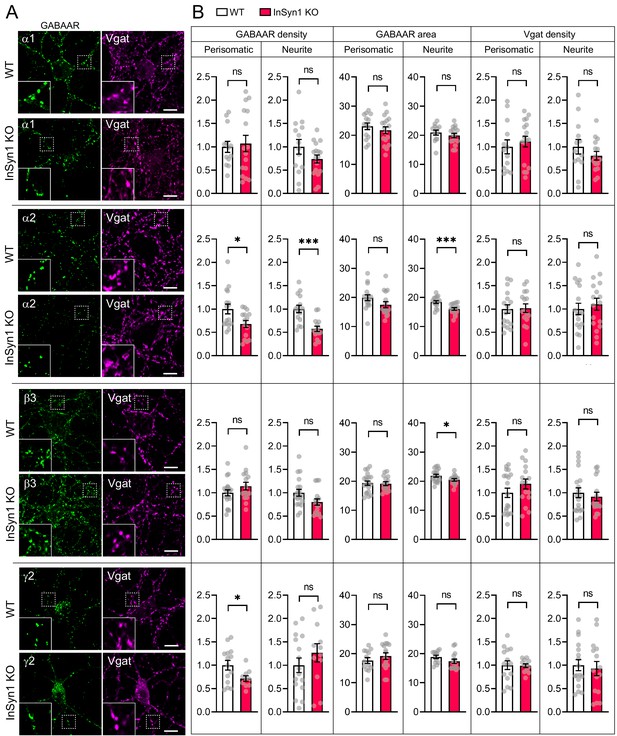
Altered distribution of GABAAR clustering in InSyn1 KO neurons.
(A) Representative images of WT and InSyn1 KO neurons at DIV13, immunostained with α1, α2, β3, and y2 GABAAR subunits (green), and Vgat (magenta). Scale bars, 10 µm. (B) Bar graphs showing the quantification of normalized cluster density and area size of each GABAAR and Vgat from perisomatic or neurite regions. GABAARα1 density (Two-tailed t test, Perisomatic; WT n = 13, KO n = 16, p=0.7421. Neurite; WT n = 14, KO n = 17, p=0.1589), and area (Two-tailed t test, Perisomatic; WT n = 14, KO n = 17, p=0.4237. Neurite; WT n = 12, KO n = 16, p=0.4012). GABAARα2 density (Two-tailed t test, Perisomatic; WT n = 16, KO n = 15, p=0.0179. Neurite; WT n = 17, KO n = 18, p=0.0021), and area size (Two-tailed t test, Perisomatic; WT n = 16, KO n = 16, p=0.1083. Neurite; WT n = 16, KO n = 16, p=0.0009). GABAARβ3 density (Two-tailed t test, Perisomatic; WT n = 19, KO n = 16, p=0.1874. Neurite; WT n = 17, KO n = 18, p=0.0522), and area size (Two-tailed t test, Perisomatic; WT n = 20, KO n = 15, p=0.8574. Neurite; WT n = 19, KO n = 15, p=0.0353). GABAARγ2 density (Two-tailed t test, Perisomatic; WT n = 14, KO n = 10, p=0.0254. Neurite; WT n = 16, KO n = 12, p=0.2996), and area size (Two-tailed t test, Perisomatic; WT n = 13, KO n = 16, p=0.3262. Neurite; WT n = 12, KO n = 15, p=0.1271). Of note, no significant difference was found in Vgat cluster quantifications (Two-tailed t-test. GABAARα1, Perisomatic; WT n = 13, KO n = 17, p=0.5510. Neurite; WT n = 13, KO n = 15, p=0.2996. GABAARα2, Perisomatic; WT n = 17, KO n = 16, p=0.8858. Neurite; WT n = 17, KO n = 17, p=0.5657. GABAARβ3, Perisomatic; WT n = 19, KO n = 16, p=0.3011. Neurite; WT n = 19, KO n = 16, p=0.5536. GABAARγ2, Perisomatic; WT n = 15, KO n = 12, p=0.9152. Neurite; WT n = 17, KO n = 15, p=0.7124). *p<0.05, ***p<0.001.
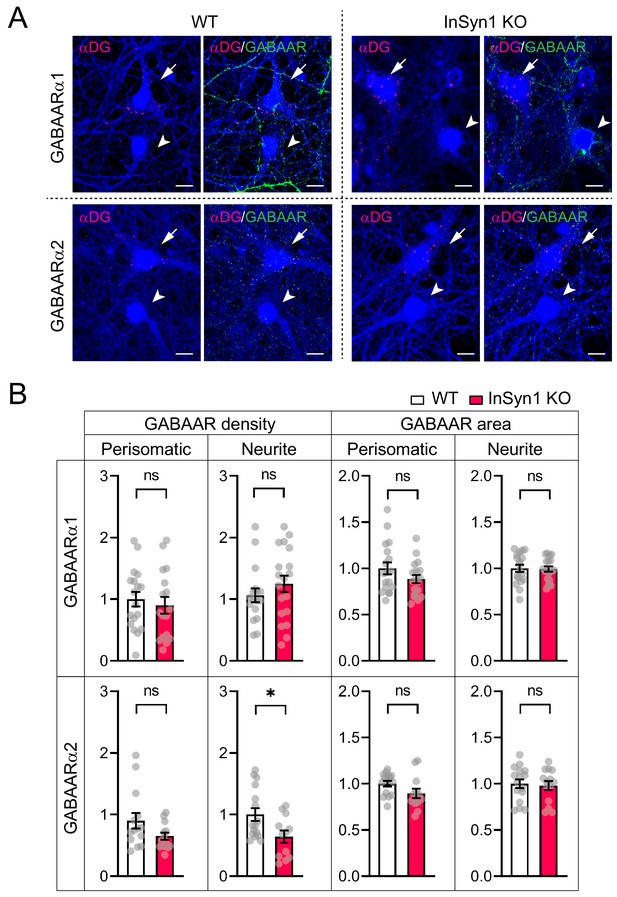
Quantification of GABAARs in αDG-negative cells.
(A) Representative images of WT and InSyn1 KO hippocampal neurons at DIV13, labeled with antibodies to αDG (red), GABAARs (green), and Tuj-1 (blue). αDG positive (arrows) and negative (arrowheads) are indicated. Scale bars, 10 µm. (B) Bar graphs showing normalized GABAARs cluster density or area size either at perisomatic or at neurite regions from αDG negative cells. GABAARα1 perisomatic cluster density (WT n = 19, KO n = 20, two-tailed t-test, p=0.0185), neurite cluster density (WT n = 20, KO n = 19, two-tailed t-test, p=0.0640), perisomatic cluster area (WT n = 19, KO n = 20, two-tailed t-test, p=0.1026), and neurite cluster area (WT n = 19, KO n = 19, two-tailed t-test, p=0.3932). GABAARα2 perisomatic cluster density (WT n = 16, KO n = 12, two-tailed t-test, p=0.3015), neurite cluster density (WT n = 16, KO n = 14, two-tailed t-test, p=0.0039), perisomatic cluster area (WT n = 17, KO n = 12, two-tailed t-test, p=0.3511), and neurite cluster area (WT n = 16, KO n = 12, two-tailed t-test, p=0.7319). *p<0.05, **p<0.001. Scale bars, 10 um.
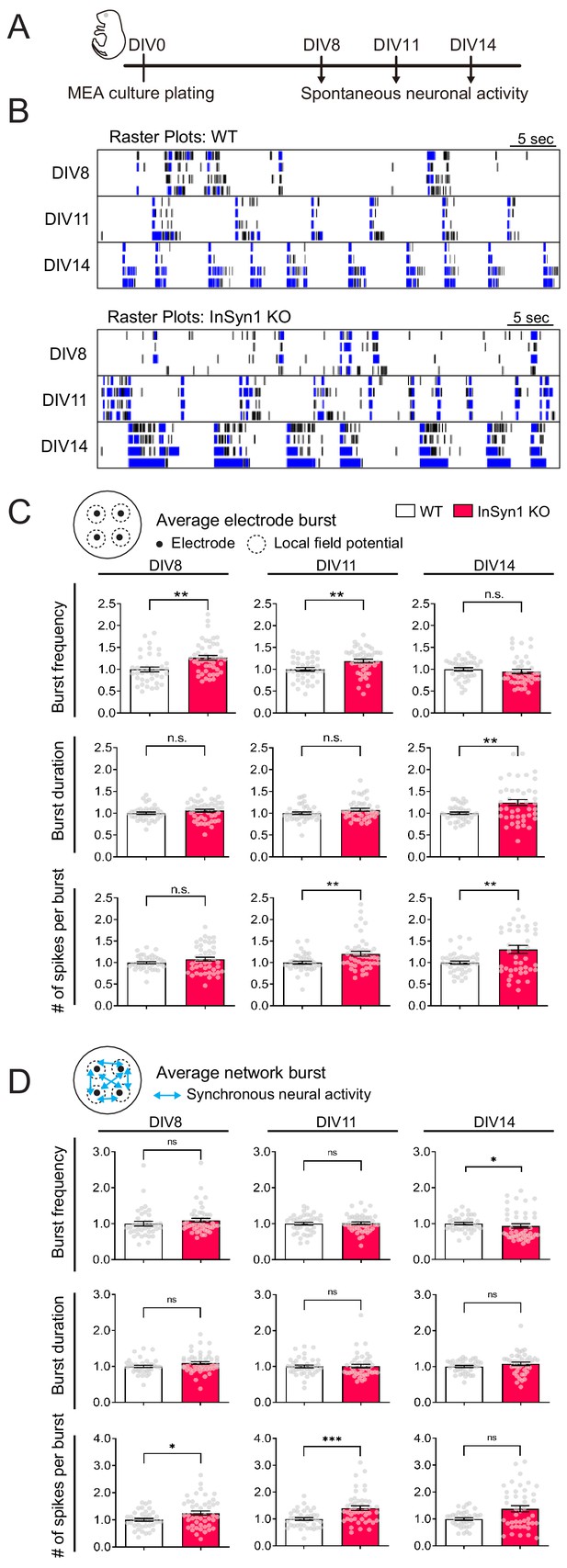
MEA recordings from InSyn1 KO neurons exhibit increased neuronal network activity.
(A) Experimental design to record spontaneous neuronal activity. Cultured cortical neurons were prepared from InSyn1 WT or KO neonatal pups at P0, and spontaneous neuronal activities were recorded at DIV8, 11, and 14. (B) Representative raster plots of four electrodes from each genotype recorded at DIV8, 11 and 14. Black ticks indicate the time of a spike occurred, and blue ticks indicate the spikes are part of single-electrode burst activity. (C) KO neurons showed increased spontaneous activity compare to WT at early time points. Bar graphs showing normalized average of burst frequency (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.0010. DIV11, WT n = 38, KO n = 45, p=0.0026. DIV14, WT n = 39, KO n = 43, p=0.1064), normalized average of burst duration (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.1689. DIV11, WT n = 38, KO n = 45, p=0.1195. DIV14, WT n = 39, KO n = 43, p=0.0021), and normalized average of number of spikes per burst (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.1460. DIV11, WT n = 38, KO n = 45, p=0.0093. DIV14, WT n = 39, KO n = 43, p=0.0038). **p<0.001. (D) Measurements of synchronous neuronal activities. Bar graphs showing the normalized average of network burst frequency (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.1253. DIV11, WT n = 38, KO n = 45, p=0.8531. DIV14, WT n = 39, KO n = 43 from three plates, p=0.0395.), normalized average of network burst duration (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.0742. DIV11, WT n = 38, KO n = 45, p=0.7676. DIV14, WT n = 39, KO n = 43, p=0.1380), and normalized average number of spikes per network burst (Two-tailed t-test. DIV8, WT n = 38, KO n = 46, p=0.0104. DIV11, WT n = 38, KO n = 45, p=0.0003. DIV14, WT n = 39, KO n = 43, p=0.0745). *p<0.05. ***p<0.001.
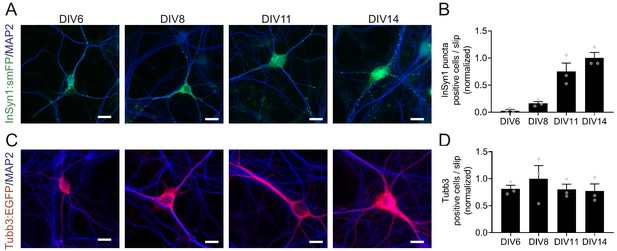
Developmental expression of InSyn1 in hippocampal neurons.
(A and C) Hippocampal neurons from H11 Cas9 neonatal pups were transduced at DIV0 either with AAV-InSyn1-HITI or AAV-mTubb3 to tag endogenous InSyn1 or β-III tubulin, respectively. Scale bars, 15 µm. (B and D) Graphs depicting the quantification of labeled neurons at each developmental time point.
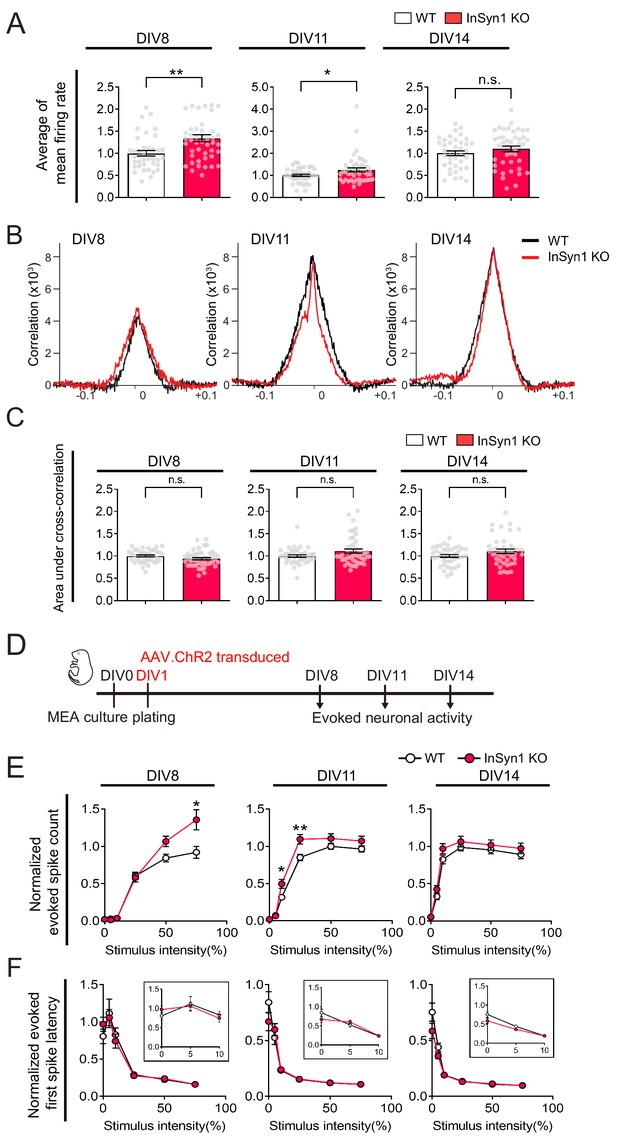
InSyn1 KO neurons showed increased network activities but no change at the network synchrony recorded from MEA.
(A) InSyn1 KO neurons exhibited an increased firing rate at early developmental stages. Bar graphs showing normalized average of mean firing rate (Two-tailed t-test, DIV8, WT n = 38, KO n = 46 from three plates, p=0.0017. DIV11, WT n = 38, KO n = 45 from three plates, p=0.0324. DIV14, WT n = 39, KO n = 43 from three plates, n.s.) *p<0.05, **p<0.001. (B) Examples of cross-correlation histogram from each time points. X-axis; time delay (s), y-axis; the strength of the correlation. (C) Graph showing cross-correlation between WT and InSyn1 KO neurons (Two-tailed t-test, DIV8, WT n = 38, KO n = 46 from three plates, p=0.0829. DIV11, WT n = 38, KO n = 45 from three plates, p=0.3555. DIV14, WT n = 39, KO n = 43 from three plates, p=0.0729). (D) Experimental design of recording network activity from optogenetically activated neurons. Cultured cortical neurons were prepared from InSyn1 WT or KO neonatal pups at P0, and light-evoked neuronal activities were recorded at DIV8, 11, and 14. (E) Graph showing normalized evoked spike count in response to light stimulation from 0% to 75%. (n = 19–49 from three independent MEA plates. Two-way ANOVA followed by Bonferroni’s comparisons test. DIV8 genotype effect, F (1, 467)=6.059, p<0.0142, 75% p<0.0001. DIV11 genotype effect, F (1, 456)=16.43, p<0.0001, 10% p=0.0287, 25% p=0.0007. DIV14 genotype effect, F (1, 482)=0.0146) *p<0.05, **p<0.0001. (F) Graph showing the normalized duration of first spike response to light stimulation (n = 17–49 from three independent MEA plates. Light stimulation from 5% to 75% was analyzed. Two-way ANOVA followed by Bonferroni’s comparisons test. DIV8 genotype effect, F (1, 387)=0.8015, p=0.3712. DIV11 genotype effect, F (1, 380)=1.139, p=0.2865. DIV14 genotype effect, F (1, 400)=2.928, p=0.0878). Inset graphs represent the light intensity response between 0 and 10%.
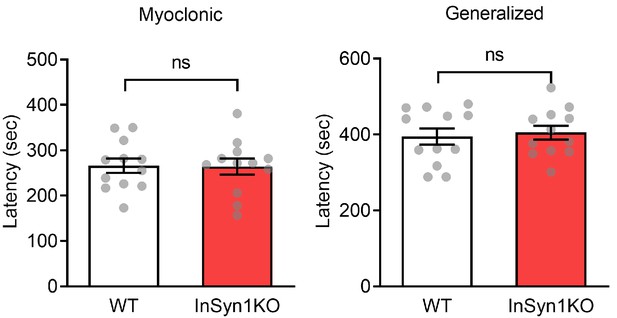
Flurothyl-induced seizure test showed no difference between WT and InSyn1 KO mice.
Graph of the latency to myoclonic seizure onset (Two-tailed t-test, WT n = 12, KO n = 12, p=0.9362) and generalized seizure (Two-tailed t-test, WT n = 12, KO n = 12, p=0.7148) were measured.
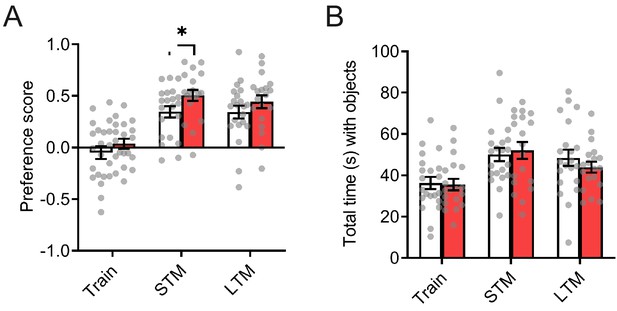
InSyn1- /- mice showed normal memory in novel object recognition test.
(A) The preference score for the novel object training (Train), short-term (STM), and long-term (LTM) tests is shown (ANOVA with repeated measure, WT n = 21, KO n = 19, no significant main effects of genotypes, F(1,38)= 4.023, p=0.052, or time x genotype interaction, F(2,76) =0.282, p=0.755). (B) Total time spent with the objects were similar between the genotypes. (ANOVA with repeated measure, WT n = 21, KO n = 19, no significant main effects of genotypes, F(1,38)= 0.180, p=0.674, or time x genotype interaction, F(2,76) =0.485, p=0.618).
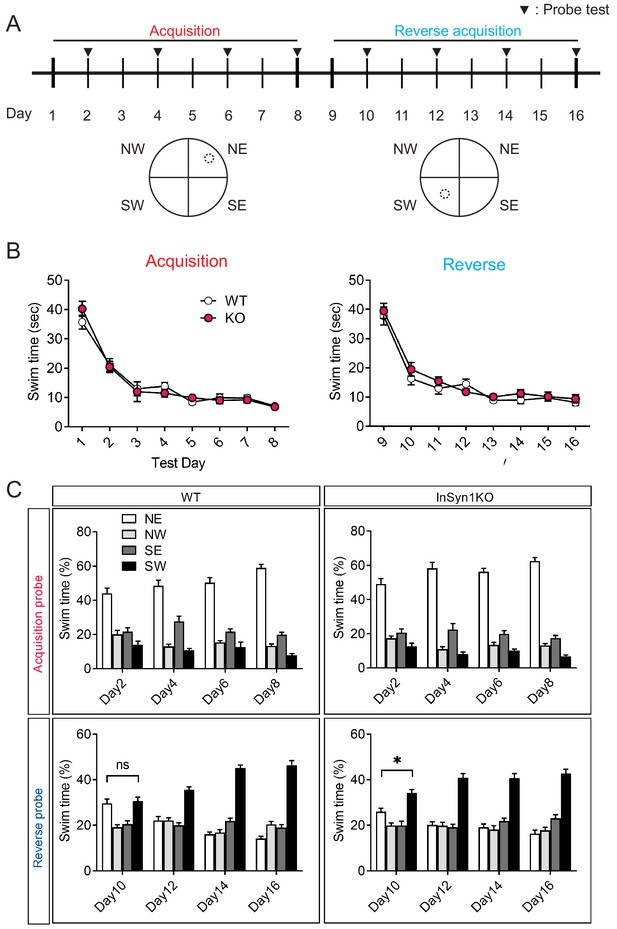
Spatial memory during the Morris Water Maze learning test of InSyn1- /- mice.
(A) A schematic of the water maze test showing the acquisition phase during days 1–8, reverse acquisition phase during days 9–16. NE, northeast; NW, northwest; SE, southeast; SW, southwest. (B) InSyn1 KO mice showed similar performance to WT mice in spatial learning as measured by total time spent to reach the platform during the acquisition phase or reverse phase. (C) Swim time in specific quadrants of the maze for WT and KO mice during acquisition probe tests (days 2, 4, 6, and 8) and reversal probe tests (days 10, 12, 14, and 16). At DIV10, KO mice showed a slight but a significant preference toward the new location (SW) compared to the previous location (NE) while WT mice did not.
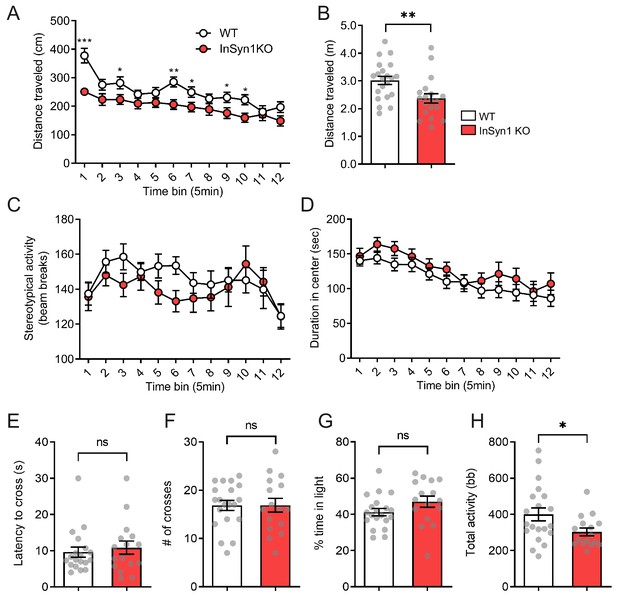
InSyn1- /- mice exhibit hypoactivity.
Analysis of open field exploration behavior (A–D). (A) Distance traveled in the open field over 1 hr shown in 5 min time blocks (ANOVA with repeated measure, WT n = 21, KO n = 21, main effects of group F(1, 38)=8.359, p=0.06, post-hoc t-test). (B) The plot of the total distance traveled (Two-tailed t-test, p=0.0066). (C) Graph of stereotypy behavior (ANOVA with repeated measure, WT n = 21, KO n = 21, no group difference for genotype F(1, 38)=1.186, p=0.283). (D) The plot of the duration spent in the center of the field (ANOVA with repeated measure, WT n = 21, KO n = 21, no group difference for genotype F(1, 38)=1.314, p=0.259). (E–F). Graphs depicting data from the light-dark transition test for: (E) Measurement of the latency to enter the light area (Two-tailed t-test, WT n = 19, KO n = 16, p=0.5970); (F) Total crosses between the areas (Two-tailed t-test, WT n = 20, KO n = 16, p=0.9888); (G) Time spent in the light area (Two-tailed t-test, WT n = 20, KO n = 16, p=0.1295); and (H) Total activity (Two-tailed t-test, WT n = 20, KO n = 16, p=0.0265). bb; beam break. *p<0.05, **p<0.01, ***p<0.001.
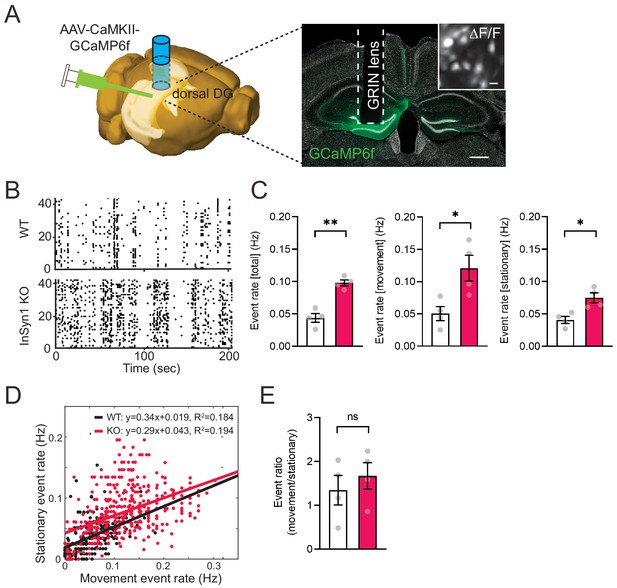
InSyn1- /- mice exhibit elevated neuronal activity in the dentate gyrus.
(A) Left; experimental design showing unilateral AAV-CaMKII-GCaMP6f injection into the DG of the dorsal hippocampus. The image was generated by the Allen Institute Brain Explorer two software (http://mouse.brain-map.org/static/brainexplorer). Right; an image of GCaMP6f expression and the position of the implanted GRIN lens. Scale bar; 500 um. A ΔF/F transformed image is inserted. Scale bar; 20 µm. (B) Representative images of a Ca2+-transient-raster plot from 40 cells. (C) Measurements of Ca2+ event rate. (Two-tailed t-test, WT n = 4, KO n = 4, total, p=0.0011. movement, p=0.0269. stationary, p=0.0143). *p<0.05, **p<0.01. (D) Scatter plot of movement and stationary state-related Ca2+-transient frequency recorded from WT and InSyn1 KO mice. (E) Bar graph showing the Ca2+-transient event ratio of movement to stationary (Two-tailed t-test, WT n = 4, KO n = 4, p=0.496).
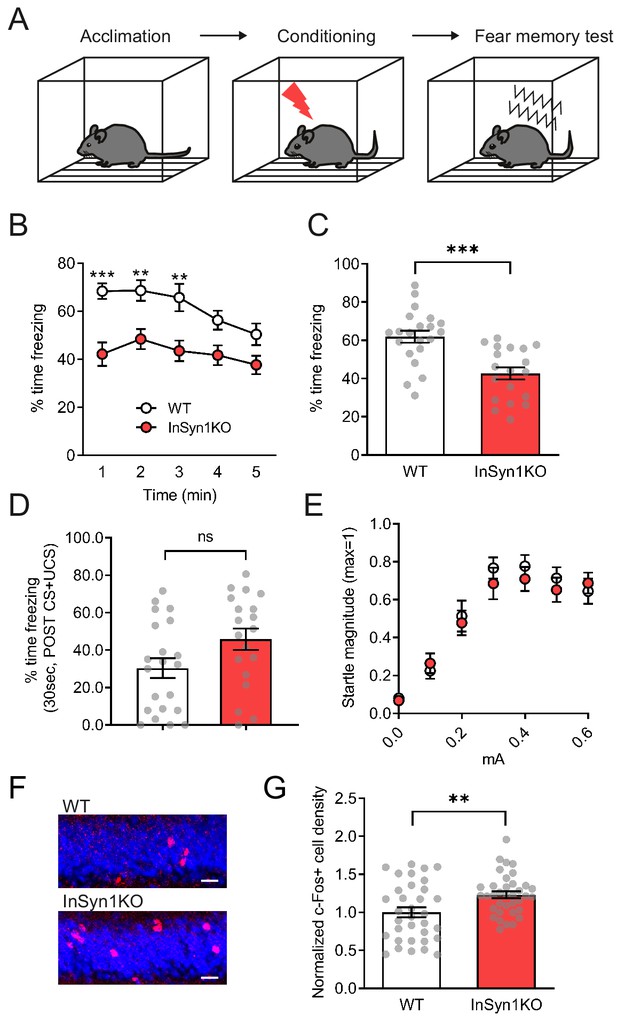
InSyn1- /- mice exhibit reduced memory recall and elevated c-Fos staining in contextual fear conditioning.
(A) Experimental scheme of contextual fear conditioning. After acclimation, mice receive a mild aversive foot-shock in a conditioning chamber. The next day, freezing upon placement in the chamber (without shock) was assessed. (B) Graph of startle responses to foot shock. Mice were exposed to different intensities of foot shock and the vertical acceleration was quantified and normalized. Two-way repeated measure ANOVA, Genotype effect, F (1, 29)=0.7095, p=0.4065. (C) Graph of the freezing response after shock stimulation between WT and KO mice. (D) Line graph representing the time of freezing in 1 min time bins during the fear memory test. Two-way repeated measure ANOVA, Genotype effect, F(1, 38)=18.38, p=0.0001. Bonferroni post-hoc analysis, 1 min p=0.0004, 2 min p=0.0085, 3 min p=0.0172, 4 min p=0.0735, 5 min p=0.1928. *p<0.05, **p<0.01, ***p<0.001. (E) Graph showing the total time of freezing is reduced more than 30% in InSyn1 KO mice compared to the WT littermates (WT n = 21, KO n = 19, two-tailed t-test, p<0.0001). (F) Representative images of hippocampus DG from WT and InSyn1KO stained for c-Fos (red) and Nissl (blue). Scale bars; 20 µm. (G) Bar graph represents c-Fos positive cell density in the DG regions of WT and KO hippocampal tissues. (Two-tailed t-test, WT n = 34 from four brains, KO n = 36 from five brains, p=0.0078). **p<0.01, ***p<0.001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Rosa26-LSL-Cas9 knockin | Jackson Laboratory | IMSR Cat# JAX:026175, RRID:IMSR_JAX:026175 | |
| Genetic reagent (M. musculus) | H11-CAG-Cas9 knockin | Jackson Laboratory | IMSR Cat# JAX:028239, RRID:IMSR_JAX:028239 | |
| Cell line (H. sapiens) | HEK293T | American Type Culture Collection | ATCC Cat# CRL-3216, RRID:CVCL_0063 | |
| Antibody | rat monoclonal anti-HA | Roche | Roche Cat# 3F10, RRID:AB_2314622 | ICC 1:1000 |
| Antibody | mouse monoclonal anti-alpha-Dystroglycan | Millipore | Millipore Cat# 05–593, RRID:AB_309828 | ICC 1:200 |
| Antibody | mouse monoclonal anti-HA | Covance | Cat# AFC-101P-1000, RRID:AB_291231 | WB 1:1000 |
| Antibody | rabbit polyclonal anti-GFP | Molecular Probes | Cat# A-11122, RRID:AB_221569 | WB 1:1000 |
| Antibody | mouse monoclonal anti-Gephyrin | Synaptic Systems | Cat# 147 011, RRID:AB_887717 | ICC 1:300 |
| Antibody | rabbit polyclonal anti-MAP2 | Synaptic Systems | Cat# 188 002, RRID:AB_2138183 | ICC 1:1000 |
| Antibody | mouse monoclonal anti-beta-Tubulin III | Sigma-Aldrich | Cat# T8660, RRID:AB_477590 | ICC 1:1000 |
| Antibody | rabbit polyclonal anti-GABAARα1 | Synaptic Systems | Cat# 224 203, RRID:AB_223218 | ICC 1:500 |
| Antibody | rabbit polyclonal anti-GABAARα2 | Synaptic Systems | Cat# 224 103, RRID:AB_2108839 | ICC 1:500 |
| Antibody | rabbit polyclonal anti-GABAARβ3 | Synaptic Systems | Cat# 224 403, RRID:AB_2619935 | ICC 1:500 |
| Antibody | rabbit polyclonal anti-GABAARγ4 | Synaptic Systems | Cat# 224 003, RRID:AB_2263066 | ICC 1:500 |
| Antibody | rabbit polyclonal anti-Vgat | Synaptic Systems | Cat# 131 002, RRID:AB_887871 | ICC 1:1000 |
| Antibody | mouse monoclonal anti-dystrophin | Abcam | Cat# ab7164, RRID:AB_305740 | ICC 1:100 |
| Antibody | rabbit polyclonal anti-c-Fos | Millipore | Cat# ABE457, RRID:AB_2631318 | IHC 1:5000 |
| Recombinant DNA reagent | pSpCas9(BB)−2A-GFP (PX458) | Addgene | RRID:Addgene_48138 | |
| Recombinant DNA reagent | pBetaActin-HA-α 1-syntrophin | This paper | See Materials and methods section | |
| Recombinant DNA reagent | pBetaActin-HA-β-dystrobrevin | This paper | See Materials and methods section | |
| Recombinant DNA reagent | pAAV-U6-sgRNA-hSyn-Cre | PMID: 27609886 | backbone of AAV CRISPR constructs | |
| Recombinant DNA reagent | pAAV-U6-InSyn1Cterm-HITI-smFP-SynI-Cre | This paper | See Materials and methods section | |
| Recombinant DNA reagent | pAAV-mTubb3 | Addgene | RRID:Addgene_87116 | HITI construct |
| Recombinant DNA reagent | pAAV-hSyn-hChR2(H134R)-EYFP | Addgene | RRID:Addgene_26973 | |
| Sequenced-based reagent | gRNAs | This paper | See Materials and methods section | |
| Chemical compound, drug | InFusion cloning kit | TaKaRa | Cat#638910 | |
| Chemical compound, drug | Bis(2,2,2-trifluoroethyl) ether/Flurothyl | Santa Cruz Bioteh | 333-36-8 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_002798 | Version 8 |
| Software, algorithm | SPSS | IBM | RRID:SCR_002865 | |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_001935 | 1.52e |
| Software, algorithm | Puncta Analyzer/ImageJ | PMID: 21113117 | 1.29 |





