Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor
Figures
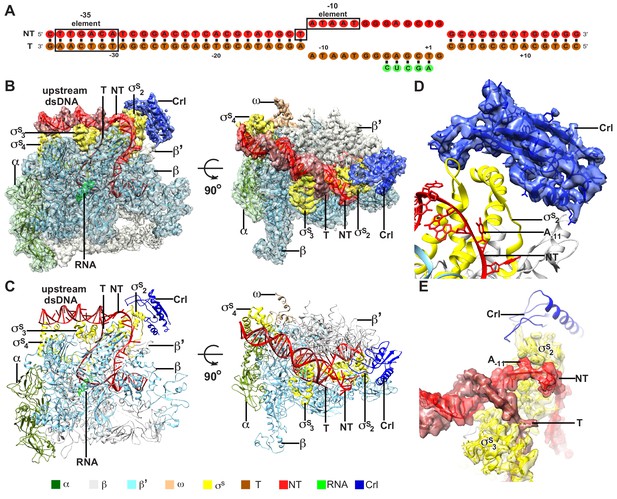
The overall structure of E. coli Crl–TAC.
(A) The scaffold used in structure determination of E. coli Crl-TAC. (B, C) The top and front view orientations of the cryo-EM density map (B) and structure model (C) of E. coli Crl–TAC. The RNAP, Crl and nucleic acids are presented as cartoon and colored as indicated in the color key. The density map is shown in gray envelop. (D) The cryo-EM density map (blue transparent surface) for Crl. (E) The cryo-EM density map (red transparent surface) for the upstream junction of the transcription bubble of promoter DNA and σS. NT, non-template-strand promoter DNA; T, template-strand promoter DNA.
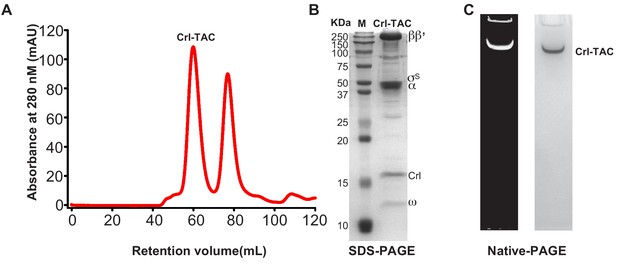
The preparation of E. coli Crl–TAC complex.
(A) Elution peaks of E. coli Crl–TAC from a size-exclusion column. (B) SDS-PAGE and (C) native-PAGE of the E. coli Crl–AC. The gel was first stained with SYBR Gold for nucleic acids and then with Coomassie Brilliant Blue for proteins.
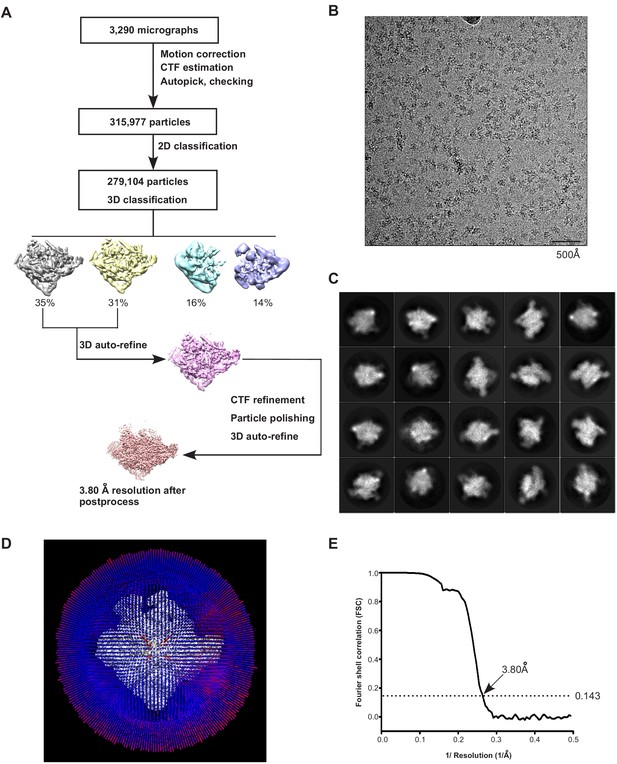
The processing pipeline for cryo-EM map construction of E. coli Crl–TAC.
(A) The flowchart of data processing for the cryo-EM structure of E. coli Crl–TAC. (B) Representative cryo-EM micrograph. (C) Representative 2D classifications of E. coli Crl–TAC single particles. (D) Angular distribution of E. coli Crl–TAC particle projections. (E) The gold-standard Fourier shell correlation (FSC) of E. coli Crl–TAC. The dotted line represents the 0.143 FSC cutoff, which indicates a nominal resolution of 3.80 Å.
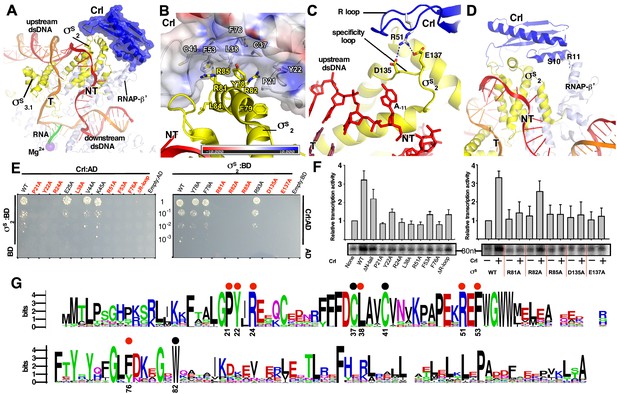
The interactions between Crl and the RNAP-holoenzyme.
(A) Crl binds to the σS2 and the RNAP-β’ subunit. Crl is represented as a blue transparent surface and cartoon. (B) σS is embedded into a shallow hydrophobic groove of Crl. The electrostatic potential surface of Crl was generated using APBS tools in Pymol. (C) The detailed salt-bridge interaction between the ‘R’ loop of Crl and the ‘specificity loop’ of σS. Salt-bridge bonds are shown as blue dashed lines. (D) The N-terminal tail of Crl makes potential weak interactions with RNAP-β’ subunit, probably through R11 and S10. The colors are as in Figure 1. (E) The yeast two-hybrid assay reveals the key interface residues (red) of Crl (left) or σS (right). Mutating the key residues disrupts interactions between Crl and σS. AD, the activation domain of GAL4; BD, the DNA-binding domain of GAL4. (F) The in vitro transcription assay shows that mutating most of the key interface residues of Crl (left) substantially impairs its transcription activation activity, and that mutating most of the key interface residues of σS resulted in reduced response to Crl. ΔR-loop, replacing residue 43–51 with a ‘GSGS’ linker. (G) Protein sequence alignment of Crl from 72 non-redundant bacterial species. Filled circles indicate residues that are involved with interactions with σS; filled red circles indicate key contact residues. The residues are numbers as in E. coli Crl. NT, non-template-strand promoter DNA; T, template-strand promoter DNA.
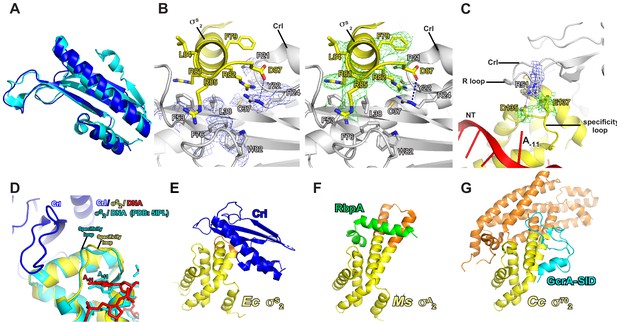
The detailed interactions between Crl and σS2.
(A) Structure superimposition of Crl proteins from crystal structure of Crl (PDB: 4Q11; cyan) and Crl in E. coli Crl-TAC (blue). (B) The σS2 inserts into a hydrophobic groove on surface of Crl. (C) The interaction between the ‘R’ loop of Crl and the ‘specificity’ loop of σS2. The cryo-EM density maps for the side chains of the interface residues of Crl and σS2 are shown as in blue and green mesh, respectively. The C, N, and O atoms of Crl residues are shown in white, blue, and red, respectively. The C, N, and O atoms of σS residues are shown in yellow, blue, and red, respectively. (D) Structure superimposition of our E. coli Crl–σS–RPo complex and E. coli σS–RPo complex (PDB 5IPM) reveals similar conformation of the specificity loop. (E) E. coli Crl interacts with σS2 in our structure. (F) Mycobacterium smegmatis RbpA interacts with σA2 in PDB 5TW1. (G) Caulobacter cresentus GcrA interacts with σ702 in PDB 5YIX. The conserved region and the non-conserved region (NCR) of σS2 in (E), (F), and (G) are shown in yellow and orange, respectively.
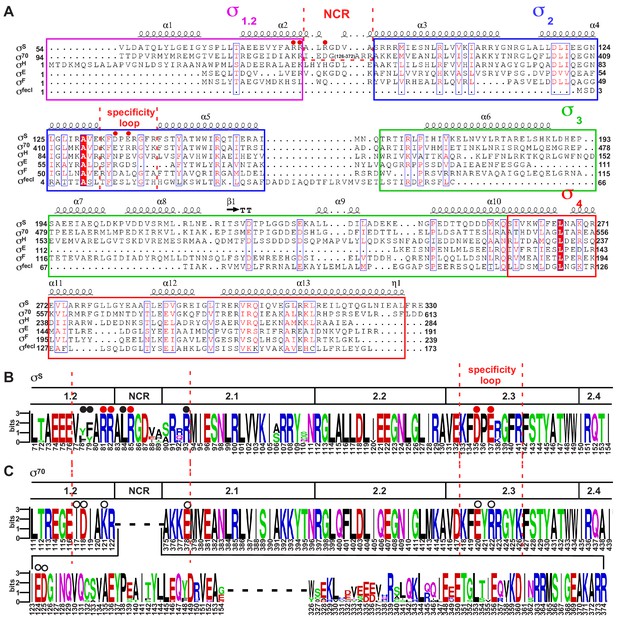
The sequence alignment of bacterial σ factors.
(A) Sequence alignment of six σ70-type E. coli σ factors. (B) The sequence alignment of the σ70subunit of ten representative bacterial species (Proteus mirabilis, Escherichia coli, Salmonella enterica serovar Typhi, Yersinia enterocolitica, Shigella flexneri, Vibrio cholerae, Dickeya dadantii, Providencia stuartii, Xenorhabdus bovienii, and Photobacterium profundum). (C) The sequence alignment of the σS subunit of the same ten bacterial species. The residues are numbered as in E. coli.
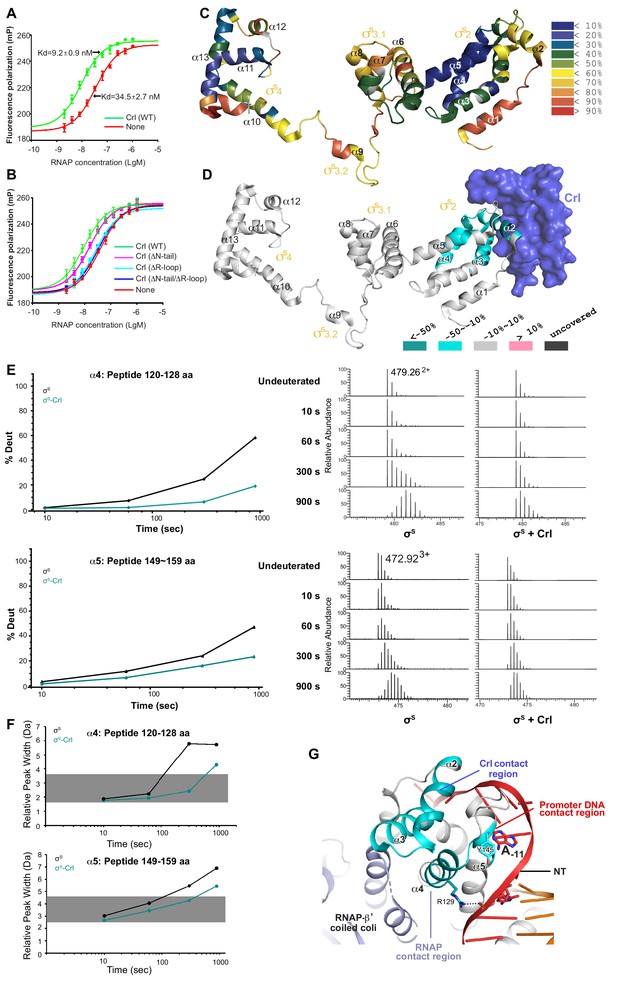
Crl promotes assembly of the σS-RNAP holoenzyme by stabilizing σS2.
(A) Crl increases binding affinity between RNAP and σS in a fluorescence polarization (FP) assay. (B) Deletion the ‘R’ loop but not the N-tail (ΔN-tail; deleting residue 1–11) abolishes the ability of Crl to promote the assembly of the σS-RNAP holoenzyme. (C) Hydrogen-to-deuterium exchange (HDX) profile of free σS after 10 s exposure to deuteron. Incorporation was mapped onto the cryo-EM structure of σS. The inset shows the color coding for different percentages of deuteron incorporation. (D) HDX changes for σS in the presence of Crl as compared to free σS, mapped on the cryo-EM structure of σS. The percentage difference in deuterium uptake values represents an average across all four time points, ranging from 10 s to 900 s. For heatmap color coding, pink, cyan, and gray indicate increase, decrease, and no significant change in HDX, respectively. Dark grey represents regions that were not consistently resolved in all HDX experiments. (E) Deuterium uptake plots and mass spectra of indicated peptides from helices α4 and α5 in the absence and presence of Crl. Left: the deuterium uptake data are plotted as percent deuterium uptake versus time on a logarithmic scale. Right: mass spectra of the indicated peptides at different labeling time points, with the mass spectra of undeuterated samples shown as controls. (F) Peak-width analysis at different time points for peptides from helices α4 and α5 reveal the existence of EX1 kinetics. (C–F) The HDX experiments were repeated at least twice. (G) σS2 regions with reduced HDX rate upon Crl binding, the colors are as in (D).
-
Figure 3—source data 1
The raw data of Figure 3A-B.
- https://cdn.elifesciences.org/articles/50928/elife-50928-fig3-data1-v1.xlsx
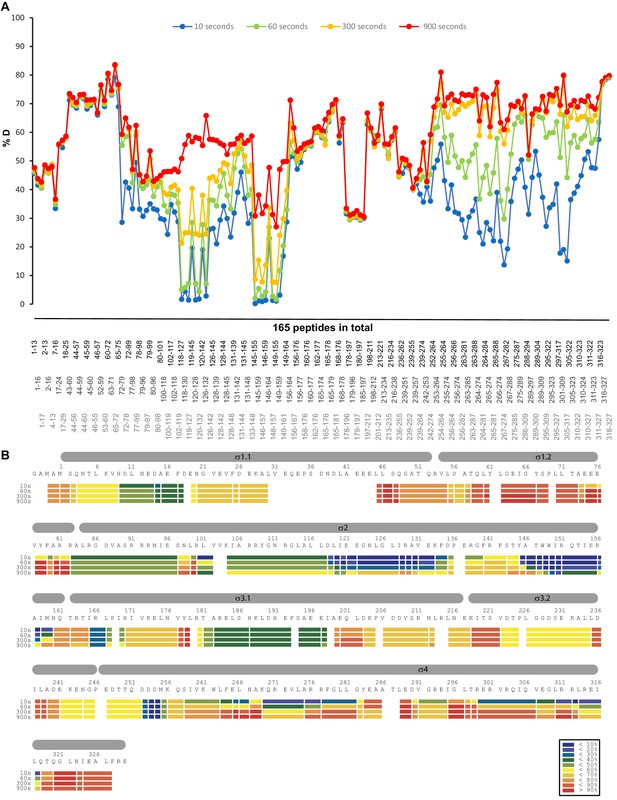
HDX profile of the σS protein in the absence of Crl.
(A) The relative deuteration levels (%D) of each peptide were calculated with the assumption that a fully deuterated sample retains 90% D in the current LC setting and plotted as a function of peptide position. The blue to red lines represent data acquired at 10 s up to 900 s deuteration. (B) Heat map of σS protein at different time points. The inset shows the color coding for different percentages of deuteron incorporation.
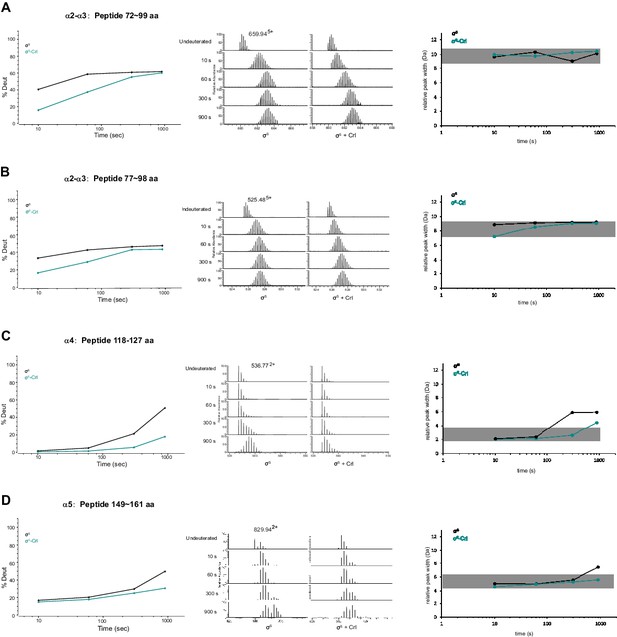
Representative peptides from helix α2, α3, α4 and α5 of σS2 whose HDX rate decreased considerably in the presence of Crl.
(A–D) Deuterium uptake plots, peak width plots and mass spectra of indicated peptides from σS2 in the absence and presence of Crl. Left: the deuterium uptake data are plotted as percent deuterium uptake versus time on a logarithmic scale. Middle: mass spectra of indicated peptides at different labeling time points, with the mass spectra of undeuterated samples shown as controls. Right: peak-width analysis at different time points for peptides covering helices α2–α5.
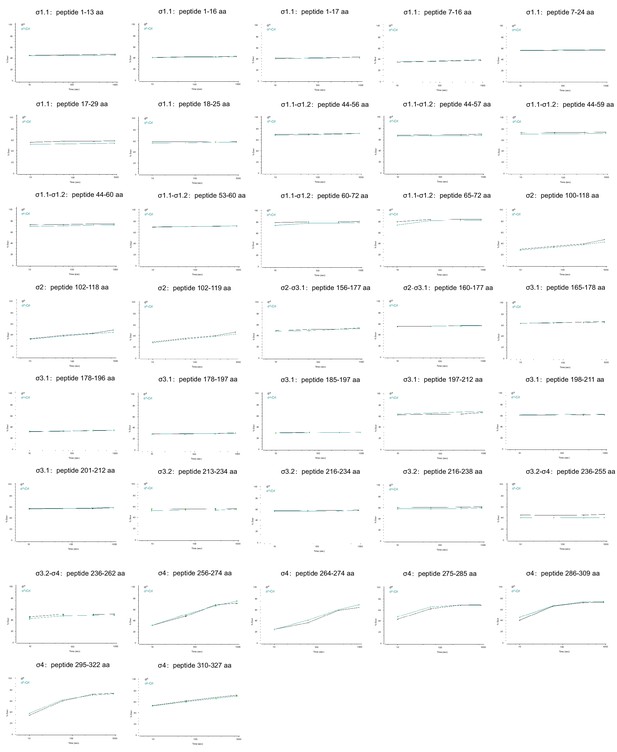
Deuterium uptake plots for additional peptides from σS whose HDX rate did not change in the presence of Crl.
The deuterium uptake data are plotted as percent deuterium uptake versus time on a logarithmic scale.
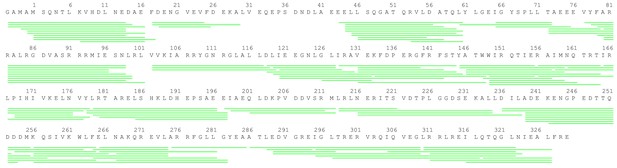
Peptide coverage of σS.
The sequence coverage map for σs in the HDX-MS study. Green lines shown below the protein sequence represent the digested peptides identified and analyzed in this study.
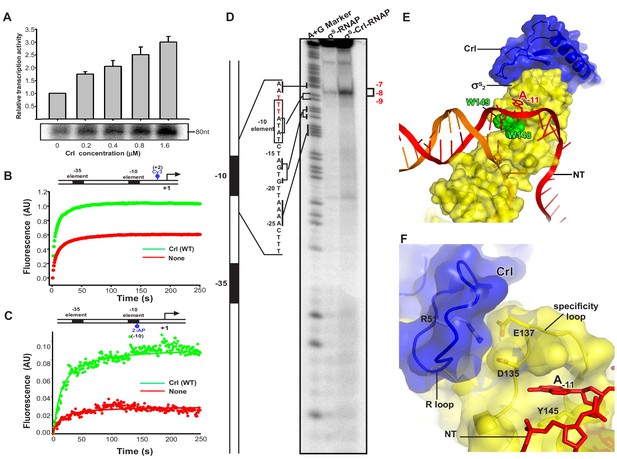
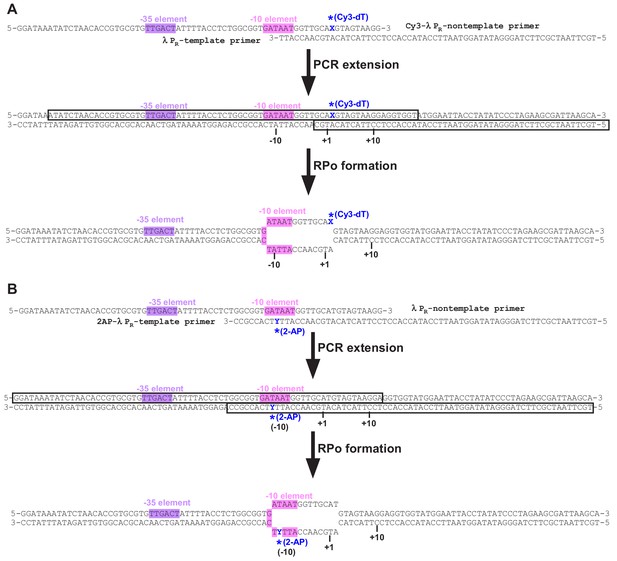
Crl facilitates RPo formation.
(A) Crl increases transcription by a pre-assembled σS-RNAP holoenzyme from the PosmY promoter in a concentration-dependent manner. (B) Crl promotes σS-RPo formation in a fluorescence stopped-flow experiment using Cy3-labeled λPR promoter DNA. (C) Crl promotes σS-RPo formation in a fluorescence stopped-flow experiment using 2AP-labeled λPR promoter DNA. (D) The KMO4 footprinting experiment result shows that Crl increases formation of the RPo complex. The osmY promoter dsDNA with 32P-labeled at the 5′-end of the non-template strand was used for the assay. The hyperactive T-stretch (−9 to −7) in the transcription bubble upon RPo formation is highlighted in red and labeled on the right. (E) The upstream promoter DNA is unwound by the W-dyad (W148 and W149), and the unwound A-11 nucleotide of the non-template strand DNA is recognized and stabilized by a protein pocket on σS2. (F) The ‘R’ loop of Crl stabilizes the conformation of the ‘specificity’ loop that forms the pocket for unwinding and recognizing the A-11 nucleotide of non-template strand promoter DNA.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Escherichia coli) | crl | Gene bank (7152411) | ||
| Strain, strain background (E. coli) | BL21(DE3) | Novo protein, China | V211-01A | Chemically competent cells |
| Recombinant DNA reagent | pET-28a-TEV-Crl (plasmid) | This paper | Protein expression vector for E. coli Crl | |
| Recombinant DNA reagent | pET-28a-TEV-EcsS (plasmid) | This paper | Protein expression vector for E. coli sS | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (P21W) (plasmid) | This paper | Protein expression vector for E. coli Crl (P21W) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (Y22A) (plasmid) | This paper | Protein expression vector for E. coli Crl (Y22A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (R24A) (plasmid) | This paper | Protein expression vector for E. coli Crl (R24A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (L38A) (plasmid) | This paper | Protein expression vector for E. coli Crl (L38A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (R51A) (plasmid) | This paper | Protein expression vector for E. coli Crl (R51A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (F53A) (plasmid) | This paper | Protein expression vector for E. coli Crl (F53A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (F76A) (plasmid) | This paper | Protein expression vector for E. coli Crl (F76A) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (ΔN-tail) (plasmid) | This paper | Protein expression vector for E. coli Crl (Δ1–11) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (ΔR-loop) (plasmid) | This paper | pET-28a-TEV-Crl (ΔR-loop; residues 43–51 of Crl by a ‘GSGS’ linker) | |
| Recombinant DNA reagent | pET-28a-TEV-Crl (ΔN-tail /ΔR-loop) (plasmid) | This paper | Protein expression vector for E. coli Crl (ΔN-tail /ΔR-loop) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS(R81A) (plasmid) | This paper | Protein expression vector for E. coli σS (R81A) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS (R82A) (plasmid) | This paper | Protein expression vector for E. coli σS (R82A) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS (R85A) (plasmid) | This paper | Protein expression vector for E. coli σS (R85A) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS (D135A) (plasmid) | This paper | Protein expression vector for E. coli σS (D135A) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS (E137A) (plasmid) | This paper | Protein expression vector for E. coliσS (E137A) | |
| Recombinant DNA reagent | pET-28a-TEV-EcσS (A239C) (plasmid) | This paper | Protein expression vector for E. coli σS (A239C) | |
| Recombinant DNA reagent | pGADT7-Crl (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (P21W) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (P21W) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (Y22A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (Y22A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (R24A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (R24A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (E25A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (E25A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (L38A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (L38A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (V44A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (V44A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (K45A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (K45A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (R51A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (R51A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (F53A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (F53A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (F76A) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (F76A) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGADT7-Crl (ΔR-loop) (plasmid) | This paper | Plasmid for yeast two-hybrid; Crl (ΔR-loop) is fused to Gal4-AD | |
| Recombinant DNA reagent | pGBKT7-σS2 (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2(53–162) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (Y78A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (Y78A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (F79A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2(F79A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (R81A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (R81A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (R82A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (R82A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (R85A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (R85A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (R93A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (R93A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (D135A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (D135A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pGBKT7-σS2 (E137A) (plasmid) | This paper | Plasmid for yeast two-hybrid; σS2 (E137A) is fused to Gal4-BD | |
| Recombinant DNA reagent | pEasyT-PosmY (plasmid) | This paper | Plasmid containing PosmY | |
| Commercial assay or kit | Ezmax one-step cloning kit | Tolo Bio-tech, China | Cat#24305–01 | |
| Other | Ni-NTA agarose | smart-lifesciences, China | Cat#SA004100 | |
| Other | C-flat CF-1.2/1.3 400 mesh | Electron Microscopy Sciences | Cat#CF413-100 |
Additional files
-
Supplementary file 1
The statistics of the cryo-EM structure of E. coli Crl–TAC.
- https://cdn.elifesciences.org/articles/50928/elife-50928-supp1-v1.docx
-
Supplementary file 2
The estimated equilibrium dissociation constants from Figure 3B of RNAP and σS in the presence of wild-type or mutant Crl.
- https://cdn.elifesciences.org/articles/50928/elife-50928-supp2-v1.docx
-
Source code 1
The map for structure of Ec-Crl–TAC.
- https://cdn.elifesciences.org/articles/50928/elife-50928-code1-v1.mrc
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/50928/elife-50928-transrepform-v1.pdf