Neonatal-derived IL-17 producing dermal γδ T cells are required to prevent spontaneous atopic dermatitis
Figures
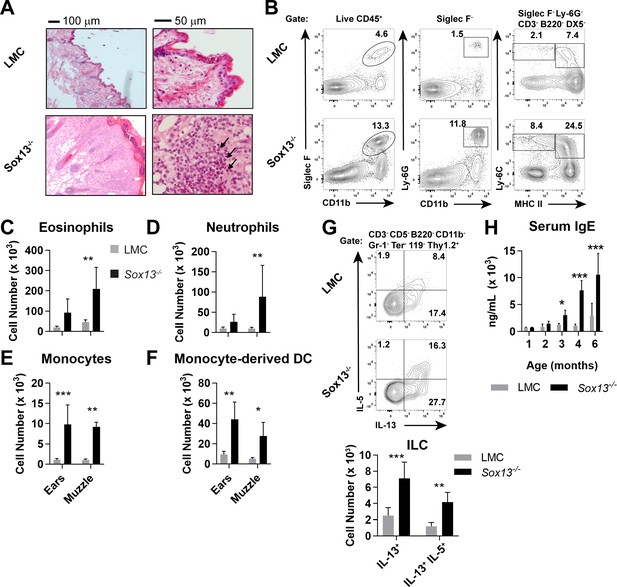
Development of AD in the absence of dermal Vγ2+ Tγδ17 cells.
(A) Biopsies of muzzle skin from 6 mo Sox13-/- and Sox13+/- littermate control (LMC) was analyzed by H and E staining. Black arrows identify numerous eosinophilic infiltrates in the epidermis. Representative of four experiments, each with minimum n = 2/group. (B) Muzzle skin was digested and analyzed via FACS for Siglec F+ eosinophils (left panels), Ly- 6G+ neutrophils (middle panels), Ly-6C+ MHC-IIlo monocytes and Ly-6C+ MHC-IIhi monocyte-derived dendritic cells (right panels). Data are representative of >6 similar experiments analyzing 2–3 mice per/group. (C–F) Enumeration of cell types examined in Panel B. n = 6/group. *, p<0.05; **p<0.01; ***, p<0.001 by ANOVA. (G) Muzzle-infiltrating cells were isolated from LMC and Sox13-/- mice and re-stimulated in vitro with PdBu/ionomycin to assess production of IL-5 and IL-13 by ILCs. ILC identified as CD45+ Thy1.2+Lineage markersneg (CD3/CD4/CD5/CD8/CD11b/DX5/Gr-1/TCRδ/TCRβ/Ter-119neg). Bottom summary graph enumerates IL-13+ and IL-5+IL-13+ ILC. N = 6/group. **p<0.01; ***p<0.001 by ANOVA. (H) Serum IgE concentration in mice of indicated genotype, aged 1–6 mo, was determined by ELISA. n = 3–6/group. *, p<0.05; ***, p<0.001 by ANOVA.
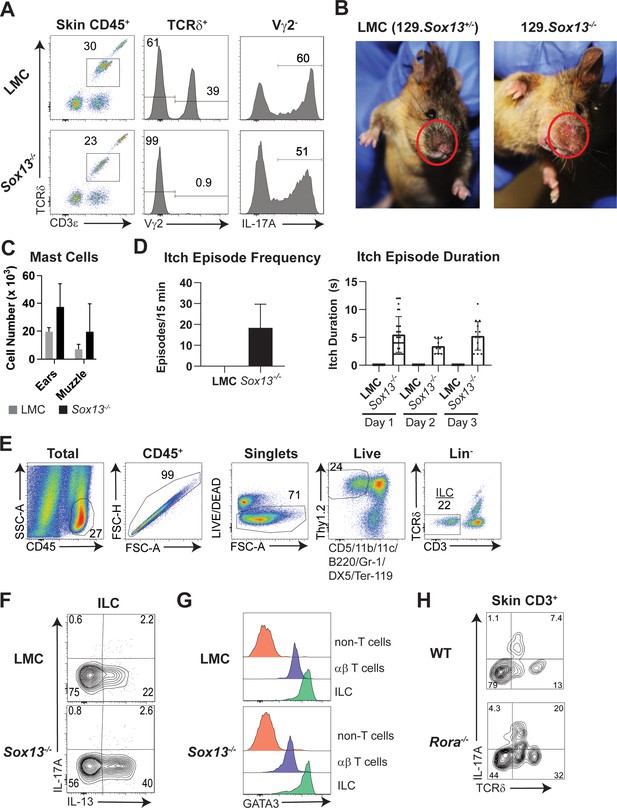
Specific loss of Vγ2+ Tγδ17 cells, scratching behaviors and reciprocally enhanced effector function of ILCs in Sox13-/- mice with dermatitis.
(A) Analysis of dermal γδ T cells, demonstrating complete loss of Vγ2+ dermal TCRδint cells and maintenance of IL-17 production by Vγ2- (Vγ4+) dermal TCRδint cells. Data are representative of >50 mice analyzed of each genotype. (B) A representative image of inflamed, raw-appearing muzzle of five months old male Sox13-/- mice, contrasted to normal skin of a male littermate control (LMC). (C) Enumeration of FcεRIα+c-Kit+ mast cells in muzzle skin. n = 6/group. (D) Summary of scratching episodes of Sox13-/- mice with AD. Female mice (three of WT and Sox13-/-, 7–8 months of age) were filmed on three consecutive days for 15 min and scored for the number of uninterrupted scratching behavior (left, as an average daily number of episodes over 3 days) and the duration (in seconds, right) of each episode per day. Also refer to associated Videos 1 and 2. (E) Gating scheme for FACS analysis of ILC in the skin. (F) Analysis of IL-17A and IL-13 production by ILCs from 3mo mice as identified in Panel A after PdBU/ionomycin stimulation. (G) Intranuclear GATA3 staining in skin ILCs. Non-T cells and T cells are presented as internal references for the high level of GATA3 expressed by ILCs. (H) Increased skin γδ T cells, including Tγδ17 cells (TCRδint), in 4–6 week old Rora-/- mice. Skin cells were stimulated with PdBU/ionomycin and IL-17 production assessed in CD3+ T cells. One of 4 mice analyzed shown. Note limited IL-17 production at this age from αβ T cells and no IL-17 production from TCRδhi Vγ3+dendritic epidermal T cells (DETCs).
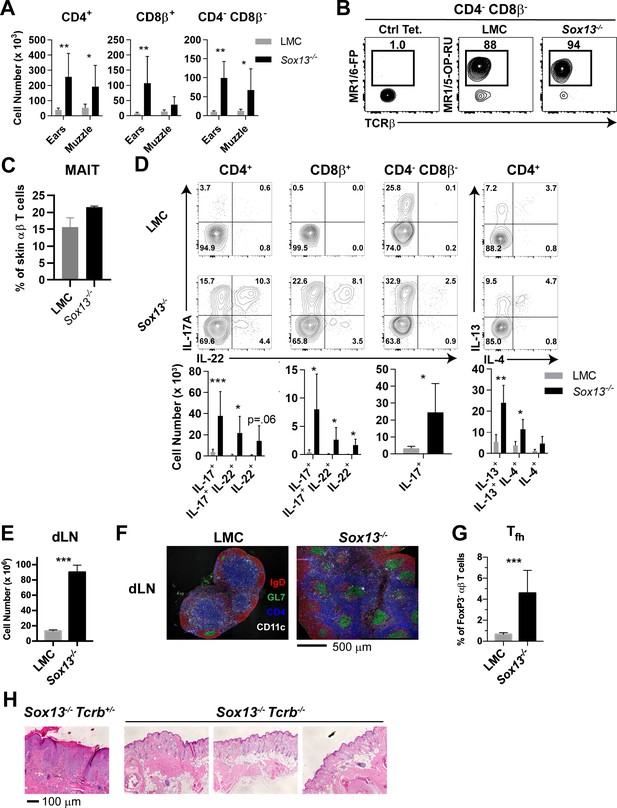
Aberrant αβ T cell activation in AD of Sox13-/- mice.
(A) Total number of the indicated T cell types recovered from skin of from 5-6mo mice were calculated using AccuCheck counting beads. n = 6/group. *, p<0.05; **, p<0.01 by ANOVA. (B) FACS analysis of CD4negCD8βneg skin T cells (gated on B220-F4/80- TCRβ+) with control MR1/6-FP or MR1/5-OP-RU tetramer to identify MAIT cells in 5 mo mice. (C) Summary data of frequency of MAIT tetramer-reactive cells among total TCRβ+ cells pooled from two independent experiments, performed as in Panel B analyzing a total of 5–6 mice/group. (D) Muzzle-infiltrating cells were isolated from indicated mice, stimulated in vitro with PdBu/ionomycin, and analyzed for αβ T cell subset-specific production of IL-17A and IL-22 and for CD4+ T cell production of IL-4, and IL-13. FACS data are representative of >5 experiments. For summary data below, n = 6/group. *, p<0.05; **p<0.01, ***, p<0.001 by ANOVA or t-test (CD4-CD8β- cells). (E) Total cell number enumeration in skin draining LNs (dLNs) of 6 mo mice of indicated genotype, n = 6/group. ***, p<0.001 by Student’s t-test. (F) Muzzle draining mandibular LN (dLN) from 5 to 6 mo mice were fixed in paraformaldehyde, frozen in OCT compound, cryosectioned, and then labeled with the indicated antibodies to visualize B cell follicles (IgD+), T cell zones (CD4+), dendritic cells (CD11c+), and germinal centers (GL7+ IgD-). Images are representative of two experiments analyzing sections from at least 3 mice per experiment. (G) Summary data of T follicular helper (Tfh) cells in dLN of 6 mo LMC and Sox13-/- mice. Tfh cells were identified as CD4+FoxP3neg PD-1hi CXCR5+Bcl6+. n = 7–8/group. ***, p<0.001 by Student’s t- test. (H) Sox13-/- and 129.Tcrb-/- mice were crossed to generate double-deficient mice, and then disease progression tracked by phenotyping and muzzle inflammation assessed by H and E staining. Sox13-/-Tcrb-/- mice do not develop overt or histological signs of AD at 6 mo. Data are representative of 10–15 mice of each genotype analyzed.
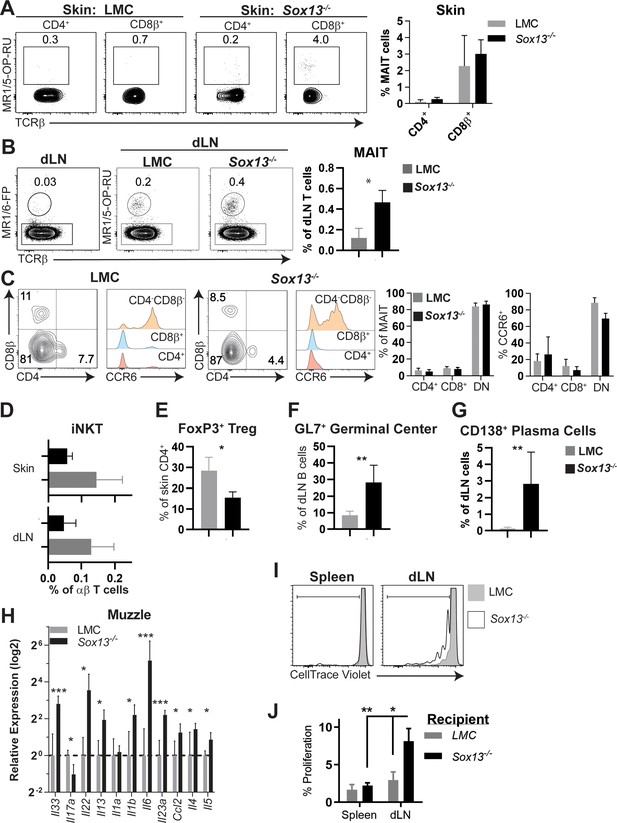
Characterization of skin and dLN MAITs, iNKT cells, regulatory T cells (Tregs), B cell subsets and melanocyte antigen-specific T cells in Sox13-/- mice.
(A) MAIT tetramer MR1/5-OP-RU (5-(2-oxopropylideneamino)−6-Dribitylaminouracil, a riboflavin biosynthetic product recognized by MAITs) staining of skin CD4+ and CD8β+ T cells reveal minimum coreceptor expressing MAITs in the skin of 5-6mo mice. Summary data, n = 5–6/group. (B) Analysis of MAIT cells in skin dLN from mice as above. Summary data, n = 5–6/group. *, p<0.05 by t-test. (C) Coreceptor and CCR6 expression, a hallmark of type 3 cytokine producing T cells, by MAIT cells in the dLN identified as in (B). Summary data, n = 5–6/group. (D) Summary of iNKT cell frequencies in the skin and dLN of mice of indicated genotype as in panel A, using CD1d/PBS57 tetramer that stains the canonical Vα14+ iNKT cells. (E) Summary of FACS analysis for FoxP3 to identify Tregs in the skin of 5–6 mo mice of indicated genotype as in panel A. n = 6/group. *, p<0.05 by t-test. (F) Summary of FACS analysis of GC B cells (GL7+ CD95+) in dLN of 5 mo mice of indicated genotype as in panel A. n = 7–8/group. **, p<0.01 by t-test. (G) Summary of FACS analysis of CD138+ B220- plasma cells in dLN of 5 mo mice of indicated genotype as in panel A. n = 7–8/group. **, p<0.01 by t-test. (H) RT-qPCR analysis of cytokine and chemokine gene expression from RNA prepared from total muzzle skin (6 mo) of mice with indicated genotype. Only those significantly altered in expression (except Il1a as an example of unchanged) are shown. LMC n = 4; Sox13-/- mice, n = 5. ***, p<0.001; *, p<0.05 by Student’s t-test. (I) Pre-melanocyte antigen 1-specific PMEL CD8+ Tg T cells were labeled with CellTrace Violet and then adoptively transferred into 6 mo LMC or Sox13-/- mice. Four days later, spleen and skin dLN were harvested for analysis of proliferation by transferred PMEL T cells (identified by Thy1.1 allomarker). Data from one of two experiments shown, n = 4/group. (J) Summary data of PMEL adoptive transfer proliferation performed in Panel H. **, p<0.01; *, p<0.05 by ANOVA.
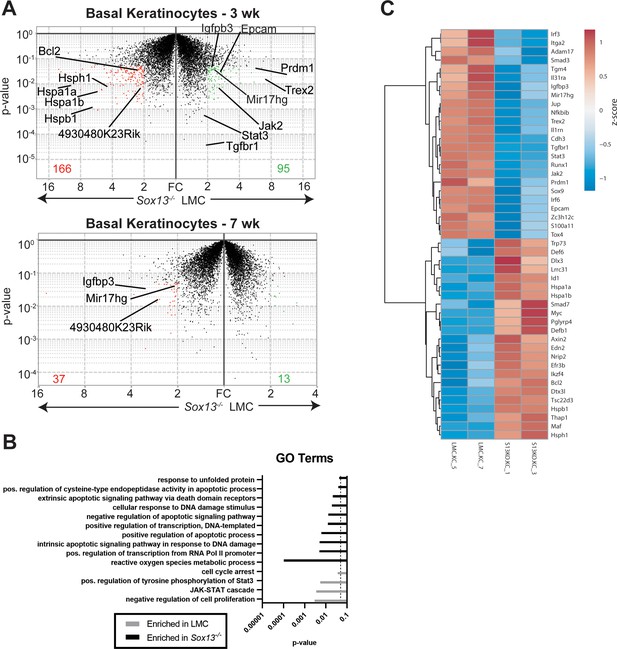
Perturbations in early basal keratinocyte transcriptome in the absence of Vγ2+ Tγδ17 cells.
(A) Epidermal basal keratinocytes were sorted from 3 and 7 wk old male LMC and Sox13-/- mice and subjected to gene expression analysis by RNA sequencing (in biological triplicates). Red and green dots represent genes with fold change (FC) >2 and p-value <. 05 and the numbers at the bottom denote number of genes whose expression was significantly altered. Select genes are annotated. (B) Differentially expressed genes from Panel A were analyzed for Gene Ontology (GO) term enrichment using DAVID. Displayed are a selection of significantly enriched (p <. 05, dashed line) GO terms. (C) Heatmap of differentially expressed genes (FC >1.5 and p-value <. 05) among male 3 wk old basal keratinocytes with genes involved in cell differentiation, barrier function, skin inflammation and stress response pathways annotated.
-
Figure 3—source data 1
RNA sequencing read count tables used to generate volcano plot in Figure 3A (upper panel) for basal keratinocytes analysis of 3 wk old mice.
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig3-data1-v1.xlsx
-
Figure 3—source data 2
RNA sequencing read count tables used to generate volcano plot in Figure 3A (lower panel) for basal keratinocytes analysis of 7 wk old mice.
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig3-data2-v1.xlsx
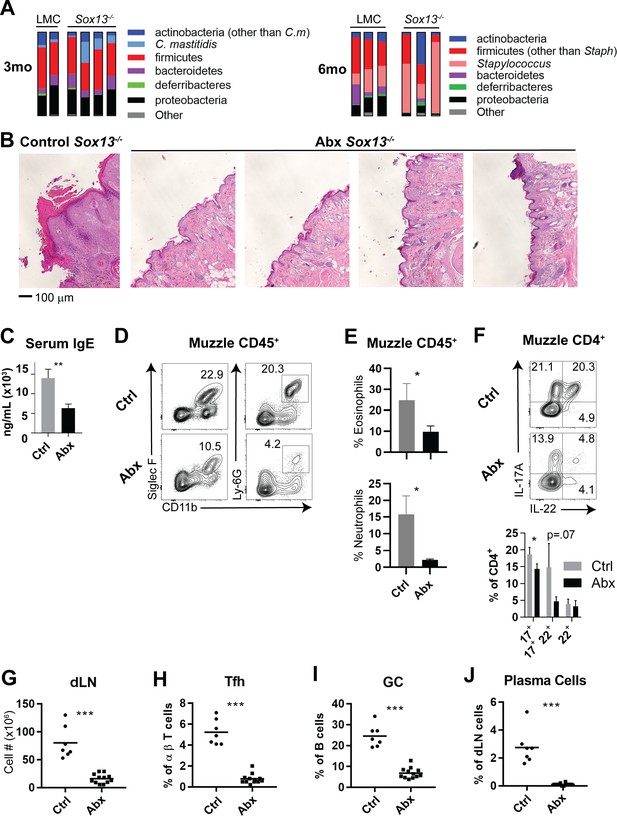
Skin commensal alterations in the absence of Vγ2+ Tγδ17 cells drive AD.
(A) Summary stacked bar charts of muzzle skin microbiome analysis of Sox13-/- and LMC mice at 3 mo and 6 mo. Species depicted are annotated on the right and their corresponding frequencies among total 16S rRNA sequences are shown. One experiment of three shown. (B) Sox13-/- mice were antibiotic (enrofloxacin and cefazolin) treated (Abx) by drinking water from 2 mo and then muzzle histology analyzed at 6 mo. Images are representative of 4 analyzed Abx-treated mice, with at least 2 sections separated by >100 microns analyzed for each mouse. (C) Serum IgE levels of Ctrl and Abx-treated Sox13-/- mice at 6 mo were assessed by ELISA. n = 6 (Ctrl) or 10 (Abx). **, p<0.01 by Student’s t-test. (D) Muzzle skin of 6 mo Ctrl and Abx Sox13-/- mice was analyzed for eosinophil and neutrophil infiltration via FACS. Data are representative of 9 analyzed Abx-treated mice from 3 independent cohorts. (E) Summary data of the frequency of Eosinophils (top) and Neutrophils (bottom) among all CD45+ muzzle skin cells. n = 3/group from 1 of 3 similar experiments. *, p<0.05 by t-test. (F) Muzzle skin of 6 mo Ctrl and Abx Sox13-/- mice was analyzed for Th17 cytokine production post PMA/ionophore reactivation. Summary data of n = 5/group, pooled from 2 independent experiments. *, p<0.05 by ANOVA. (G–J) Mandibular and parotid dLN cells from Ctrl and Abx Sox13-/- mice were analyzed for total cell number (G), and the frequency of Tfh cells (H), GC B cells (I), and CD138+ plasma cells (J). n = 7–12/group pooled from 4 independent cohorts. ***, p<0.001 by Student’s t-test.
-
Figure 4—source data 1
Bacterial species abundance from the muzzle skin of 3 mo and 6 mo LMC and Sox13-/- mice as summarized in Figure 4A.
Values indicate percent abundance per sample (total = 100).
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig4-data1-v1.xlsx
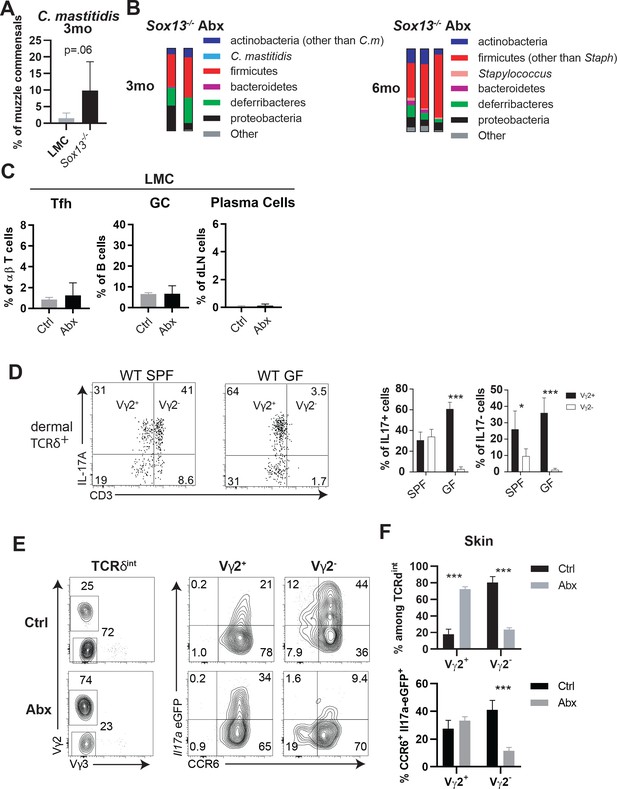
Skin microbiome of antibiotic-treated (Abx) Sox13-/- mice and independence of dermal Vγ2+ Tγδ17 from CB for their development and maintenance.
(A) Summary of the relative abundance of C. mastitidis on the muzzle skin of LMC and Sox13-/- mice. n = 6/group from 2 separate sequencing analyses. (B) Muzzle skin microbiome analyses of Abx Sox13-/- mice at 3 and 6 mo show near complete depletion of Corynebaceria mastitidis and decreased Staphylocccus, compared to the age-dependent blooming of these species in Sox13-/- mice shown in Figure 5, and correlating with the prevention of AD in Abx Sox13-/- mice. (C) Summary of FACS analysis of the frequency of Tfh CD4+ T cells, GC B cells, and Plasma cells in Ctrl (gray bars) and Abx-treated LMC mice, demonstrating no alteration in these populations due to Abx treatment. Data are summarized from 2 independent treatment cohorts. (D) Loss of dermal Vγ4+ Tγδ17 cells (CD3hi, among TCRδ+ cells, with residual Vγ3+ DETCs gated out) in 6 wk old germ free (GF) B6 mice skin, relative to those housed in the SFP condition, as reported previously. However, Vγ2+ Tγδ17 (CD3int) cells are maintained and fully capable of IL-17 production in age-matched GF mice. Summary data, n = 4 (GF) and n = 5 mice (SPF) from two independent experiments. *, p<0.05; ***p<0.001 by ANOVA. (E) Abx Il17a-Egfp mice show that in vivo Il17a transcription in Vγ2+ Tγδ17 cells occurs independent of the Abx sensitive CB in panel A. Mice were treated with Abx for 4 weeks, and then ex vivo dermal γδ cells in the skin (TCRδint) were analyzed for expression of Vγ chain (left panel, in the dermis Vγ2neg cells express Vγ4) and CCR6 and EGFP (right two panels) expression in dermal Vγ2+ Tγδ17 and Vγ2- (Vγ4+) Tγδ17 cells. (F) Summary of data from experiments depicted in Panel E show the loss of Vγ4+ Tγδ17 cells and compensatory increase in Vγ2+ Tγδ17 cells in one month Abx mice (Top) and the preserved capacity to express Il17a in Vγ2+ Tγδ17, but not in residual Vγ4+ Tγδ17, cells. n = 3/group. ***, p<0.001 by ANOVA.
-
Figure 4—figure supplement 1—source data 1
Bacterial species abundance from the muzzle skin of 3 mo and 6 mo LMC and Sox13-/- mice treated with Abx as summarized in Figure 4—figure supplement 1B.
Values indicate percent abundance per sample (total = 100).
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig4-figsupp1-data1-v1.xlsx
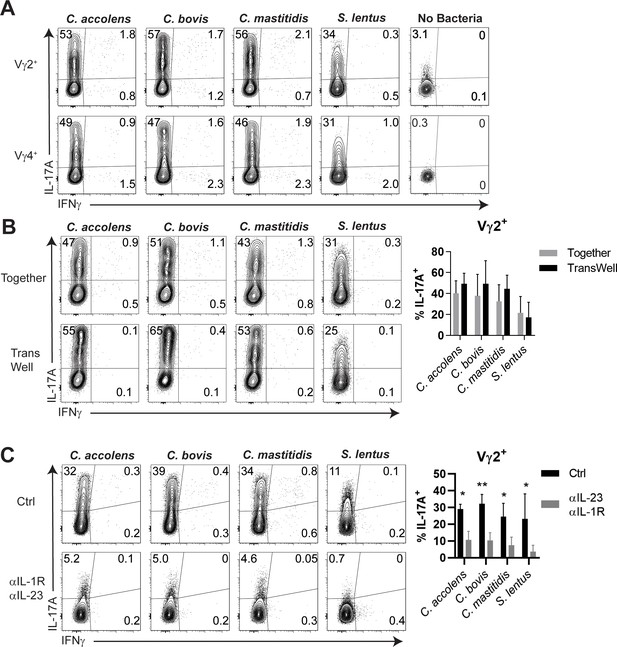
Cytokine-dependent, contact-independent Tγδ17 responses to skin commensals.
(A) Total γδ cells were enriched from skin-draining LN and co-cultured with antigen presenting cells and the indicated heat-killed commensal bacteria at a 1:1:10 ratio for 16–18 hr, and then cultured for an additional 4 hr in the presence of Golgi Stop and Plug. IL-17A and IFNγ production was assessed by intracellular cytokine staining. Data are representative of 2 independent experiments. (B) Total γδ cells, splenic DC, and the indicated commensal bacteria were cultured at a 1:1:10 ratio as in (A) Together in a well or in a 0.4 micron TransWell apparatus in which DC and bacteria were placed in the top chamber and γδ cells were placed in the bottom chamber. Summary data are pooled from 2 independent experiments. (C) Cultures as above with 10 ug/mL each of anti-IL-1R and anti-IL-23 neutralizing Abs or isotype control Abs. Intracellular production of IL-17A and IFNγ was then assessed by FACS. Summary data are pooled from 4 independent experiments. *, p<0.05; **, p<0.01 by ANOVA.
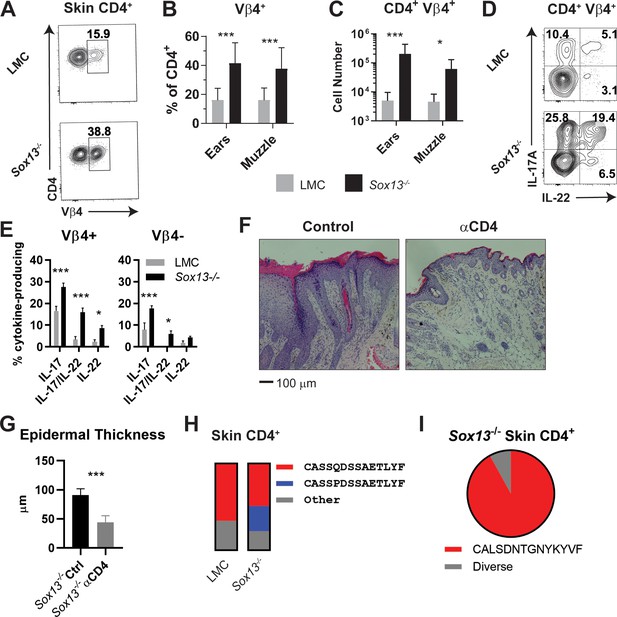
Expansion of dominant CD4+ clonotypes in Sox13-/- skin.
(A) Muzzle-infiltrating cells were isolated from 5-6mo LMC and Sox13-/- mice and analyzed for Vβ usage by CD4+ T cells. (B) Summary data of Vβ4+ frequency among skin-infiltrating CD4+ cells in LMC and Sox13-/- mice. n = 13–17 mice. ***, p<0.001 by ANOVA. (C) Enumeration of CD4+ Vβ4+ cells in LMC and Sox13-/- skin. n = 6/group. ***, p<0.001; *, p<0.05 by ANOVA. (D) Skin-infiltrating cells were isolated from 5 mo mice, restimulated in vitro with PdBu/ionomycin, and IL-17 and IL-22 production by Vβ4+ and Vβ4- CD4+ T cells assessed via FACS. Data are representative of >4 experiments analyzing 2–3 mice/genotype/experiment. (E) Summary data of multiple experiments performed as in Panel D. n = 5–6 pooled from 3 independent experiments. ***, p<0.001; *, p<0.05 by ANOVA. (F) Starting at 3 months of age, Sox13-/- mice were treated with control Ab (Ctrl) or a cell depleting Ab targeting CD4 antigen (αCD4) until 6 mo. AD disease severity was then assessed by H and E staining of muzzle skin. Data are representative of 10 mice treated with αCD4 Ab across 2 independent experiments. (G) Epidermal thickness in Ctrl and αCD4 Ab treated Sox13-/- mice as assessed by analysis of histology images. n = 5 mice/group. ***, p<0.001 by Student’s t-test. (H) Summary stacked bar charts of TCR Vβ4 CDR3 clonotype analysis of skin (ear and muzzle combined) infiltrating CD4+ T cells in LMC and Sox13-/- mice by deep sequencing, focusing on the two major clonotypes. Minimal 1 million reads/sample. Each stack reports proportion of each class on the right amongst total Vβ4 CDR3 sequence reads. (I) Summary of TCR Vα4 CDR3 clonotype analysis by pie chart of skin-infiltrating CD4+ T cells in Sox13-/- mice. LMC control not shown as there were insufficient reads.
-
Figure 6—source data 1
TCR Vβ4 (TRBV2 by IMGT nomenclature) CDR3 sequencing analysis of CD4+ non-Treg cells from the skin of LMC and Sox13-/- mice as summarized in Figure 6H.
Values indicate percent of all reads per sample (total = 1).
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig6-data1-v1.xlsx
-
Figure 6—source data 2
TCR Vα4 (TRAV6 by IMGT nomenclature) CDR3 sequencing analysis of CD4+ non-Treg from the skin of Sox13-/- mice as summarized in Figure 6I.
Values indicate percent of all reads per sample (total = 1).
- https://cdn.elifesciences.org/articles/51188/elife-51188-fig6-data2-v1.xlsx
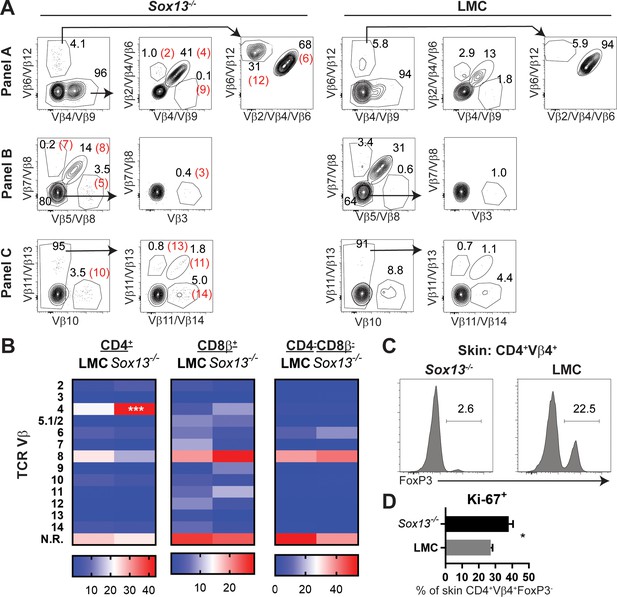
Expansion of CD4+Vβ4+ T cells in the skin of Sox13-/- mice.
(A) Representative multiplex analysis of TCR Vβ usage in skin T cells of 13 Vβ for which commercial Abs are available that bind TCR haplotypes expressed by 129/Sv mice. In this illustration, in Panel A αVβ4 and αVβ9 are both FITC conjugates; αVβ6 and Vβ12 are PE conjugates; αVβ2, αVβ4, and αVβ6 are biotin conjugates. This combinatorial staining strategy allows for deconvolution of which TCR Vβ is expressed by a given population via the gating shown. Inset red numbers in parentheses indicate the specific Vβ identified by the gated population. (B) Heatmap of TCR Vβ expression by T cells in the skin of LMC and Sox13-/- mice. n = 9, pooled from 3 independent experiments using 6mo mice. From this analysis, only Vβ4 in CD4+ cells was consistently and significantly altered across multiple experiments. N.R., non-reactive to Abs in panel. Color coding at the bottom indicates % positive among αβ T cell subsets. ***, p<0.001 by ANOVA (C) Intranuclear FoxP3 staining in skin CD4+Vβ4+ T cells from 5-6mo mice. Compared to Figure 2—figure supplement 1E, data indicate that CD4+Vβ4+ T cells are underrepresented in the skin Treg pool. (D) Intranuclear expression of Ki-67 in skin CD4+Vβ4+ T cells from 5-6mo mice, indicating their enhanced proliferation in Sox13-/- mice. n = 3 from 1 of 2 similar experiments. *, p<0.05 by Student’s t-test.
Videos
Comparative Scratching behavior of Sox13-/- and WT control (tails painted solid white) mice.
Isolated scratching episode typical of Sox13-/- mice.
Tables
PCR Primers used in this study.
| Sequence | F/R | Description |
|---|---|---|
| CCTGGACTCTCCACCGCAA | F | Il17a |
| TTCCCTCCGCATTGACACAG | R | Il17a |
| TTTCCTGTCTGTATTGAGAAACCT | F | Il33 |
| TATTTTGCAAGGCGGGACCA | R | Il33 |
| CGCTTGAGTCGGCAAAGAAAT | F | Il1a |
| TGGCAGAACTGTAGTCTTCGT | R | Il1a |
| GCCACCTTTTGACAGTGATGAG | F | Il1b |
| GACAGCCCAGGTCAAAGGTT | R | Il1b |
| TCCTCTCTGCAAGAGACTTCC | F | Il6 |
| TTGTGAAGTAGGGAAGGCCG | R | Il6 |
| AGCTGTAGTTTTTGTCACCAAGC | F | Ccl2 |
| GTGCTGAAGACCTTAGGGCA | R | Ccl2 |
| TCACAGCAACGAAGAACACCA | F | Il4 |
| CAGGCATCGAAAAGCCCGAA | R | Il4 |
| CAAGCAATGAGACGATGAGGC | F | Il5 |
| GCATTTCCACAGTACCCCCA | R | Il5 |
| CACTACGGTCTCCAGCCTCC | F | Il13 |
| CCAGGGATGGTCTCTCCTCA | R | Il13 |
| CACCAGCGGGACATATGAATCT | F | Il23a |
| CACTGGATACGGGGCACATT | R | Il23a |
| TTGAGGTGTCCAACTTCCAGCA | F | Il22 |
| AGCCGGACGTCTGTGTTGTTA | R | Il22 |
| AGAGTTTGATCCTGGCTCAG | F | 16S V1 Universal Primer 27F |
| ATTACCGCGGCTGCTGG | R | 16S V3 Universal Primer 534R |
| AAGCCTGATGACTCGGCCACA | F | Vb4 TCR deep seq |
| CTTGGGTGGAGTCACATTTCTCAGATCCTC | R | Cbeta TCR deep seq |
| AACTGTACTTATTCAACCACA | F | Va4 TCR deep seq |
| CTGTGAACTGTTCCTATGAAACC | F | Va4 TCR deep seq |
| TAAACTGTACTTATTCAACCACA | F | Va4 TCR deep seq |
| CCTGATAATAAATTGCACGTATTCA | F | Va4 TCR deep seq |
| GGTACACAGCAGGTTCTGGGTTCTGGATG | R | Calpha TCR deep seq |





