Biallelic TANGO1 mutations cause a novel syndromal disease due to hampered cellular collagen secretion
Figures
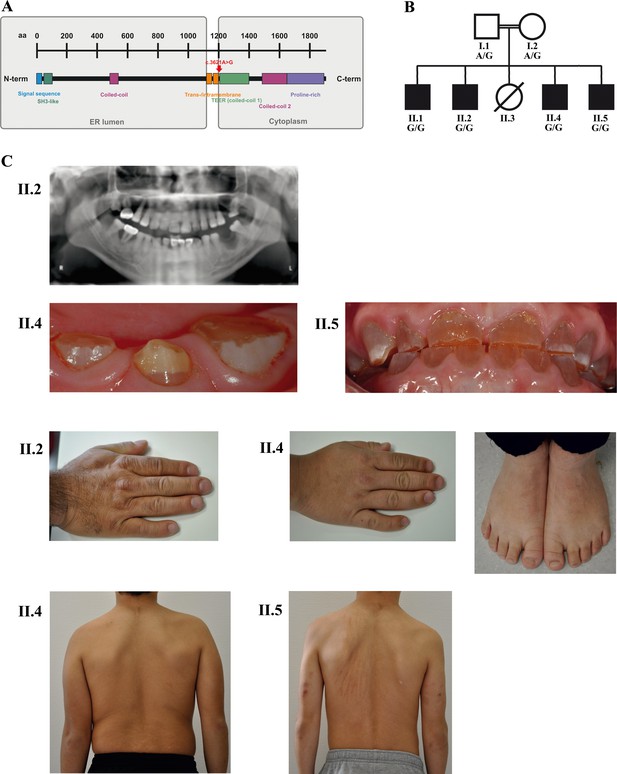
A novel syndrome caused by biallelic TANGO1 mutations in a consanguineous family.
(A) Structure of TANGO1 protein. The lumenal portion contains an N-terminal signal sequence followed by an SH3-like domain required for cargo binding, as well as a coiled-coil domain. A trans- and intramembrane domain anchors TANGO1 within the ER membrane. The cytoplasmic portion consists of two coiled-coil domains (CC1, also named TEER, and CC2) and a proline-rich domain at the C-terminus. The identified mutation affects residue 1207 (p.(Arg1207=)) between the intramembrane and the CC1 domain at the beginning of the cytoplasmic portion. (B) Pedigree of the studied family. Filled or clear symbols represent affected or unaffected individuals, respectively. The parents (I.1 and I.2) are first cousins. The four affected sons (II.1, II.2, II.4, and II.5) share a homozygous TANGO1 (c.3621A > G) variant. The healthy child II.3 died in a household accident at the age of 16. (C) Dental and skeletal abnormalities of the affected brothers II.2, II.4, and II.5. Note the brachydactyly of hands and feet, clinodactyly of the fifth finger, dentinogenesis imperfecta (including an opalescent tooth discoloration with severe attrition affecting the primary and permanent dentition, as well as juvenile periodontitis, bulbous crowns, long and tapered roots, and obliteration of the pulp chamber and canals in the permanent dentition), the skin lesions due to pruritus in all affected children; and the scoliosis in II.4 and II.5.
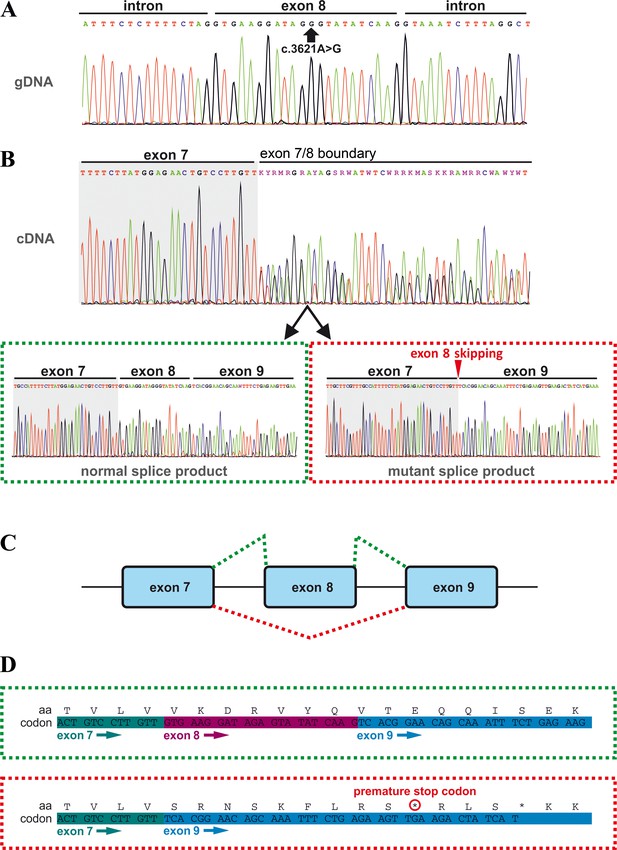
Effects of the TANGO1 (c.3621A > G) mutation on pre-mRNA splicing.
(A) The synonymous variant, which was identified by WES and validated by Sanger sequencing in all family members, resides in exon 8 of TANGO1 at genomic position 222,822,182 (GRCh37/hg19). It is predicted to disrupt an exon splice enhancer (ESE) motif recognized by the human SR protein SC35. (B) Electropherograms of the TANGO1 cDNA sequence of one affected child. Note the splitting of the sequence starting at the exon 7/8 boundary. Sequencing of individual bands after gel electrophoretic separation revealed TANGO1 wild-type cDNA and cDNA lacking exon 8 (c.3610_3631delins30). For the cDNA sequencing of TANGO1 splice products, at least two (up to 6) technical replicates were performed for each family member. (C) Schematic representation of the alternatively used splice sites resulting in the normal TANGO1 mRNA (green dotted lines) and in exon eight skipping (red dotted lines). (D) Consequences of TANGO1 exon eight skipping on the reading frame and the amino acid level. Exclusion of exon eight causes a premature stop codon.
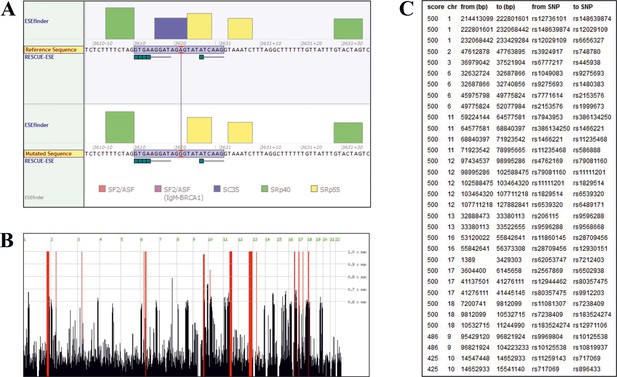
Sequence analysis.
(A) Splicing of TANGO1 exon 8, as predicted by Alamut Visual. The window shows the binding sites of various SR proteins (color code at the bottom) required for correct splicing. The height of the boxes reflects the probability of binding. The upper diagram shows the WT, the lower half the mutated sequence. The mutation is predicted to disrupt the consensus sequence for the SR protein SC35. (B) Homozygous intervals (red bars) shared by patients II.1 and II.2 of the investigated family. The identified TANGO1 mutation lies within a ~ 19 Mb homozygous interval on chromosome 1. (C) Genomic localisation (GRCh37/hg19) of homozygous intervals, detected by Homozygosity Mapper.
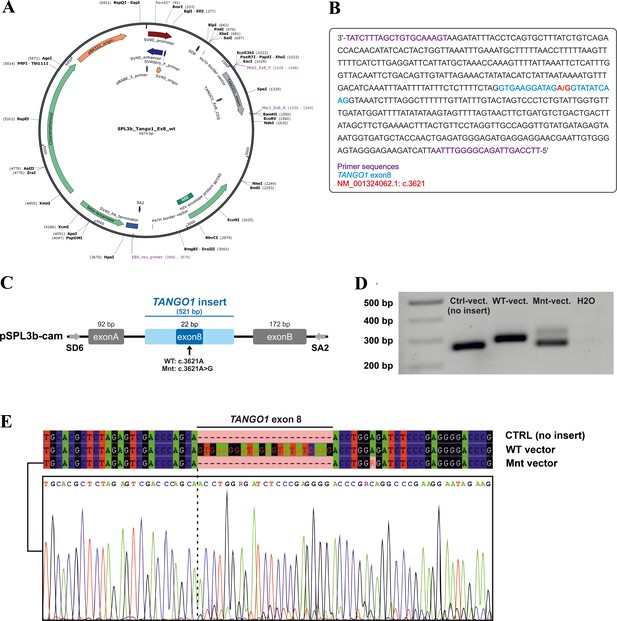
Minigene assay.
(A) Map of the pSPL3b-cam vector. (B) Sequence of the 521 bp amplicon (TANGO1 exon eight and ~250 bp flanking intronic sequences) inserted into pSPL3b-cam. (C) Vector constructs for the minigene assay. An amplicon containing either wild-type (WT vector) or mutated exon 8 (Mnt vector) was inserted between the vector-specific exons A and B into pSPL3b-cam. (D) Gel electrophoresis of cDNA PCR products from HEK293T cells transfected with either WT, Mnt, or CTRL vector (without insert). Transfection with the CTRL and WT vector resulted in cDNA PCR products of 264 bp and 286 bp, respectively. Cells transfected with the Mnt vector produced two splice products. (E) Sanger cDNA sequencing of the CTRL-, WT-, and Mnt-vector splice products. The Mnt vector yielded two separate TANGO1 splice products, the majority of which did not contain exon 8, indicative of exon skipping and a small portion of correctly spliced products. Following TA cloning of individual cDNA molecules, 15 of 22 (68%) splice products lacked and 7 (32%) contained exon 8.
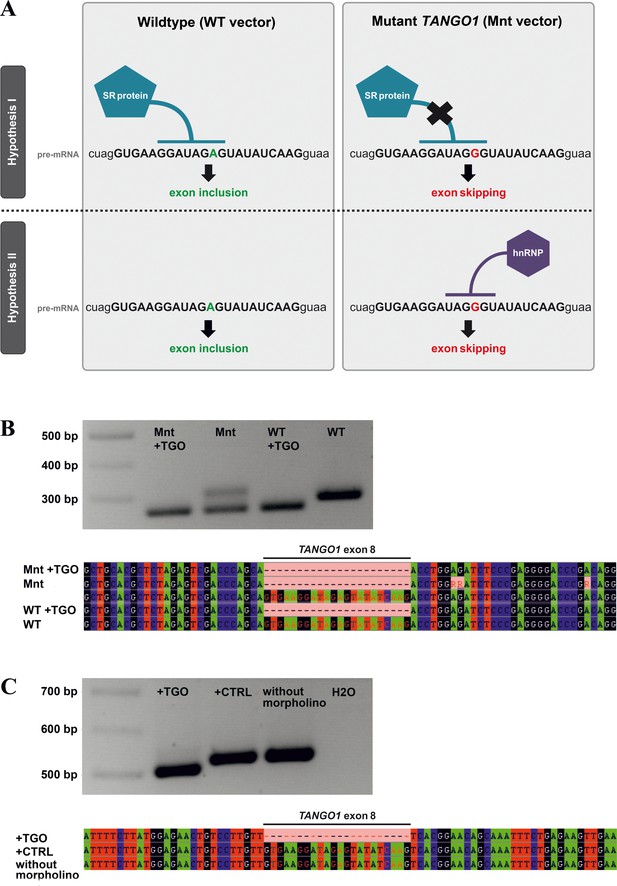
Possible mechanisms underlying TANGO1 exon eight skipping.
(A) Exon skipping during pre-mRNA splicing could be due to disruption of an ESE motif, which prevents the human SR protein SC35 from binding (hypothesis I). On the other hand, the mutation creates a sequence motif (UAGGGU) that is recognised by hnRNPA1, which may repress splicing (hypothesis II). Most morpholinos alter splicing by sterically blocking the snRNP binding sites utilised by the spliceosome, which are usually located in the intron near the splice donor/acceptor junctions. The TGO morpholino used here targets the entire TANGO1 exon eight but not enough intronic sequences to block snRNP binding sites. It rather obstructs the consensus sequence for SC35 binding. If disruption of a splice enhancer motif is the underlying mechanism, TGO treatment of WT-transfected cells should also prevent SC35 binding and thus induce exon eight skipping. If a splice inhibitor is recruited by the mutated sequence, TGO treatment of WT-transfected cells should not affect TANGO1 exon eight splicing. In cells expressing the mutation from a transfected vector, addition of the TGO morpholino would prevent the binding of hnRNPA1 and induce exon eight skipping. (B) Effects of the TANGO1 exon eight morpholino (TGO) on HeLa cells transfected with mutated TANGO1 exon 8 (Mnt vector) or wild-type (WT vector). To discriminate between vector-derived and endogenous splice products, vector-specific primers were used for cDNA sequencing. The vector-derived splice products of TGO treated HeLa cells transfected with either mutated (Mnt) or wild-type (WT) TANGO1 both showed exon skipping. HeLa cells transfected with the Mnt vector but not with TGO were endowed with two splice products, one lacking the entire TANGO1 exon eight and one presenting the normal cDNA. Cells transfected with the WT vector but not with TGO demonstrated only the normal TANGO1 splice product. (C) To prove that the exon skipping effect is not caused by morpholino treatment alone, HeLa cells were either treated with TGO or a standard control morpholino (CTRL). As expected, cDNA from TGO-treated cells lacked TANGO1 exon 8, whereas cDNAs from CTRL-treated or untreated cells included exon 8. Collectively, these results suggest that the identified TANGO1 mutation leads to the disruption of an exon splice enhancer, which prevents SC35 binding.
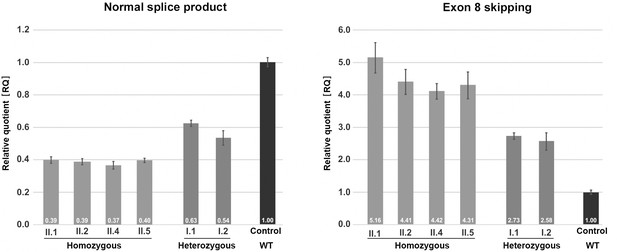
Quantification of TANGO1 splice products in homozygous and heterozygous mutation carriers, compared to a control individual (without mutation).
The right bar diagram shows the relative amounts of the normally spliced TANGO1 cDNA and the left diagram of splice products lacking exon 8. The standard deviation of each bar represents the results of triplicate measurements. A control cDNA sample was used for normalisation and relative comparison (RQ = 1). By qRT-PCR the control sample used was representative for three other control individuals.
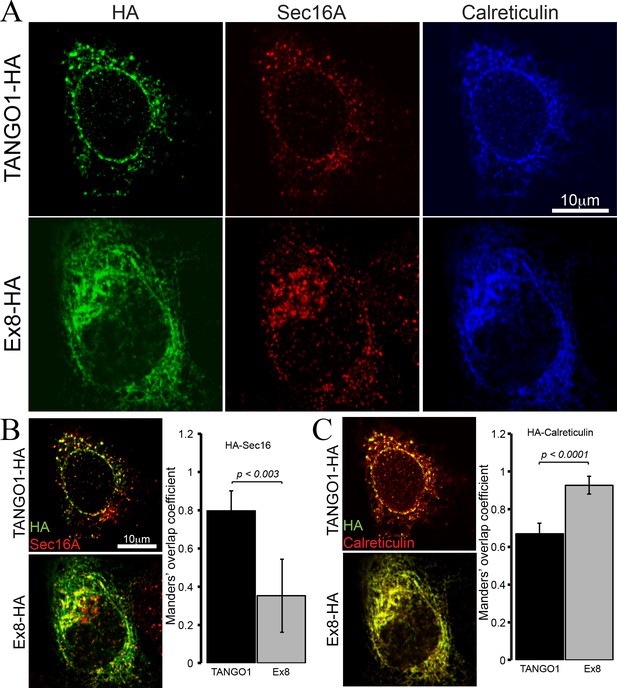
Ex8 mutant does not localise to ER exit sites.
Immunofluorescence images of U2OS cells, transiently transfected with WT TANGO1-HA or Ex8-HA. Representative images of three independent experiments. (A) Cells were probed with anti-HA (green), anti-Sec16A (red) and anti-Calreticulin (blue) antibodies. Scale bar 10 μm. (B) Merged images of TANGO1-HA or Ex8-HA (green) and sec16A (red) and a plot comparing Manders’ overlap coefficient of HA with sec16A in TANGO1- or Ex8-expressing U2OS cells. (C) Merged images of TANGO1-HA or Ex8-HA (green) with calreticulin (red) and a plot comparing Manders’ overlap coefficient of HA and calreticulin in TANGO1- or Ex8-expressing U2OS cells. Student’s t test was performed to compare the Manders’ overlap coefficients, p values are shown.
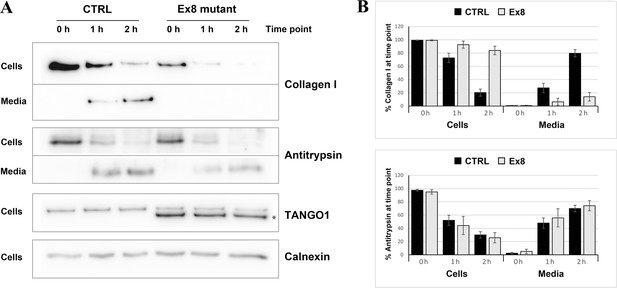
Ex8 mutant expression reduces collagen I secretion in U2OS cells.
(A) Media of U2OS control cells (CTRL) or stably expressing Ex8-HA mutant (Ex8) were replaced with OptiMEM media containing 0.25 mM ascorbic acid and 50 μM cycloheximide to block protein synthesis and follow collagen secretion. Cell extracts and media were collected at the indicated time points and analysed by SDS-PAGE followed by Western blotting with antibodies raised against Collagen I, TANGO1, and Calnexin (loading control). (*) indicates Ex8-HA. Representative images of four independent experiments. (B) For each time point, the band intensities of collagen I (upper panel) or antitrypsin (lower panel) were measured for the cell extract and media samples and expressed as percentage of the total (cells plus media). Each graph represents the average quantification of four experiments and corresponding standard deviations.
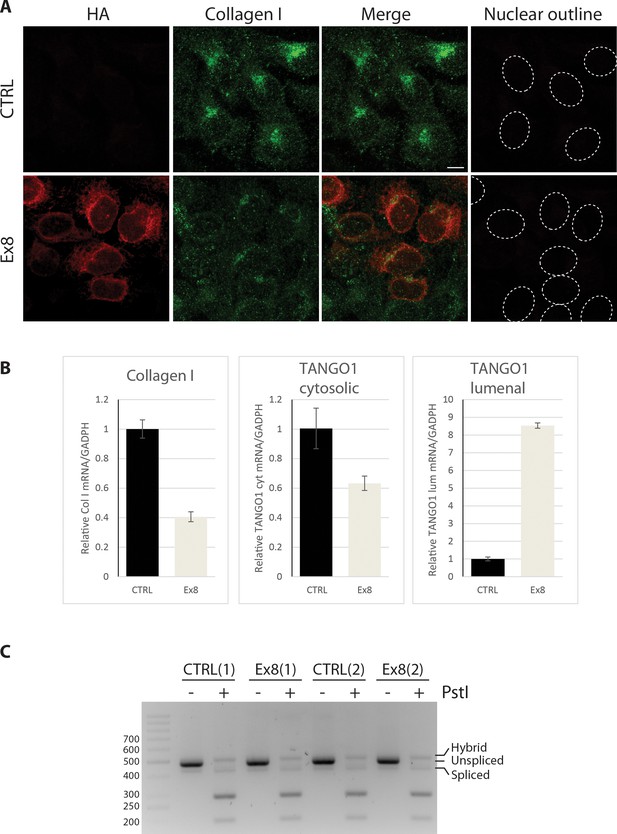
TANGO1 exon eight mutant (Ex8) expression reduces collagen I expression in U2OS cells without inducing UPR.
(A) Immunofluorescence Z-stack projections of control or Ex8-HA expressing U2OS cells, probed with anti-HA antibody (red) and anti-Collagen I antibody (green). Nuclear borders were traced from DIC images. Scale bar = 10 μm. Representative images of three independent experiments. (B) RNA levels from control or Ex8-HA expressing U2OS cells normalised by GAPDH values. Primers were designed to amplify two different portions of TANGO1 mRNA corresponding to the cytosolic portion (to quantify mRNA of endogenous protein only) or to the lumenal portion of TANGO1 protein (to quantify mRNA of endogenous TANGO1 and overexpressed Ex8-HA). Collagen I primers were specific for mRNA encoding for pro-α1(I) chain. Two samples of WT and Ex8-HA cells were used for RNA extraction. From each sample three technical replicas were used for qPCR quantification. Graphs show the average relative mRNA quantifications and standard deviations. (C) To test UPR activation, XBP-1 mRNA splicing was analyzed by PCR. PCR products were subjected to PstI restriction digestion, to specifically digest unspliced XBP-1, and separated in 3% agarose gel alongside undigested control products. Fragments corresponding to spliced (processed) products were not affected, since the PstI restriction site is lost after mRNA splicing. The results show that expression of Ex8-HA does not change the level of processed/spliced XBP-1 mRNA compared to control cells. Shown are two independent experiments.
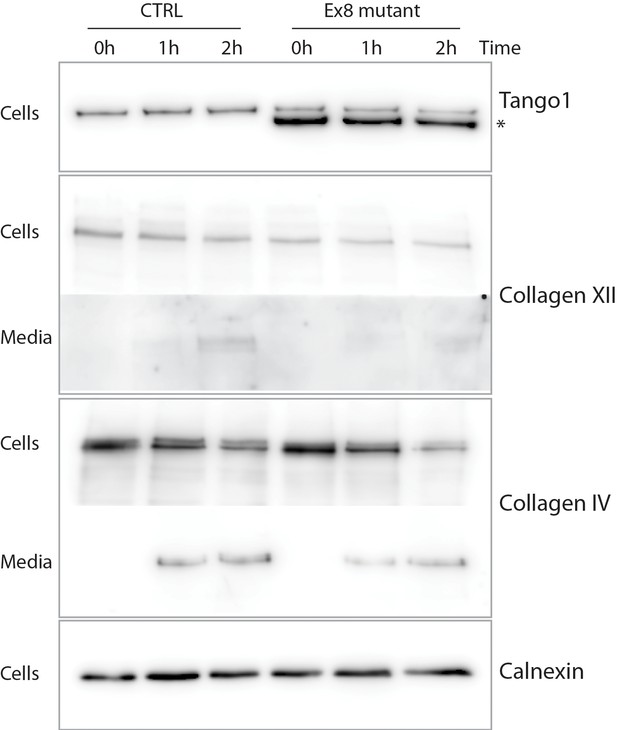
Ex8 mutant expression reduces collagen XII and collagen IV secretion in U2OS cells.
Media of U2OS control cells (CTRL) or stably expressing Ex8-HA mutant (Ex8) were replaced with OptiMEM media containing 0.25 mM ascorbic acid and 50 μM cycloheximide to block protein synthesis and follow collagen secretion. Cell extracts and media were collected at the indicated time points and analysed by SDS-PAGE followed by Western blotting with antibodies raised against collagen XII or collagen IV, TANGO1, and calnexin (loading control). For panels corresponding to media samples, longer exposures are showed compared to cell sample panels, due to low levels of secreted collagen IV and XII. (*) indicates Ex8-HA.
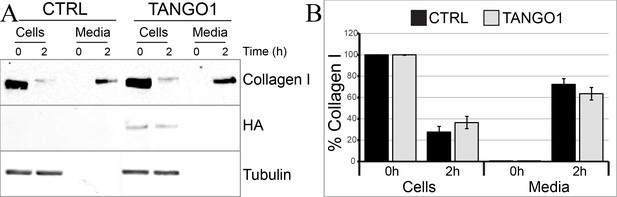
TANGO1 full-length overexpression has no effect on collagen I secretion in U2OS cells.
(A) Media of U2OS control cells (CTRL) or stably expressing TANGO1-HA were replaced with OptiMEM media containing 0.25 mM ascorbic acid and 50 μM cycloheximide to block protein synthesis and follow collagen secretion. Cell extracts and media were collected at the indicated time points and analysed by SDS-PAGE followed by Western blotting with antibodies raised against collagen I, HA, and beta-tubulin (loading control). Representative images of five independent experiments. (B) For each time point, the band intensities of collagen I were measured for the cell extract and media samples and expressed as percentage of the total (cells plus media). Each graph represents the average quantification of five experiments and corresponding standard deviations.
Tables
Clinical symptoms in four affected brothers
| II.1 | II.2 | II.4 | II.5 | |
|---|---|---|---|---|
| Dentinogenesis imperfecta | x | x | x | x |
| Delayed eruption of permanent teeth | x | x | x | x |
| Juvenile periodontitis with early tooth loss | x | x | ||
| Growth retardation | x | x | x | x |
| Proportionate short stature | x | x | x | x |
| High nasal bridge | x | x | x | x |
| Retrognathia | x | |||
| Phalangeal brachydactyly of fingers | x | x | x | x |
| Clinodactyly of 5th finger | x | x | x | x |
| Cone-shaped epiphyses in the hands | x | x | x | |
| Brachydactyly of toes | x | x | ||
| Platyspondyly (flattened vertebral corpora) | x | x | x | x |
| Scoliosis | x | x | ||
| Prominent knees | x | x | x | x |
| Mild intellectual disability | x | x | x | x |
| Sensorineural hearing loss | x | x | x | x |
| Mild retinopathy | x | x | ||
| Insulin-dependent diabetes mellitus | x | x | x | x |
| Primary obesity | x | x | x | x |
| Early onset puberty | x | |||
| Pruritus | x | x | x | x |
| Asthma | x | x | x | x |
| Osteopenia | x | x | ||
| Hydronephrosis (junctional stenosis) | x | |||
| Nephropathy (microalbuminuria) | x |
Additional files
-
Supplementary file 1
Whole Exon Sequencing (WES) was performed in the four affected bothers and their parents.
Shown are the 10 variants found to be homozygous in all affected children and heterozygous in both parents.
- https://cdn.elifesciences.org/articles/51319/elife-51319-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51319/elife-51319-transrepform-v3.docx





