Tritrophic metabolism of plant chemical defenses and its effects on herbivore and predator performance
Figures
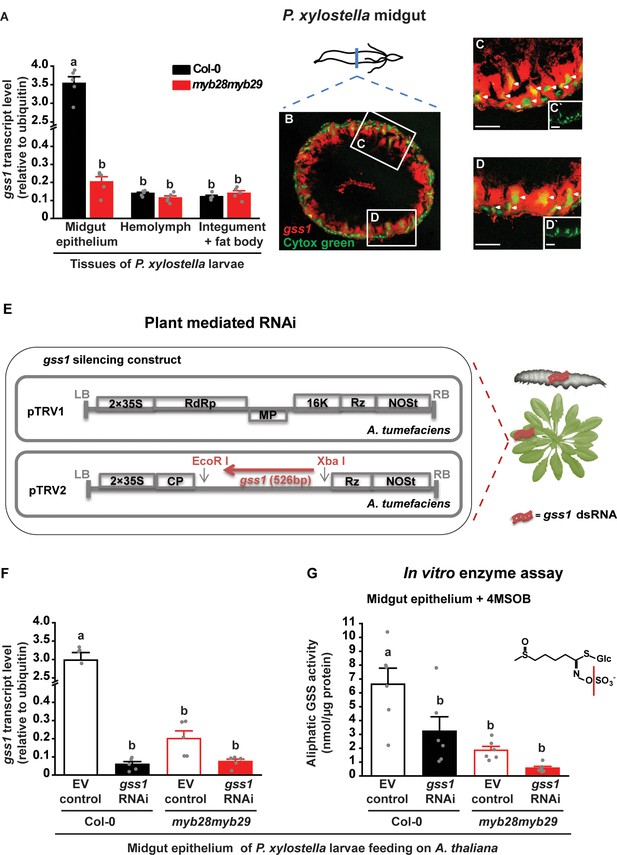
Localization and plant-mediated RNAi silencing of gss1 gene expression in P. xylostella larvae.
(A) Levels of gss1 transcripts (expressed relative to ubiquitin) are higher in midgut epithelium than in hemolymph, integument and fat bodies of fourth-instar larvae feeding on A. thaliana Col-0 (wild-type glucosinolates) and myb28myb29 (no aliphatic glucosinolates) plants (Plants, F1,24 = 338.032, p≤0.0001; Tissues, F2,24 = 367.522, p≤0.0001; Plants*Tissues, F2,24 = 336.042, p≤0.0001; n = 5 for all bars). (B–D) Visualization of gss1-expressing cells in the midgut of P. xylostella as seen in a transverse section. Cells containing gss1 transcripts were strongly stained by means of fluorescent in situ hybridization (FISH) using a gss1-specific antisense riboprobe labeled by digoxigenin (DIG) (in red). Counter staining of nuclei of midgut cells employed Cytox green (in green). Two selected areas in B (C and D) are magnified further. Both the columnar cells and the basal midgut cells are gss1-positive, with white arrows pointing to nuclei of different morphologies. C` and D` are lower magnification views of the nuclear staining presented in C and D, respectively. Scale bar, 50 µm. (E) Silencing strategy for gss1 employed a virus-based dsRNA-producing system in the host plant A. thaliana created by infiltration of tobacco rattle virus (TRV, detail structure described in Ratcliff et al., 2001) engineered to express a 526 bp fragment of gss1 dsRNA. (F) Levels of gss1 transcripts (expressed relative to ubiquitin) in larval midgut epithelium (F3,16 = 185.508, p≤0.0001, n = 5 for all bars) and (G) levels of GSS activity measured in vitro in extracts of midgut epithelium (F3,20 = 10.697, p≤0.0001, n = 6 for all bars) of fourth-instar larvae feeding on empty vector (EV) control and gss1-RNAi plants in backgrounds of Col-0 and myb28myb29 A. thaliana. The aliphatic glucosinolate 4MSOB (5 mM) was used as a substrate. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with a two-way ANOVA in A and with one-way ANOVA in F and G.
-
Figure 1—source data 1
gss1 gene transcript levels and GSS in vitro enzyme assays with 4MSOB glucosinolate.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig1-data1-v2.xlsx
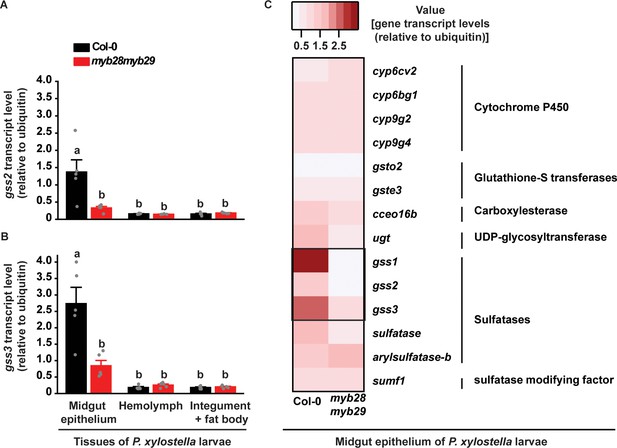
Transcripts of gss genes are localized in the P. xylostella larval midgut epithelium and are induced by dietary ingestion of glucosinolates.
Transcript levels of gss2 (A) and gss3 (B) (expressed relative to ubiquitin) in midgut epithelium, hemolymph, and integument and fat body of fourth-instar larvae (gss2: Plants, F1,24 = 8.244, p≤0.01; Tissues, F2,24 = 14.942, p≤0.0001; Plants*Tissues, F2,24 = 8.684, p≤0.001. gss3: Plants, F1,24 = 11.740, p≤0.01; Tissues, F2,24 = 36.609, p≤0.0001; Plants*Tissues, F2,24 = 13.685, p≤0.001. n = 5 in all bars). (C) Heat map of detoxification-related gene transcripts (selected members of P450, GST, CoE, UGT, sulfatase and SUMF families) in the midgut of P. xylostella fourth-instar larvae feeding on A. thaliana Col-0 and myb28myb29 plants. Values represent gene transcripts (expressed relative to ubiquitin). Gene accession numbers are provided in (Supplementary file 1). See (Figure 1—figure supplement 4A,C) for detailed expression of other sulfatase-related genes. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with two-way ANOVA in A and B.
-
Figure 1—figure supplement 1—source data 1
Transcript levels of gss2, gss3 and other detoxification-related genes.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig1-figsupp1-data1-v2.xlsx
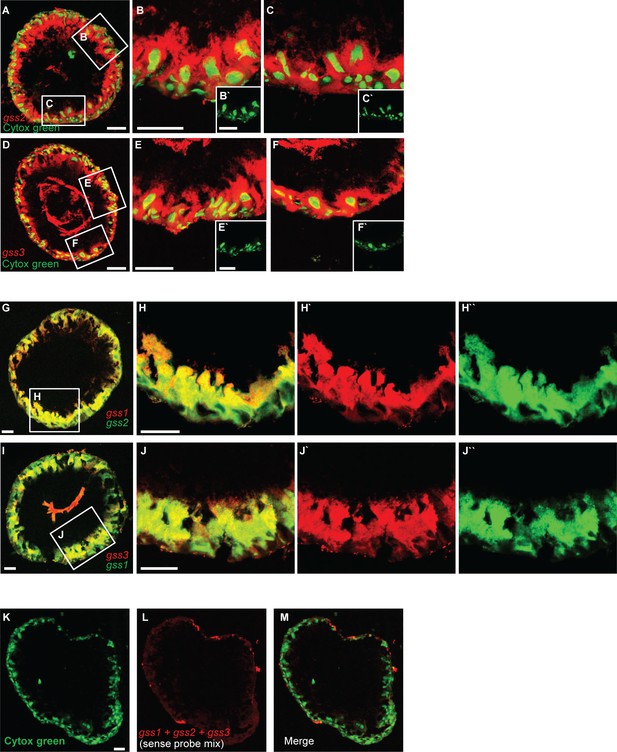
Transcripts of gss genes are localized in midgut cell types of P. xylostella fourth-instar larvae.
(A–F) Cells containing gss2- and gss3 transcripts were visualized by means of FISH using gss2- and gss3-specific antisense riboprobes labeled by DIG (in red), respectively. Counter staining of nuclei of midgut cells employed Cytox green (in green). Selected areas in A and D are further magnified in B, C, E and F. B` to F` are lower magnification views of nuclear staining from A and D. (G) Transcripts of gss1 and gss2 were co-expressed in the same set of cells as visualized by means of two-color FISH using specific antisense riboprobes labeled by DIG (gss1, in red) and biotin (BIO) (gss2, in green). A selected area (H) in G is further magnified on the right to demonstrate the co-expression in more detail. H` and H``, represent the red and green fluorescent channels, respectively, of H. (I) Transcripts of gss1 and gss3 were largely co-expressed in the same set of cells as visualized by means of two-color FISH using specific antisense riboprobes labeled by DIG (gss3, in red) and BIO (gss1, in green). A selected area (J) in I is further magnified on the right to demonstrate the co-expression pattern in more detail. J` and J``, represent the red and green fluorescent channels, respectively, of I. (K–M) Demonstration of labeling specificity of individual gss-specific antisense riboprobes. No apparent red labeling was observed in a FISH experiment using a mixture of gss1, gss2 and gss3 sense probes labeled by DIG in conjunction with nuclear counter staining with Cytox green (green). Scale bar, 50 µm.
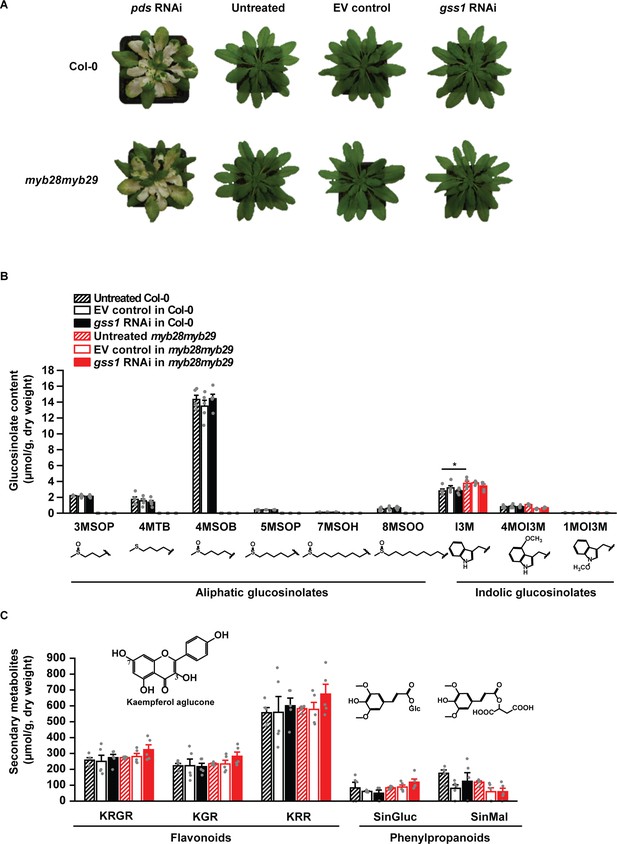
Infiltration of A.thaliana Col-0 and myb28myb29 lines with gss1 RNAi and empty vector (EV) control constructs does not alter their morphology and chemical phenotypes.
(A) Morphology of untreated, EV control and gss1 RNAi plants in Col-0 and myb28myb29 backgrounds. Phytoene desaturase (pds) RNAi constructs were used as a positive control of virus induced gene silencing. Photographs were taken 4 weeks after infiltration. (B) Glucosinolate profiles of untreated, EV control, and gss1 RNAi plants in Col-0 and myb28myb29 backgrounds (I3M, F5,24 = 4.031, p≤0.01, n = 5 in all bars). 3MSOP: 3-methylsulfinylpropyl; 4MSOB: 4-methylsulfinylbutyl; 5MSOP: 5-methylsulfinylpentyl; 7MSOH: 7-methylsulfinylheptyl; 4MTB: 4-methylthiobutyl; 8MSOO: 8-methylsulfinyloctyl; I3M: Indol-3-ylmethyl; 4MOI3M: 4-methoxyindol-3-ylmethyl; 1MOI3M: 1-methoxyindol-3-ylmethyl glucosinolate. (C) Concentrations of selected secondary metabolites (flavonoids and phenylpropanoids) in untreated, EV control, and gss1 RNAi plants in Col-0 and myb28myb29 backgrounds. KRGR, kaempferol 3-O-glucoside-2’’-rhamnoside-7-O-rhamnoside; KGR, kaempferol 3-O-glucoside-7-O-rhamnoside; KRR, kaempferol 3-O-rhamnoside-7-O-rhamnoside; SinGluc, 1-O-sinapoyl-β-D-glucose; SinMal, sinapoyl malate. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in B and C.
-
Figure 1—figure supplement 3—source data 1
Glucosinolate, flavonoid, and phenylpropanoid profiles in A. thaliana plants.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig1-figsupp3-data1-v2.xlsx
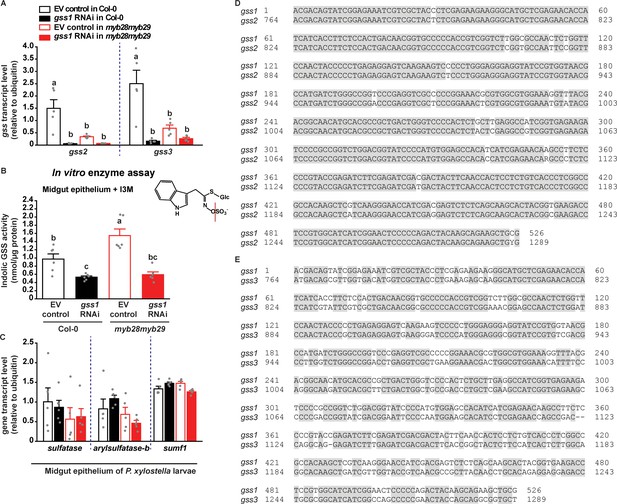
Plant-mediated RNAi of gss1 co-silences gss2 and gss3 due to high sequence similarity, and suppresses the desulfation of indolic glucosinolates as well.
(A) Levels of gss2 and gss3 transcripts (expressed relative to ubiquitin) (gss2, F3,16 = 14.831, p≤0.0001; gss3, F3,16 = 14.810, p≤0.0001; n = 5 for all bars) in midgut epithelium of P. xylostella fourth-instar larvae show silencing after RNAi of gss1. (B) Levels of indolic glucosinolate sulfatase activity (F3,20 = 18.696, p≤0.0001, n = 6 for all bars) measured in vitro in extracts of midgut epithelium of fourth-instar larvae using 5 mM I3M as substrate show suppression upon gss1 silencing. (C) Levels of other sulfatase gene transcripts, including sulfatase, arylsulfatase-b, and sulfatase modifying factor 1 (sumf1) (expressed relative to ubiquitin) were not silenced by gss1 RNAi in midgut epithelium of fourth-instar larvae. All insects fed on empty vector (EV) control and gss1 RNAi plants in backgrounds of Col-0 (wild-type glucosinolates) and myb28myb29 (no aliphatic glucosinolates). Sequence alignments of gss1 526 bp cDNA fragment selected for RNAi silencing experiments are given with the homologous sequences of (D) gss2 and (E) gss3. The sequence of gss1 shows 95% identity with gss2% and 75% with gss3, and possesses identical homologous regions of >21 nt (similarities were determined using nucleotide BLAST). Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in A–C).
-
Figure 1—figure supplement 4—source data 1
gss2 and gss3 gene transcript levels, and GSS in vitro enzyme assays with I3M glucosinolate.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig1-figsupp4-data1-v2.xlsx
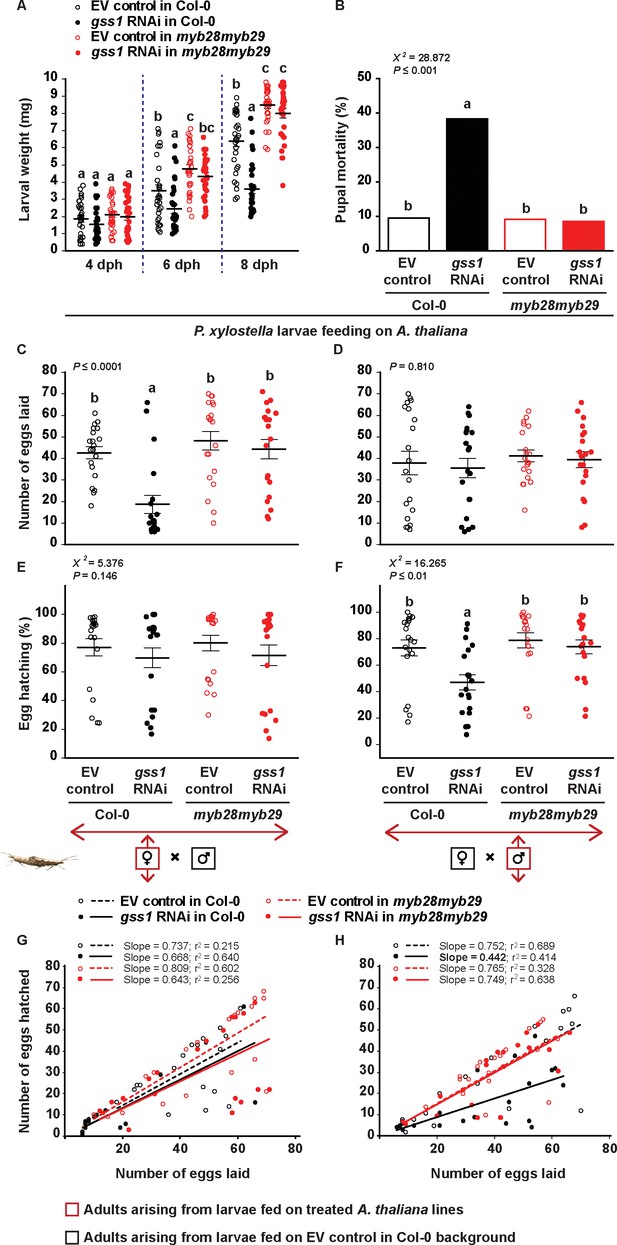
Silencing of gss affects P. xylostella growth and fitness in the presence of aliphatic glucosinolates.
Larvae were fed on empty vector (EV) control and gss1 RNAi plants from either Col-0 (wild-type glucosinolates) or myb28myb29 (no aliphatic glucosinolates) backgrounds, and various parameters of larval performance were measured: (A) Weights of larvae after four dph (days post hatching), six dph, and eight dph were reduced by silencing but only in the presence of wild-type glucosinolates (six dph, Plants F1,116 = 33.471, p≤0.0001; RNAi F1,116 = 7.440, p≤0.01; Plants*RNAi F1,116 = 1.322, p=0.253. eight dph, Plants F1,116 = 145.33, p≤0.0001; RNAi F1,116 = 37.083, p≤0.0001; Plants*RNAi F1,116 = 18.340, p≤0.0001. n = 30 for all treatments); (B) Mortality (%) in pupae was highest in those pupae arising from silenced larvae fed on wild-type glucosinolates (χ2 = 228.872, df = 3, p≤0.001; n = 63, 60, 66 and 59, respectively). The number of eggs laid by females was measured in crosses between (C) females arising from larvae raised on the four plant lines mated with males arising from larvae fed on EV control Col-0 plants and (D) males arising from larvae raised on the four plant lines mated with females arising from larvae fed on EV control Col-0 plants. Only crosses with females arising from gss-silenced larvae fed on Col-0 showed a decline in egg production (F3,76 = 11.157, p≤0.0001, n = 20 for all treatments). Hatching (%) of eggs laid by females (E, F), and the correlation between the numbers of eggs hatched and the numbers of eggs laid (G, H) were recorded from crosses between (E, G) females arising from larvae raised on the four plant lines mated with males arising from larvae fed on EV control Col-0 plants, and (F, H) from males arising from larvae raised on the four plant lines mated with females arising from larvae fed on EV control Col-0 plants. Only crosses with males arising from gss-silenced larvae fed on Col-0 showed a decline in egg hatching rate (χ2 = 216.265, df = 3, p≤0.01; n = 20 for all treatments). Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with two-way ANOVA in A, a proportions test with pairwise comparisons in B, Tukey HSD tests in conjunction with one-way ANOVA in C–D, and Dunn's post hoc tests in conjunction with non-parametric Kruskal-Wallis tests in E–F.
-
Figure 2—source data 1
P. xylostella larval weight, pupal mortality, and adult egg laying capacity.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig2-data1-v2.xlsx
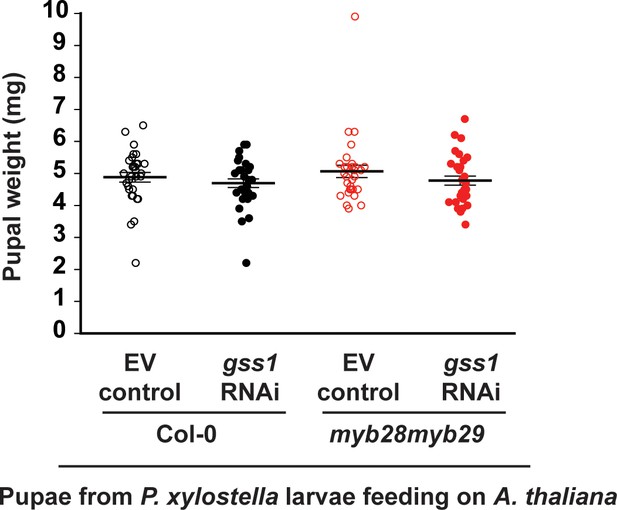
Silencing of gss has no effect on weights of P. xylostella pupae.
Weights of pupae were measured on the second day post pupation. Significant difference (p≤0.05) between means (±s.e.) was determined by Tukey HSD tests in conjunction with a one-way ANOVA.
-
Figure 2—figure supplement 1—source data 1
P. xylostella pupal weight.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig2-figsupp1-data1-v2.xlsx
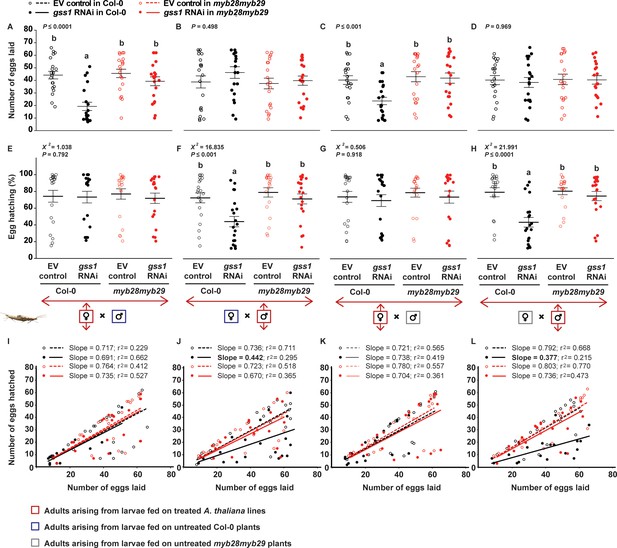
Silencing of gss reduces number of eggs and their hatching percentage.
Number of eggs laid by (A) P. xylostella females arising from larvae raised on the four plant lines (empty vector (EV) control and gss1 RNAi in both Col-0 and myb28myb29 backgrounds) mated with males arising from larvae feeding on untreated Col-0 plants (F3,76 = 13.560, p≤0.0001; n = 20), (B) females arising from larvae feeding on untreated Col-0 plants mated with males arising from larvae raised on the four plant lines, (C) females arising from larvae raised on the four plant lines mated with males arising from larvae feeding on untreated myb28myb29 plants (F3,76 = 6.179, p≤0.001; n = 20), and (D) females arising from larvae feeding on untreated myb28myb29 plants mated with males arising from larvae raised on the four plant lines. Hatching (%) of eggs laid by (E) females arising from larvae raised on the four plant lines mated with males arising from larvae feeding on untreated Col-0 plants, (F) females arising from larvae feeding on untreated Col-0 plants mated with males arising from larvae raised on the four plant lines (χ2 = 216.835, df = 3, p≤0.001; n = 20), (G) females arising from larvae raised on the four plant lines mated with males arising from larvae feeding on untreated myb28myb29 plants, and (H) females arising from larvae feeding on untreated myb28myb29 plants mated with males arising from larvae raised on the four plant lines (χ2 = 221.991, df = 3, p≤0.0001; n = 20). The correlations between the numbers of eggs hatched and the numbers of eggs laid were recorded from crosses between (I) females arising from larvae raised on the four plant lines mated with males arising from larvae feeding on untreated Col-0 plants, (J) females arising from larvae feeding on untreated Col-0 plants mated with males arising from larvae raised on the four plant lines, (K) females arising from larvae raised on the four plant lines mated with males arising from larvae feeding on untreated myb28myb29 plants, and (L) females arising from larvae feeding on untreated myb28myb29 plants mated with males arising from larvae raised on the four plant lines. The number of eggs laid was reduced only in matings involving females arising from gss-silenced larvae fed on plants with wild-type glucosinolate content (Col-0). The percentage of eggs hatching was reduced only in matings involving males arising from gss-silenced larvae fed on plants with wild-type glucosinolate content (Col-0). Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in A–D, and Dunn's post hoc tests in conjunction with non-parametric Kruskal-Wallis tests in E–H.
-
Figure 2—figure supplement 2—source data 1
P. xylostella adult egg laying capacity.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig2-figsupp2-data1-v2.xlsx
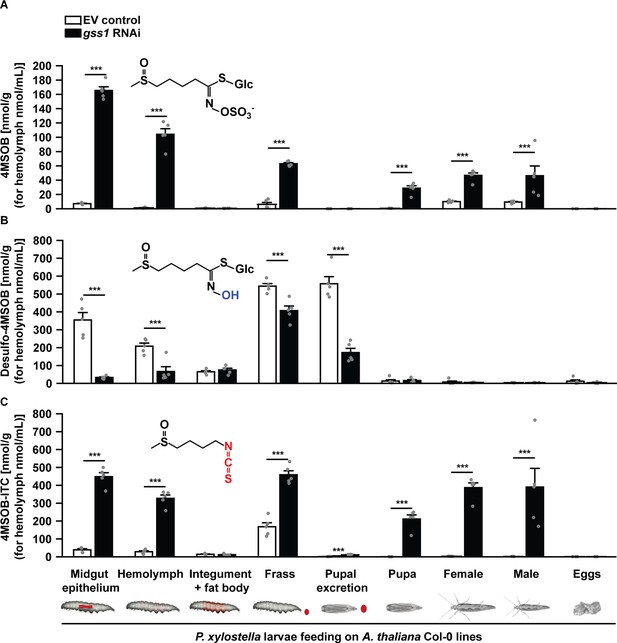
Silencing of gss decreases desulfo-4MSOB and increases 4MSOB-ITC concentrations in P. xylostella.
(A) Intact 4MSOB (Midgut epithelium, F3,16 = 931.79, p≤0.0001; Hemolymph, F3,16 = 172.90, p≤0.0001; Frass, F3,16 = 391.817, p≤0.0001; Pupa, F3,16 = 68.59, p≤0.0001; Female, F3,16 = 150.48, p≤0.0001; Male, F3,16 = 9.64, p≤0.001; n = 5 for all bars), (B) desulfo-4MSOB (Midgut epithelium, F3,16 = 66.94, p≤0.0001; Hemolymph, F3,16 = 34.84, p≤0.0001; Frass, F3,16 = 329.97, p≤0.0001; Pupal excretion, F3,16 = 129.825, p≤0.0001; n = 5 for all bars), and (C) 4MSOB-ITC (Midgut epithelium, F3,16 = 356.30, p≤0.0001; Hemolymph, F3,16 = 269.44, p≤0.0001; Frass, F3,16 = 182.96, p≤0.0001; Pupal excretion, F3,16 = 81.248, p≤0.0001; Pupa, F3,16 = 78.44, p≤0.0001; Female, F3,16 = 211.63, p≤0.0001; Male, F3,16 = 13.83, p≤0.0001; n = 5 for all bars) were measured in various larval tissues, excretions, pupae, and adult moths developed from gss-silenced and non-silenced larvae feeding on A. thaliana Col-0 (wild-type glucosinolates). The concentrations of 4MSOB and its metabolites were nearly undetectable in P. xylostella fed on myb28myb29 (no aliphatic glucosinolates) plants, and these data are not shown in the figure, but given in Figure 3—source data 1. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA.
-
Figure 3—source data 1
The concentration of 4MSOB glucosinolate metabolites in P. xylostella.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig3-data1-v2.xlsx
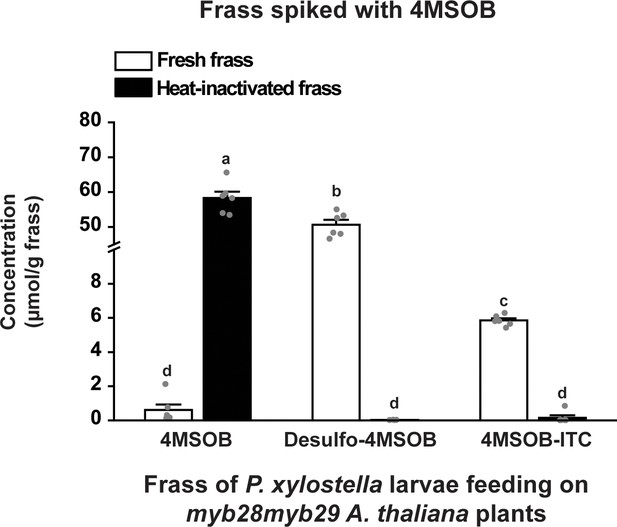
Enzymatic conversion of 4MSOB to desulfo-4MSOB and 4MSOB-ITC is enzymatic and occurs in fresh frass.
Depicted are the concentrations of 4MSOB, desulfo-4MSOB and 4MSOB-ITC in fresh and heat-inactivated larval frass spiked with 4MSOB (Metabolites, F2,30= 456.246, p≤0.0001; Treatments, F1,30= 0.359, p=0.554; Metabolites*Treatments, F2,30 = 1667.024, p≤0.0001; n = 6 for all bars). Frass was collected from P. xylostella larvae feeding on A. thaliana myb29myb29 leaves to ensure the absence of 4MSOB. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with a two-way ANOVA.
-
Figure 3—figure supplement 1—source data 1
Enzyme assay of P. xylostella larval frass with 4MSOB glucosinolate.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig3-figsupp1-data1-v2.xlsx
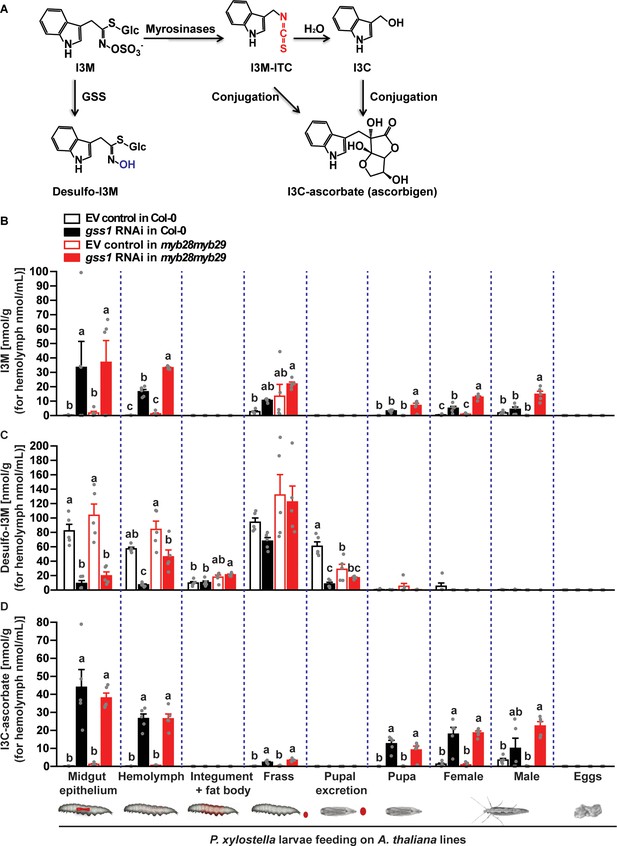
Silencing of gss decreases desulfo-indol-3-ylmethyl glucosinolate (desulfo-I3M) and increases a metabolite of indol-3-ylmethyl glucosinolate isothiocyanate (I3M-ITC) in P. xylostella.
(A) The pathways for formation of desulfo-I3M and indol-3-ylmethyl carbinol (I3C)-ascorbate (ascorbigen) in P. xylostella. When GSS activity in the midgut of P. xylostella is suppressed, plant myrosinases hydrolyze I3M to form I3M-ITC. However, I3M-ITC is unstable and is readily converted to I3C, which can conjugate with ascorbate to form I3C-ascorbate. In the presence of GSS, I3M is desulfated to form desulfo-I3M, reducing the substrate for myrosinases. (B) I3M (Midgut epithelium, F3,16 = 17.095, p≤0.001; Hemolymph, F3,16 = 308.329, p≤0.0001; Pupa, F3,16 = 15.081, p≤0.001; Female, F3,16 = 53.596, p≤0.0001; Male, F3,16 = 24.462, p≤0.0001; n = 5 for all bars), (C) desulfo-I3M (Midgut epithelium, F3,16 = 23.338, p≤0.0001; Hemolymph, F3,16 = 17.750, p≤0.0001; Pupal excretion, F3,16 = 22.218, p≤0.0001; n = 5 for all bars), and (D) I3C-ascorbate (Midgut epithelium, F3,16 = 20.907, p≤0.0001; Hemolymph, F3,16 = 67.127, p≤0.0001; Frass, F3,16 = 34.41, p≤0.0001; Pupa, F3,16 = 19.899, p≤0.0001; Female, F3,16 = 25.107, p≤0.0001; Male, F3,16 = 10.13, p≤0.001; n = 5 for all bars) were measured in various larval tissues, excretions, pupae and adult moths developed from gss-silenced and non-silenced larvae feeding on Col-0 and myb28myb29 plants (Data are shown in Figure 3—figure supplement 2—source data 1). Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in B–D.
-
Figure 3—figure supplement 2—source data 1
The concentration of I3M glucosinolate metabolites in P. xylostella.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig3-figsupp2-data1-v2.xlsx
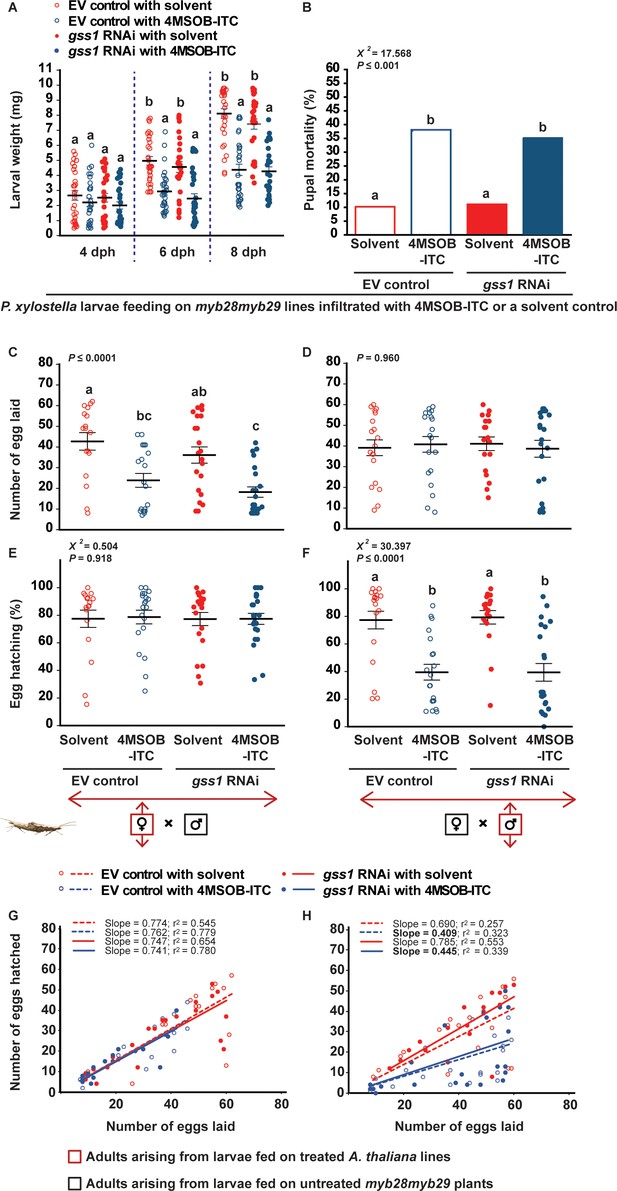
Negative effects on P. xylostella fitness after gss silencing are attributed to the increased 4MSOB-ITC concentrations in the insect body.
Larvae that were gss-silenced and non-silenced were fed on myb28myb29 leaves infiltrated with 4MSOB-ITC (dissolved in a 0.4% aqueous ethanol solvent) or solvent-infiltrated control leaves and the following parameters were measured: (A) weights of larvae four dph (days post hatching), six dph and eight dph, (B) mortality (%) in pupae formed from treated larvae, (C) number of eggs laid by females arising from treated larvae mated with males developed from control larvae feeding on untreated myb28myb29 plants, (D) number of eggs laid by female control larvae mated with males arising from treated larvae, (E) hatching (%) of eggs laid by females arising from treated larvae mated with males developed from control larvae, (F) hatching (%) of eggs laid by females arising from control larvae mated with males developed from treated larvae, and the correlations between the numbers of eggs hatched and the numbers of eggs laid (G) by females arising from treated larvae mated with males arising from control larvae, and (H) by females arising from control larvae mated with males arising from treated larvae. Feeding on 4MSOB-ITC caused a significant decline in larval weight (six dph, RNAi F1,116 = 2.130, p=0.147; Infiltration F1,116 = 46.287, p≤0.0001; RNAi*Infiltration F1,116 = 0.012, p=0.913. eight dph, RNAi F1,116 = 1.394, p=0.240; Infiltration F1,116 = 103.860, p≤0.0001; RNAi*Infiltration F1,116 = 0.765, p=0.384. n = 30 for all treatments), a significant increase in pupal mortality (χ2 = 217.568, df = 3, p≤0.001; n = 49, 42, 45 and 51, respectively), a significant decline in egg laying in crosses when females arise from larvae fed on 4MSOB-ITC (F3,76 = 10.046, p≤0.0001, n = 17, 20, 21 and 22, respectively), and a significant decline in egg hatching rate when males arose from larvae fed on 4MSOB-ITC (χ2 = 230.397, df = 3, p≤0.0001; n = 19, 20, 18 and 22, respectively). Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with two-way ANOVA in A, a proportions test with pairwise comparisons in B, Tukey HSD tests in conjunction with one-way ANOVA in C–D, and Dunn's post hoc tests in conjunction with non-parametric Kruskal-Wallis tests in E–F.
-
Figure 4—source data 1
P. xylostella larval weight, pupal mortality, and adult egg laying capacity in complementation experiment.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig4-data1-v2.xlsx
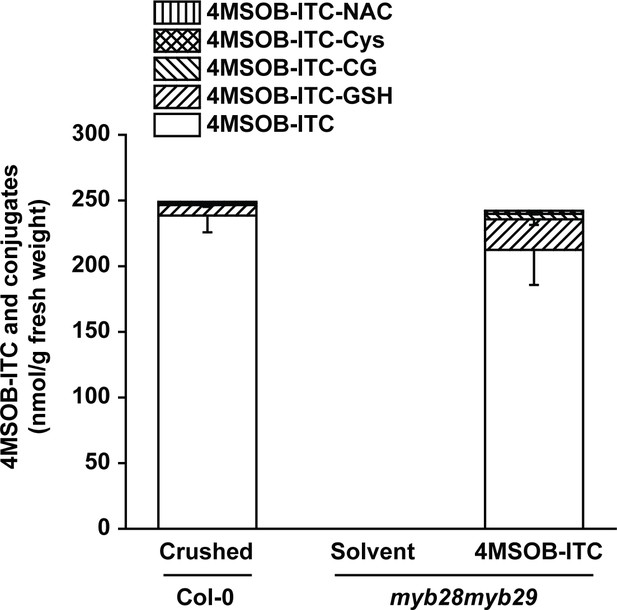
Concentrations of 4MSOB-ITC and conjugates formed in crushed leaves of Col-0 A. thaliana plants, and in myb28myb29 leaves infiltrated with 4MSOB-ITC in aqueous ethanol or with pure aqueous ethanol as a control.
4MSOB-ITC and conjugates were extracted with 60% aqueous methanol and analyzed by LC-MS/MS. NAC, N-acetylcysteine; CG, cysteinylglycine; Cys, cysteine; GSH, glutathione. (see Figure 5A for structures).
-
Figure 4—figure supplement 1—source data 1
The concentration of 4MSOB-ITC derivates in A. thaliana plants.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig4-figsupp1-data1-v2.xlsx
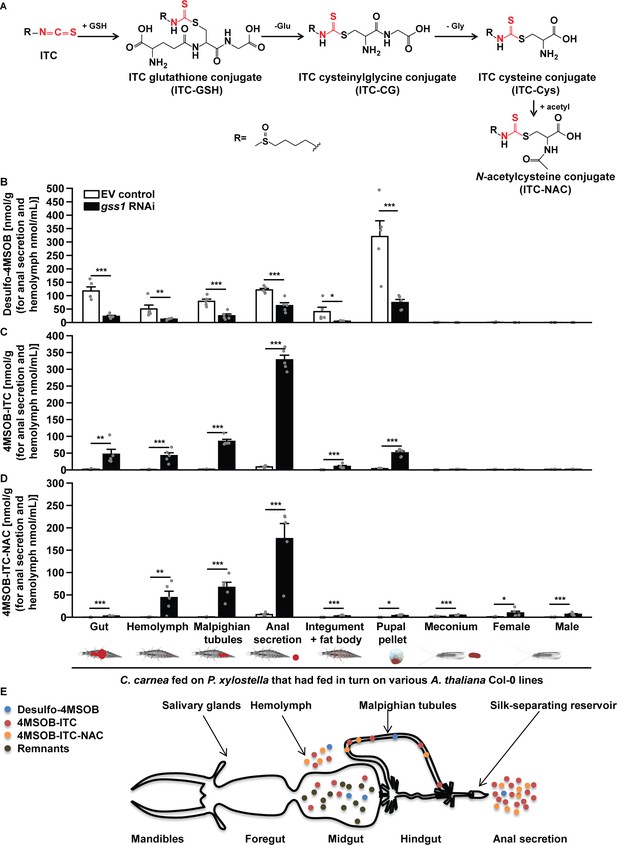
The lacewing C. carnea circumvents the toxicity of glucosinolate hydrolysis products by conjugation and mobilization.
(A) General mercapturic acid pathway for detoxification of 4MSOB-ITC in various insects: ingested 4MSOB-ITC is detoxified by conjugation with glutathione (GSH), followed by cleavage to recover glutamate (Glu) and glycine (Gly) and further modification of cysteine (Cys) to form the final N-acetylcysteine conjugate (4MSOB-ITC-NAC). Distribution of (B) desulfo-4MSOB, (C) 4MSOB-ITC, and (D) 4MSOB-ITC-NAC conjugate in various tissues and excretions of C. carnea larvae and adults arising from larvae that predated upon gss-silenced and non-silenced P. xylostella larvae feeding on A. thaliana Col-0 (wild-type glucosinolates) and myb28myb29 (no aliphatic glucosinolates) plants. Predation on gss-silenced P. xylostella larvae caused a significant reduction in desulfated 4MSOB (Gut, F3,16 = 32.378, p≤0.0001; Hemolymph, F3,16 = 10.23, p≤0.001; Malpighian tubules, F3,16 = 50.855, p≤0.0001; Anal secretion, F3,16 = 199.006, p≤0.0001; Integument and fat body, F3,16 = 5.959, p≤0.01; Pupal pellet, F3,16 = 24.907, p≤0.001; n = 5 for all bars), a significant increase in the toxic hydrolysis product, 4MSOB-ITC (Gut, F3,16 = 9.895, p≤0.001; Hemolymph, F3,16 = 22.967, p≤0.0001; Malpighian tubules, F3,16 = 180.333, p≤0.0001; Anal secretion, F3,16 = 110.697, p≤0.0001; Integument and fat body, F3,16 = 13.919, p≤0.001; Pupal pellet, F3,16 = 14.571, p≤0.0001; n = 5 for all bars), and a significant increase in the detoxification product 4MSOB-ITC-NAC (Gut, F3,16 = 29.098, p≤0.0001; Hemolymph, F3,16 = 9.242, p≤0.001; Malpighian tubules, F3,16 = 33.688, p≤0.0001; Anal secretion, F3,16 = 32.659, p≤0.0001; Integument and fat body, F3,16 = 14.981, p≤0.0001; Pupal pellet, F3,16 = 6.544, p≤0.01; Meconium, F3,16 = 18.232, p≤0.0001; Female, F3,16 = 5.547, p≤0.01; Male, F3,16 = 16.777, p≤0.0001; n = 5 for all bars). Since the concentrations of 4MSOB and its metabolites were nearly undetectable in C. carnea preying on P. xylostella whose larvae fed on myb28myb29 plants, these data are not shown in the figure, but given in Figure 5—source data 1. (E) A schematic representation of the alimentary tract of a C. carnea larva preying on P. xylostella gss-silenced larvae feeding on Col-0 (redrawn from McEwen et al., 2001) showing storage of the majority of free and conjugated isothiocyanates and derivatives in the anal secretion reservoir. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in B–D).
-
Figure 5—source data 1
The concentration of 4MSOB glucosinolate metabolites in C. carnea.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig5-data1-v2.xlsx
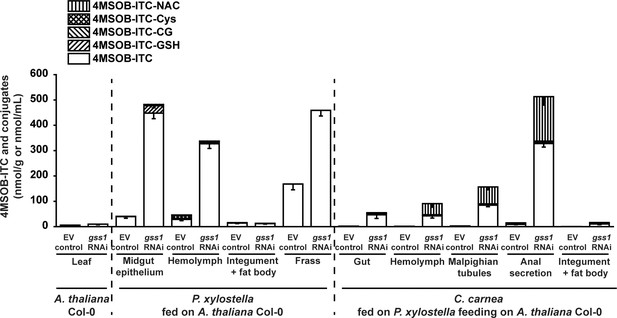
C. carnea detoxifies 4MSOB-ITC by forming the 4MSOB-ITC-NAC conjugate.
Depicted are levels of 4MSOB-ITC and its mercapturic acid pathway conjugates in gss1 RNAi and empty vector (EV) control infiltrated Col-0 plants, in P. xylostella feeding on those plants, and in C. carnea after preying on P. xylostella larvae.
-
Figure 5—figure supplement 1—source data 1
The concentration of 4MSOB-ITC derivates in third-trophic level.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig5-figsupp1-data1-v2.xlsx
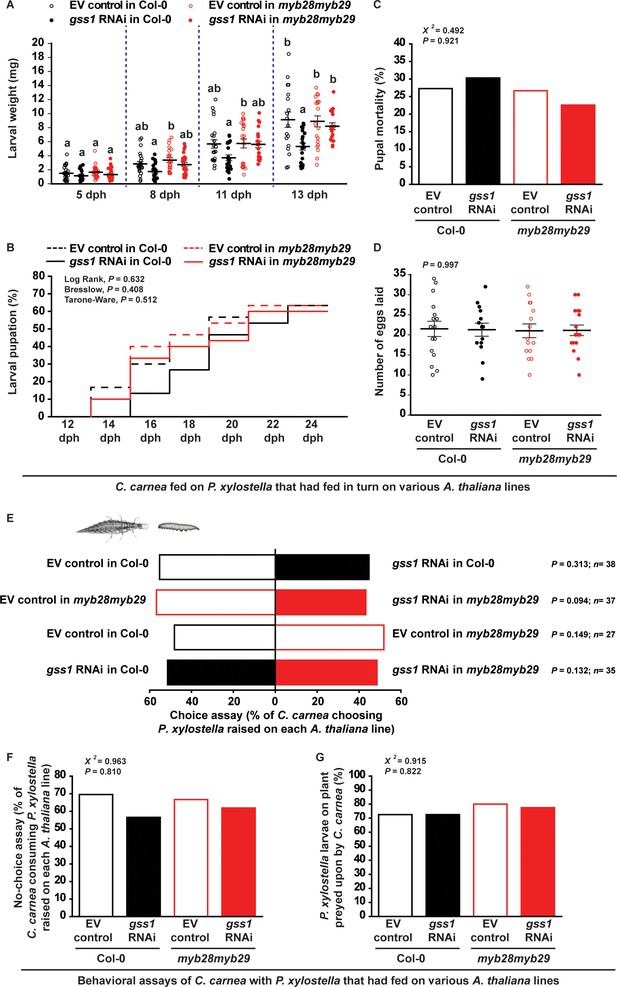
C. carnea larval development is slowed by glucosinolate metabolites in its prey, but behavior and adult fitness are not affected.
(A) C. carnea larval weights (eight dph, days post hatching, F3,90 = 5.164, p≤0.05, n = 24, 23, 23 and 24, respectively; eleven dph, F3,82 = 3.235, p≤0.05, n = 22, 21, 22 and 21, respectively; thirteen dph, F3,73 = 5.767, p≤0.01, n = 18, 20, 20 and 19, respectively) were decreased when predating on gss-silenced P. xylostella larvae feeding on A. thaliana Col-0 with wild-type glucosinolate levels in comparison to predation on non-silenced larvae or larvae feeding on plants without aliphatic glucosinolates. (B) Although the duration of the pupal stage (EV control in Col-0: 18.3 day; gss1 RNAi in Col-0: 20.9 day; EV control in myb28myb29: 18.6 day; gss1 RNAi in myb28myb29: 19.1 day; see Figure 6—source data 1) was influenced by the prey, the final percentages of pupation between 12 to 24 dph (Log Rank, df = 3, p=0.632; Breslow, df = 3, p=0.408; Tarone-Ware, df = 3, p=0.512; n = 30 for all treatments) were not affected. (C) C. carnea pupal mortality (%) (χ2 = 20.492, df = 3, p=0.921; n = 33, 33, 30 and 31, respectively) and (D) adult egg-laying capacity (F3,58 = 0.018, p=0.997, n = 16, 14, 15, 17, respectively) were not significantly affected by predation upon gss-silenced and non-silenced P. xylostella larvae feeding on Col-0 (wild-type glucosinolates) or myb28myb29 (no aliphatic glucosinolates) plants. (E) Choice assays, (F) no-choice assays (χ2 = 20.963, df = 3, p=0.810; n = 23, 23, 21 and 21, respectively), and (G) predation trials on plants (χ2 = 20.915, df = 3, p=0.822; n = 40 in all bars) showed that C. carnea did not avoid gss-silenced P. xylostella larvae, in spite of their higher 4MSOB-ITC concentrations. Significant differences (p≤0.05) between means (±s.e.) were determined by Tukey HSD tests in conjunction with one-way ANOVA in A,D, Kaplan-Meier survival analysis test in B, two-sided proportions test in C,F,G, and C. carnea prey choices were analyzed with a two-sided binomial test (between two P. xylostella larvae) and GLM with a binomial distribution and a logit link function (between each treatment) in E.
-
Figure 6—source data 1
C. carnea larval weight, the percentages of pupation and pupal mortality, adult egg laying capacity, and behavior assays.
- https://cdn.elifesciences.org/articles/51029/elife-51029-fig6-data1-v2.xlsx
Additional files
-
Supplementary file 1
Primer sets for gene cloning and qRT-PCR validation.
- https://cdn.elifesciences.org/articles/51029/elife-51029-supp1-v2.xlsx
-
Supplementary file 2
The sources of Plutella xylostella moths for adult fecundity experiments.
- https://cdn.elifesciences.org/articles/51029/elife-51029-supp2-v2.xlsx
-
Supplementary file 3
LC-MS/MS parameters used for the multiple reaction monitoring (MRM) analyses of I3C derivatives.
Q1, quadrupole one voltage; Q3, quadrupole three voltage; DP, declustering potential; EP, entrance potential; CEP, collision cell entrance potential; CE, collision energy; CXP, collision cell exit potential.
- https://cdn.elifesciences.org/articles/51029/elife-51029-supp3-v2.xlsx
-
Supplementary file 4
External standards used for quantification.
- https://cdn.elifesciences.org/articles/51029/elife-51029-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51029/elife-51029-transrepform-v2.docx





