Multifaceted roles of microRNAs: From motor neuron generation in embryos to degeneration in spinal muscular atrophy
Figures
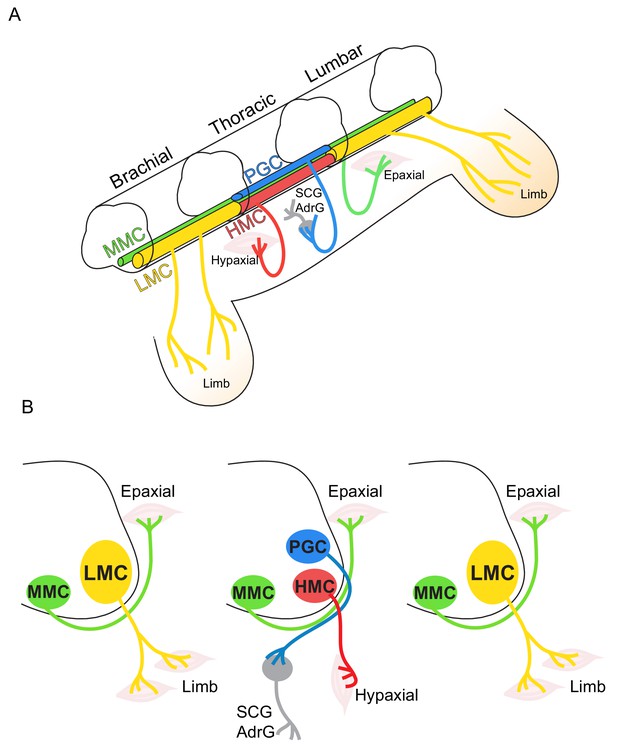
Topographic distribution of motor neuron (MN) columnar identities along the rostrocaudal axis of the spinal cord.
(A) Schematic illustration of the three-dimensional arrangement of motor columns in the spinal cord. Limb-innervating MNs are located in the lateral motor columns (LMC) at brachial and lumbar levels, whereas preganglionic (PGC) and hypaxial (HMC) neurons at thoracic levels innervate the sympathetic ganglia and hypaxial muscles, respectively. Medial motor (MMC) neurons are distributed at all rostrocaudal levels and project dorsally to epaxial muscles. (B) Diagram illustrating transverse sections through each segment, depicting the directions of MN axonal projection to the muscle targets.
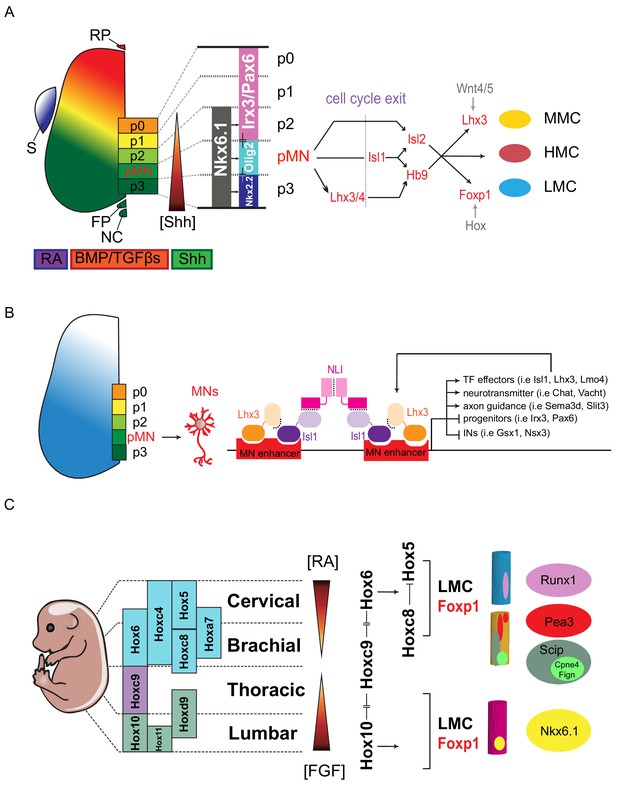
Transcription factor-based dorsoventral (DV) and rostrocaudal (RC) pattering of the spinal cord.
(A) Upon neural tube closure, a gradient of Sonic hedgehog protein (Shh) emanating from the notochord (NC) and floor plate (FP), together with BMP/TGFβ signaling from the roof plate (RP), mediates repression of class I homeodomain proteins (e.g. Irx3 and Pax6) and induction of class II protein expression (e.g. Nkx6.1 and Nkx2.2) at different threshold concentrations. Retinoic acid (RA) is expressed by the paraxial mesoderm to induce expression of class I proteins, which are turned off more ventrally by threshold levels of Shh. Class I and class II proteins that abut each other to define progenitor domain boundary respectively. Shh signaling defines five progenitors (p0-p3 and pMN) that give rise to V0-V3 interneurons and motor neurons. S, somite; RP, roof plate; BMP, bone morphogenetic proteins; TFGβs, transforming growth factor beta. (B) Isl1–Lhx3 forms MN-hexamer complexes to direct the differentiation of MNs by binding to MN enhancers. Isl1–Lhx3 induces the expression of a battery of MN genes that give rise to functional hallmarks of MNs, while suppressing key interneuron genes. Furthermore, the Isl1-Lhx3 complex amplifies its own expression through a potent autoregulatory feedback loop and simultaneously enhances the transcription of Lmo4 to promote MN differentiation and maturation. (C) Fibroblast growth factor (FGF) signals maintain a caudal progenitor zone during axis extension, and down-regulation of FGF signaling by RA is required for neuronal patterning and differentiation at more rostral levels. The RA and FGF gradient elicits expression of Hox genes along the rostrocaudal axis. Similar to patterning along the dorsoventral axis, the coordinated interactions between Hox family members allow regional boundaries to be delineated. Specific expression of Hox accessory factors, such as Foxp1, can further specify MN columnar subtypes. Within lateral motor columns, MN pools that innervate different limb muscle types can be defined by hallmark transcription factors (TFs) such as Runx1, Pea3, Scip, Nkx6.1, as well as digit-innervating MNs that manifest Fign, Cpne4, or other TFs.
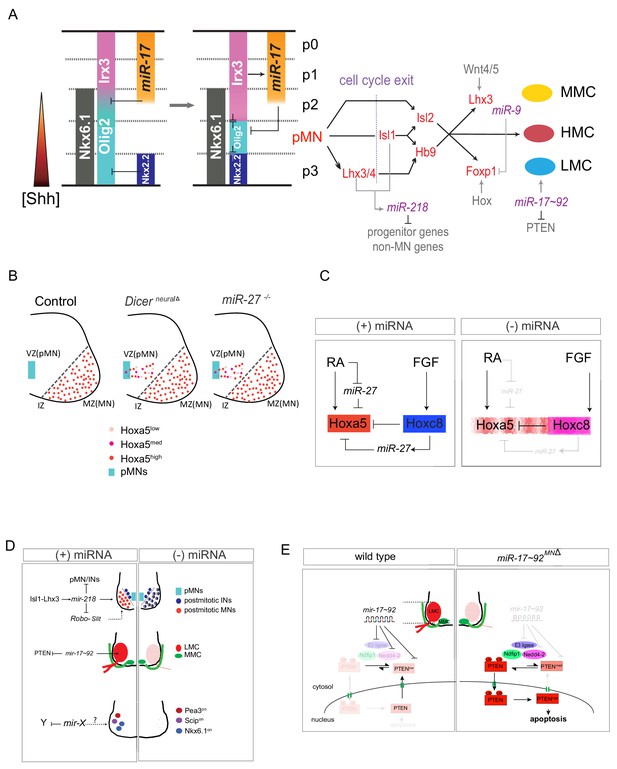
MicroRNA-mediated dorsoventral (DV) and rostrocaudal (RC) pattering of the spinal cord.
(A) In early embryonic stages (E8.5~E9), Nkx6.1 and Olig2 are co-expressed in a broad ventral domain spanning the prospective p3, pMN and part of the p2 domains. Subsequently, Nkx2.2 induced by sustained Shh signaling represses Olig2 in the p3 domain and miR-17–3p induced by Irx3 silences Olig2 in the p2 domain, forming the normal p2, pMN, and p3 progenitor domains. After MN generation, miR-218 acts as a downstream effector of the Isl1-Lhx3 complex to establish and maintain MN identity. Among MN subtypes, miR-9 seems to be involved in regulating MMC/LMC balance, whereas miR-17∼92 directly targets components of E3 ubiquitin ligases to impact subcellular localization of PTEN through monoubiquitination in the LMC-MNs. (B and C) Hoxa5 transcription in progenitor cells fluctuates, and translation of fluctuating transcripts at this time propagates noise, leading to strong stochastic variability. Two critical coherent forward loops involving miR-27 are capable of preventing precocious Hoxa5 protein expression, thereby maintaining the critically sharp boundary between Hoxa5 and Hoxc8 protein expression in embryonic spinal cords. Dicer nerualΔ: Sox1Cre+/-; Dicer floxed, pMN: motor neuron progenitors, VZ: ventricular zone, IZ: intermediate zone, MZ: mantle zone. (D) MN-miRNA working models: miR-218 is expressed in mature MNs to inhibit alternative neighboring genes. The potential miR-218-Slit-Robo autoregulatory loop is depicted. Subsequently, numbers of LMC motor neurons are reduced whereas MMC neurons are spared in Dicer MNΔ and miR-17 ~92 MNΔ spinal cords. Whereas surviving caudal brachial LMC motor neurons in Dicer MNΔ mutants correctly express Hoxc6 and Hoxc8 factors, expression of motor pool markers (i.e. Pea3, Scip, and Nkx6.1) is significantly eroded, resulting in near absence of defined motor pool subtype identity. The miRNA candidates needed to maintain motor pool identity have yet to be identified. (E) Illustration of how miR-17∼92 controls PTEN subcellular localization in developing LMC-MNs. This miRNA-mediated regulation modulates both target expression and target subcellular localization, providing LMC-MNs with an intricate defensive mechanism that controls their survival.
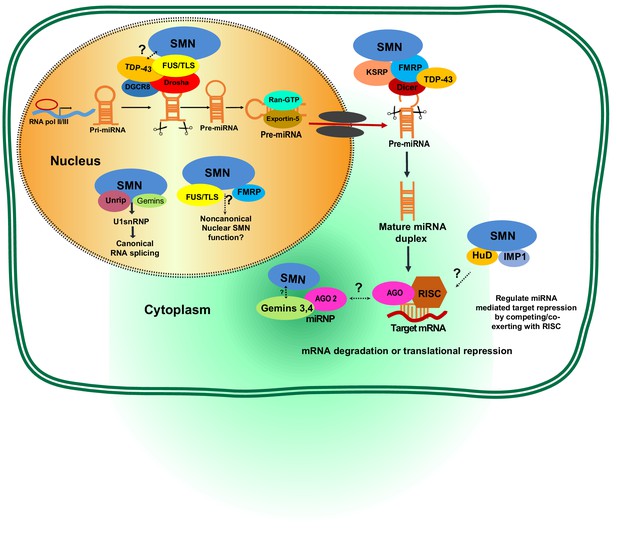
Potential involvement of SMN RNA-binding proteins (RBPs) in regulating microRNA biogenesis and function.
During the first step of miRNA biogenesis in the nucleus, pri-miRNA is processed by a microprocessor complex comprising Drosha and DiGeorge syndrome chromosomal region 8 (DGCR8) to produce hairpin-shaped pre-miRNA. Pre-miRNA is then exported to the cytoplasm by exportin-5 and further cleaved by Dicer. During nuclear and cytoplasmic processing of primary and precursor levels of miRNA, several RBPs—including TAR DNA-Binding Protein 43 (TDP-43), fused sarcoma/translocated in liposarcoma (FUS/TLS), KH-type splicing regulatory protein (KSRP), and Fragile X mental retardation protein (FMRP)— are known to link the SMN complex to the Drosha/DGRC8 complex and/or Dicer. Subsequently, the miRNA duplex is unwound into a single strand that is then loaded into argonaute proteins (AGO) to form the RNA-induced silencing complex (RISC). The miRNA-RISC complex plays a crucial role in post-transcriptional repression of target mRNA expression. Notably, during maturation and action of functional miRNAs either in the cytoplasm or nucleus, the SMN complex is known to interact with several RBPs including FMRP, HuD, and insulin-like growth factor 2 mRNA-binding protein 1 (IMP1). However, it remains to be established if SMN-RBP is involved in miRNA-mediated silencing.
Tables
Proposed functions of microRNAs during spinal motor neuron (sMN) development
https://doi.org/10.7554/eLife.50848.005| miRNA | Proposed roles in sMNs | Targets/interaction | Organism/cell models | Methods of genetic analyses | References |
|---|---|---|---|---|---|
| miR-9 | miR-9 promotes the switch from early-born to late-born motor neuron populations as well as MMC/LMC subtypes | Foxp1 | Chicken | transient decoy of miR-9 and overexpression of miR-9 in ovo | (Luxenhofer et al., 2014; Otaegi et al., 2011) |
| miR-17 ~ 92 | 1. miR-17–3p carves p2/pMN boundary at progenitor stage 2. miR-17 ~ 92 governs LMC-MN survival during development and degeneration in ALS | Olig2 PTEN/Ndfip1/Nedd4-2 | Mouse Mouse/human induced pluripotent stem cells | CAGGS:CreER; Dicer floxed and miR-17 ~ 92-/- embryos Olig2:Cre; miR-17 ~ 92 floxed embryos and miR-17 ~ 92 overexpression in human ALS-iPSC derived MNs | (Chen et al., 2011) (Tung et al., 2015; Tung et al., 2019) |
| miR-27 | miR-27 filters Hox temporal transcription noise to confer boundary formation in the spinal cord | Hoxa5 | Mouse and chicken | Sox1:Cre; Dicer floxed, transient decoy of miR-27 in ovo, and mir-23 ~ 27 ~ 24-/- embryos | (Li et al., 2017) |
| miR-196 | miR-196 confines the rostrocaudal axis in the neural tube | Hoxb8 | Chicken | Transient decoy of miR-196 in ovo | (Asli and Kessel, 2010) |
| miR-218 | miR-218 defines a neuronal gene network that is selectively tuned down in MNs to prevent neuromuscular failure and neurodegeneration. | Progenitor genes (Sox21, Tead1, etc.) Neighboring genes (Slc6a1, Bcl11a, Foxp2, etc.) | Mouse and chicken | Transient decoy of miR-218 in ovo and miR-218-/- embryos | (Amin et al., 2015; Thiebes et al., 2015) |
Proposed spinal muscular atrophy (SMA)-microRNA relationships
https://doi.org/10.7554/eLife.50848.006| miRNA | Role in MN disease | Targets | Organism/cell models and expression profiles | Findings and proposed mechanism of SMA pathogenesis | References |
|---|---|---|---|---|---|
| miR-2 | Neuronal development and function; correct NMJ functioning | CHRM2, m2R | C. elegans model, SMA mouse model. Decreased | Alters NMJ function | (O'Hern et al., 2017) |
| miR-9 (1) | MN dendritic outgrowth and synaptic function | Neurofilament heavy subunit (NEFH), REST, Map1b, MCPIP1 | Mouse, patient fibroblasts, patient serum. Decreased in spinal cord, but increased in skeletal muscle | Dysregulated expression in MNs differentiated from ESCs. Regulation of MN subtype determination (FOXP1). miR-9 can delay neurite outgrowth in vitro and impair radial neuronal migration in embryonic mouse neocortex in vivo. | (Catapano et al., 2016; Haramati et al., 2010) |
| miR-23a | Neuroprotective properties; regulate axonal development; suppress skeletal muscle atrophy | Atrogin1, MuRF1 (maybe, no direct target experiment was verified by luciferase assay) | Patient induced pluripotent stem cells, SMA mouse model. Decreased | miR-23a can prevent the astrocyte-conditioned media-induced MN loss in vitro. In mice model, enhanced miR-23a expression via virus vector increased MN soma size and muscle fiber area, and reduced NMJ defects | (Kaifer et al., 2019) |
| miR-100–5p | Abnormal proliferation of neural progenitors; aberrant cell cycle | Potentially insulin-like growth factor one receptor (IGF1R) | SMN∆7 mouse neural stem cells from spinal cord. Decreased | Decreased miR-100–5p in SMAΔ7 mice neural stem cells induces high IGF1R, excessive proliferation of neural progenitors, and prevents appropriate exit of the cell cycle. | (Luchetti et al., 2015) |
| miR-132 (possible) | Neuron dendritic outgrowth and synaptic plasticity; neovascularization, may cause ischemic pathology in both skeletal muscle and spinal cord of SMA model | Dysregulated expression due to TDP43 interaction with Dicer (amyotrophic lateral sclerosis data), p250GAP | SMA mice, patient serum. Decreased in spinal cord, but increased in skeletal muscle | Expression is dysregulated in TDP-43-deficient amyotrophic lateral sclerosis (ALS). Neuronal morphology and cognition in ALS. miR-132 can delay neurite outgrowth in vitro and impair radial neuronal migration in embryonic mouse neocortex in vivo. Involved in synaptic plasticity. Process of neovascularization. Recent reports have highlighted vascular defects in both skeletal muscle and spinal cord of SMA patients. | (Catapano et al., 2016) |
| miR-146 | MN loss caused by astrocyte-mediated pathology through NFκB signaling | GDNF, NOTCH2, GATA transcription factors | Patient induced pluripotent stem cells. Increased | miR-146 levels are influenced both directly and indirectly by SMN1 levels. SMN re-expression decreases miR-146a levels nearly to control levels. The NFκB pathway is an inducer of miR-146a. | (Sison et al., 2017) |
| miR-183 | Protein synthesis; axonal outgrowth | mTOR pathway | Mouse, cortical neurons, patient fibroblasts. Increased | Increased miR-183 and reduced local axonal translation of mTOR in SMN-deficient neurons. | (Kye et al., 2014) |
| miR-206 (2) (muscle-specific) | Myofiber formation; satellite cell differentiation; neuroprotective role in re-innervation of muscle endplates after acute nerve injury | Axis of HDAC4-FGFBP1, Pola1, BDNF | SMA mouse, patient serum. Increased in both spinal cord and skeletal muscle | Endogenously increased miR-206, with HDAC4 protein reduction and increased FGFBP1 mRNA, activates neuroprotective mechanism in muscle cells to increase re-innervation of muscle endplates. | (Catapano et al., 2016; Valsecchi et al., 2015) |
| miR-335–5p | Control of differentiation or self-renewal of mouse ESCs | MEST, OCT4, RB1 | SMN∆7 mouse neural stem cells, human induced pluripotent stem cells. Decreased | Possible epigenetic regulation through methylation to affect cell differentiation. | (Luchetti et al., 2015; Murdocca et al., 2016) |
| miR-375 | Neurogenesis and protects neurons from apoptosis in response to DNA damage. | P53, PAX6, CCND2 | Human neural progenitor cell cultures. Decreased | MNs from an SMA patient have shown reduced levels of miR-375, elevated p53 protein levels, and higher susceptibility to DNA damage-induced apoptosis. | (Bhinge et al., 2016) |
| miR-431 | Regulation of motor neuron axon neurite outgrowth | Chondrolectin (Chodl): a type one transmembrane protein and member of the c-type lectin domain-containing family | Mouse MN culture, patient fibroblasts/induced pluripotent stem cells. Increased | Increased miR-431 regulates motor neuron neurite length by targeting chondrolectin involved in motor neuron axon outgrowth. | (Wertz et al., 2016) |
-
1 miR-9/9* is another microRNA potentially implicated in motor neuron disease. It has been linked to the loss of spinal motor neurons that leads to SMA.
2 Since miR-206 is required for efficient regeneration of neuromuscular synapses after acute nerve injury, this scenario probably accounts for its salutary effects in SMA.
Proposed SMN-associated RNA-binding proteins involved in microRNA pathways.
https://doi.org/10.7554/eLife.50848.008| SMN-associated RBPs | RBP interaction assay | General functions in posttranscriptional regulation of mRNA | Proposed roles in miRNA metabolism/processing | Neuronal modeling mechanism | References |
|---|---|---|---|---|---|
| TAR DNA-binding protein-43 (TDP-43)a,b | FLAG-Drosha or FLAG-TDP-43 interacts in nuclear extracts from HEK293T cells. Partial colocalization of TDP-43 and SMN in MNs. | Transcriptional regulation, pre-mRNA splicing, mRNA transport and translation | 1. Integrating with the Dicer and Drosha complexes 2. Regulating pre-miRNA biogenesis | Neuron differentiation, neuronal plasticity, synapse formation, neurite outgrowth | (Kawahara and Mieda-Sato, 2012; Wang et al., 2002) |
| Fused sarcoma/translocated in liposarcoma (FUS/TLS)a,c | IP and GST pulldown SMN in HeLa nuclear extracts and mouse neuroblastoma (N2a) using FUS antibody. | Transcriptional regulation, pre-mRNA splicing, mRNA transport and translation | Recruiting the Drosha complex | Synapse formation, neuronal plasticity, neurite outgrowth, neuronal differentiation and proliferation | (Morlando et al., 2012; Sun et al., 2015; Yamazaki et al., 2012) |
| Fragile X mental retardation protein (FMRP) | IP and GST pulldown SMN complex in human neuroblastoma SH-SY5Y cells using FLAG-tagged FMRP antibody. IP antibodies specific to human FMRP were used to pull down Dicer and associated miRNAs in an EBV-transformed human B cell line. IP was performed from total HeLa cell lysates with a monoclonal antibody to eIF2C2. | mRNA stability, translation inhibition | Integrating with AGO and Dicer in RNA-induced silencing complex (RISC)d | Synapse formation, neuronal plasticity, neurite outgrowth, neuronal differentiation and proliferation | (Edbauer et al., 2010; Jin et al., 2004; Piazzon et al., 2008) |
| KH-type splicing regulatory protein (KSRP) | Endogenous KSRP was IP from N2a cells with SMN Tudor domain. KSRP and SMN colocalize in neuronal processes foci | mRNA stability, translation inhibition | Interacting with Drosha and Dicer complexes to regulate miRNA maturation and mRNA decay | KSRP is expressed in the nervous system in both neurons and glia and plays a role in control of neuronal mRNA stability and rate of axonal outgrowth | (Amirouche et al., 2014; Tadesse et al., 2008; Trabucchi et al., 2009) |
| HuD (ELAV-like protein 4) | SMN and the neuron-specific RBP HuD interact biochemically in cultured primary cortical neurons, MN1-cells, cultured mouse MNs, rat brain extracts. Genetic evidence from zebrafish embryos support that the interaction between SMN and HuD is critical for MN development. | mRNA stability, transport, translation, mTOR pathway | Interacting with RISCd to regulate miRNA function | Axon development, maintenance, and plasticity | (Fukao et al., 2014; Hao le et al., 2017; Loffreda et al., 2015; Sosanya et al., 2013) |
| IMP1 (IGF2BP1; ZBP1), insulin-like growth factor 2 mRNA-binding protein 1 | IP and GST pulldown SMN in brain lysates extracts using IMP1 antibody. | Promotes mRNA stability by preventing miRNA-mediated silencing | Interacting with AGO2 to regulate miRNA function | Axon development | (Fallini et al., 2014; Gardiner et al., 2015) (Degrauwe et al., 2016) |
-
a. TDP-43 and FUS/TLS proteins bind to pre-mRNA molecules and determine their fates by regulating splicing, transport, stability and translation.
b. The RNA-binding protein, TDP43, biochemically interacts with the miRNA processing enzyme Drosha, raising the possibility that TDP-43 may play a role in miRNA processing.
-
c. FUS/TLS promotes biogenesis of specific miRNAs, including miR-132, miR-134, and miR-9.
d. Additional proteins associated with RISC include MOV10 and Hu-AntigenR.





