Structural basis of substrate recognition by a polypeptide processing and secretion transporter
Figures
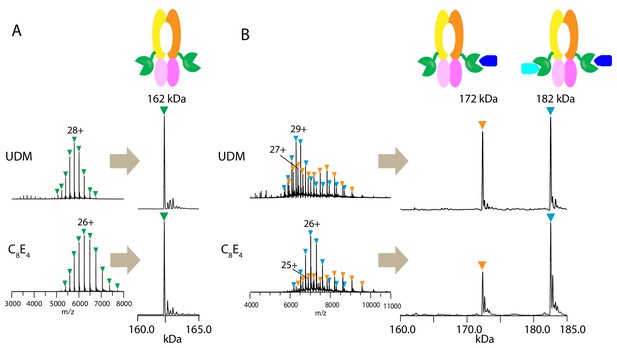
Native mass spectrometry analysis of wt PCAT1 and PCAT1(C21A)-CtA complexes.
(A) wt PCAT1 and (B) PCAT1(C21A)-CtA complex. The samples were analyzed in 200 mM ammonium acetate containing either UDM or C8E4 at 2X critical micelle concentration (CMC). The peak series for the protein complex were at lower charge states in C8E4 (charged-reduced) than in UDM, consistent with previous native MS results of other membrane protein complexes (Reading et al., 2015). Note that the peak intensities for the 172 kDa complex in the deconvoluted spectra in (B) includes both the complex present in-solution and the subcomplex resulting from the gas-phase dissociation of the 182 kDa complex upon collision activation (for estimation of the % relative abundances of the complexes with correction for gas-phase dissociation, see Figure 1—figure supplement 2).
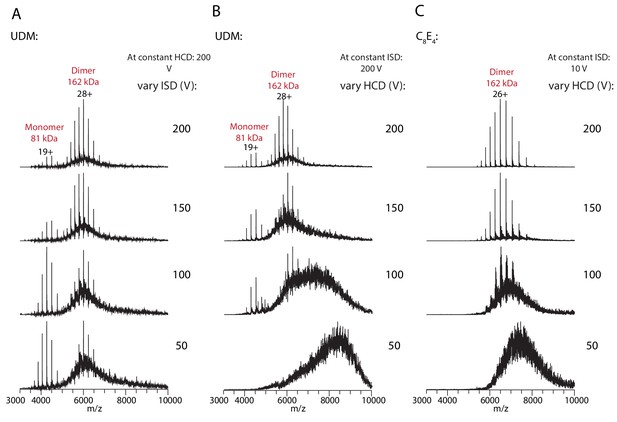
Native MS analysis of the wt PCAT1 at varying activation energies.
Native MS spectra for PCAT1 with CtA in UDM with variable (A) in-source dissociation (ISD) and (B) higher-energy collisional dissociation (HCD), and for C21A-PCAT1 in C8E4 with variable (C) HCD. The activation parameters were screened to determine the minimal collision activation energy required for detergent clean-up that leads to a well-resolved, near-baseline spectrum without dissociating the protein complex in the gas phase. Higher collision activation energies were needed for optimal native MS analysis of samples in UDM (ISD: 150–200 V and HCD: 150–200 V) than in C8E4 (ISD: 10 V and HCD: 150–200 V). Consequently, a peak series for the 81 kDa PCAT1 monomer can be observed at the lower m/z range for samples in UDM but not in C8E4.
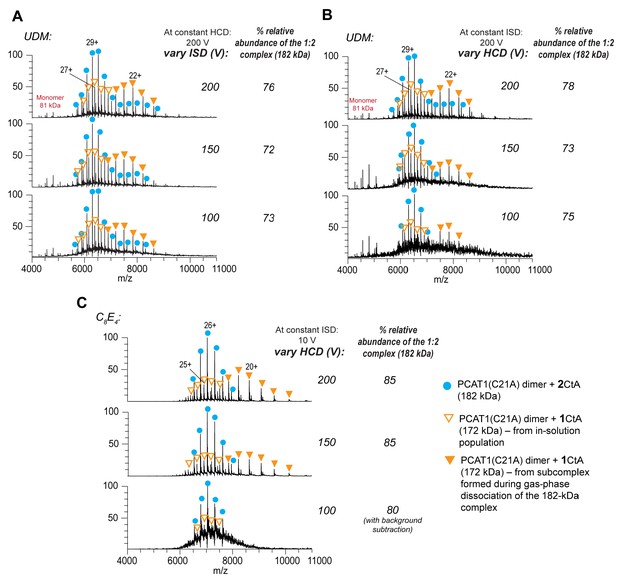
Native MS analysis of the PCAT1(C21A) transporter in complex with CtA at varying activation energies.
Native MS spectra for PCAT1(C21A) with CtA in UDM with variable (A) ISD at constant HCD of 200 V, (B) HCD at constant ISD of 200 V, and (C) HCD at constant ISD of 10 V for samples in C8E4. The predominant peak series in all the spectra has a mass of 182 kDa (blue filled circles) corresponding to the PCAT(C21A) dimer + 2 CtA complex (1:2 complex). Note that most spectra had well-resolved, near-baseline spectra at various activation energy settings due to the removal of the detergent background. However, these parameters also led to gas-phase activation of the 182 kDa complex and subsequent ejection of one of the bound CtA yielding a charge-stripped subcomplex (172 kDa, PCAT(C21) dimer + 1 CtA complex). Hence, the charge-state series for the 172 kDa complex exhibits a bimodal distribution. The subgroup with higher charge states corresponds to a population that is present in-solution (hollow triangles) and another subgroup with lower charge states (filled triangles) corresponds to the population from gas-phase dissociation of the 182 kDa complex. To estimate the relative abundance of the 1:2 complex in the sample, the peak intensities for the 182 kDa complex and from the charge-stripped 172 kDa subcomplex (at z = 20+ to 24+ for UDM and z = 18+ to 22+ for C8E4) were combined and divided by the total peak intensities for all of the PCAT1 assemblies within each MS spectrum. The ionization and transmission efficiencies as well as MS response factors for the observed PCAT1 complexes might differ. Overall, the calculated relative abundance for the 182 kDa complex remained consistent for all samples within each detergent type at the activation energies that were tested. The lower abundance of the 182 kDa complex in UDM relative to C8E4 is mainly due to the higher collision activation energy settings required to remove the UDM detergent from the complex (i.e., 100–200 V ISD in UDM versus 10 V ISD in C8E4). When indicated, background subtraction was performed using the UniDec software with curved background and smoothing correction.
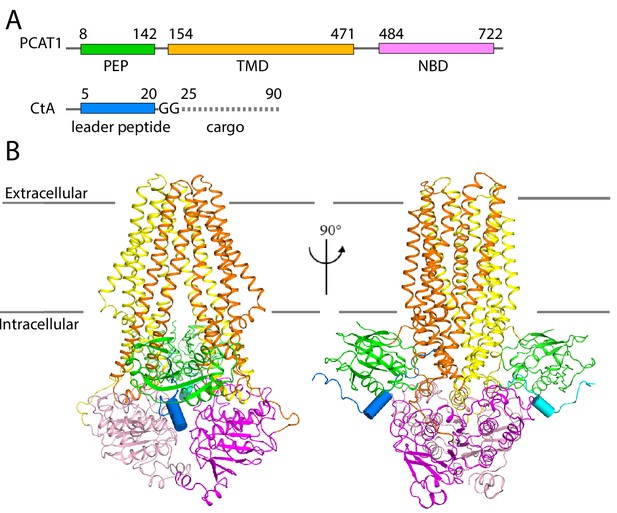
The cryo-EM structure of PCAT1-CtA complex.
(A) Cartoon illustration depicting domain organization of PCAT1 and its substrate, CtA. The symbol GG denotes the double glycine motif. The dotted line represents the unstructured cargo region. (B) Two orthogonal views of the PCAT1-CtA complex structure, the leader peptide helix of CtA is shown as a cylinder. The cartoon is color-coded by domains. Blue, CtA; magenta, NBD; yellow, TMD; green, PEP.

Summary of image processing workflow.
(A) A representative cryo-EM micrograph. (B) Representative 2D classes. (C) Flowchart of cryo-EM reconstruction.
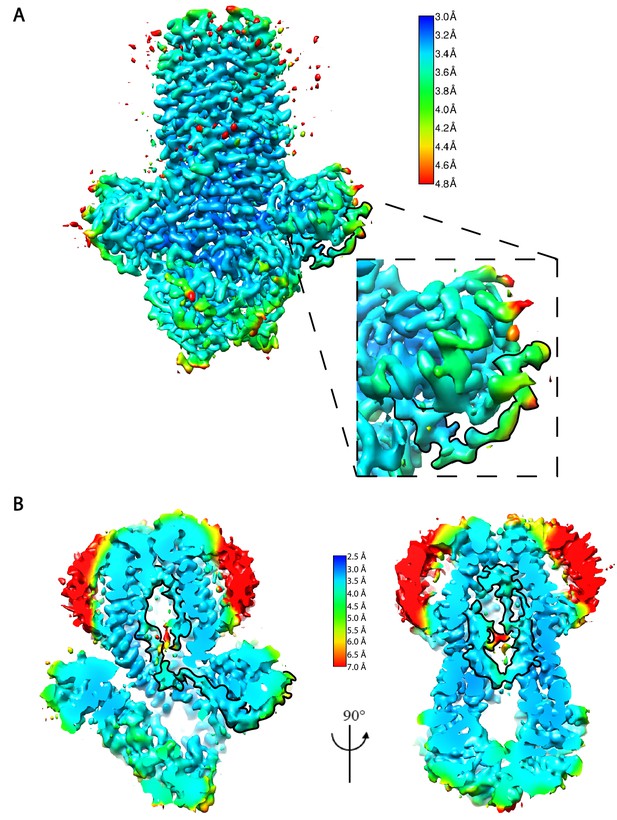
Local resolution estimation of the cryo-EM density map.
(A) The local resolution estimation of the sharpened map with a b-factor correction of −100 Å2. The map is contoured at 0.7 in Chimera. The box shows the magnified view of the substrate density highlighted in thick boundary. (B) Two orthogonal views of the unsharpened cryo-EM map shown at a crossed-section through the translocation pathway. The map is contoured at 0.3 in Chimera. The substrate density is highlighted in thick boundary.
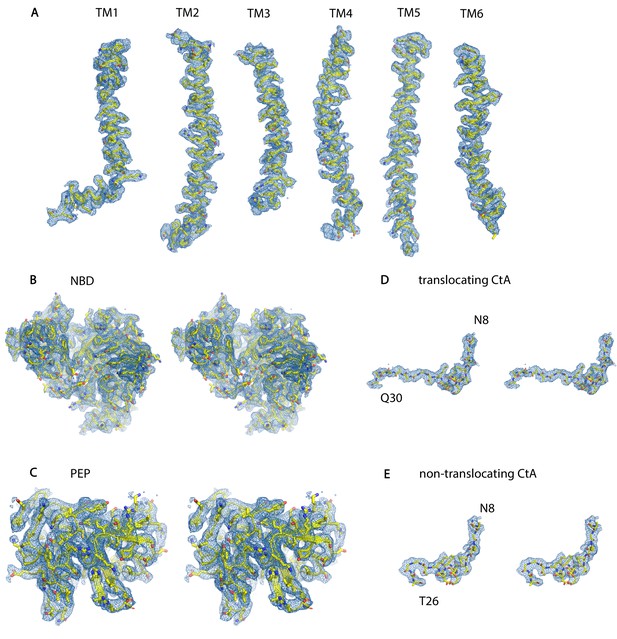
The cryo-EM density of different parts of the PCAT1-CtA complex.
Map shown in (A–C) was sharpened with a b-factor correction of −100 Å2, in (D) and (E) was sharpened with a b-factor of −50 Å2. All maps were generated in Chimera with a box size of 300 Å3. (A) TM helices, map contoured at 0.8. (B) Stereoview of the NBDs, map contoured at 0.7. (C) Stereoview of the PEP, map contoured at 0.6. (D–E) Stereoviews of the leader peptides, map contoured at 0.3.
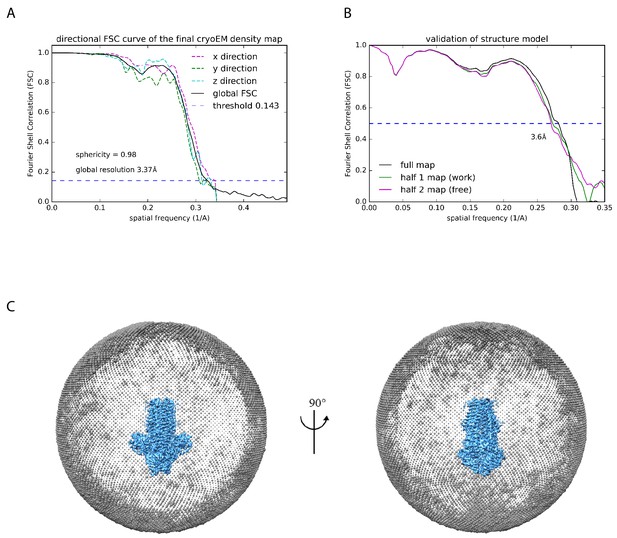
Resolution estimation, structure validation, and particle angular distribution.
(A) Anisotropy analysis using the 3D Fourier shell correlation (FSC) plot along with the global resolution estimation (Tan et al., 2017). (B) FSC curves between the refined structure and the half map used for refinement (working, green), the other half map (free, magenta), and the full map (black). (C) Angular distribution plot. Each sphere indicates the particles images oriented in that angle, and the size of the spheres corresponds to the relative number of particles in that angle.
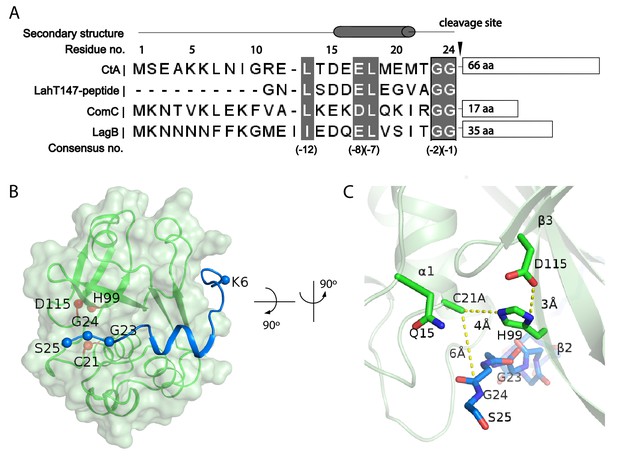
Substrate-PEP interaction.
(A) Sequence alignment of different leader peptides. Consensus residues are highlighted in grey. (B) Docking of the leader peptide (blue) on the PEP surface (green). The PEP catalytic triad residues are indicated as red spheres and the CtA double glycine motif are indicated as blue spheres. (C) A zoomed-in view of the catalytic site.
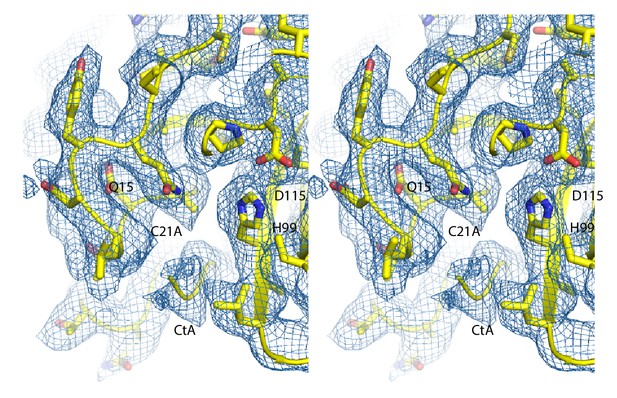
Stereoview of the catalytic site.
The cryo-EM map was sharpened with a b-factor of −100 Å2. The catalytic residues and CtA are labeled.
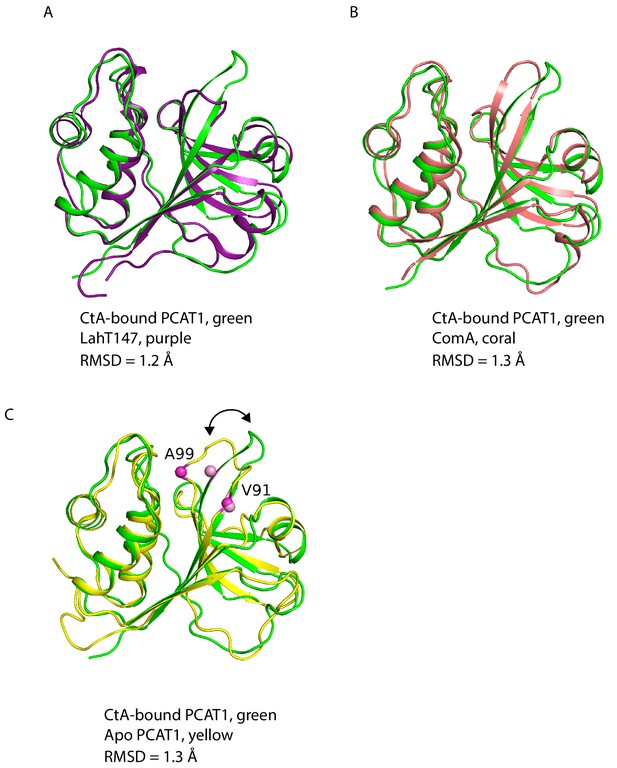
Comparison of PEP domains among PCATs.
Superpositions of CtA-bound PCAT1 (green) with (A) LahT147, (B) ComA, and (C) apo PCAT1. Residue V91 and A98 are indicated by spheres. The arrows indicate conformational change upon CtA binding. The root-mean square deviation (RMSD) of the backbone atoms are also shown.
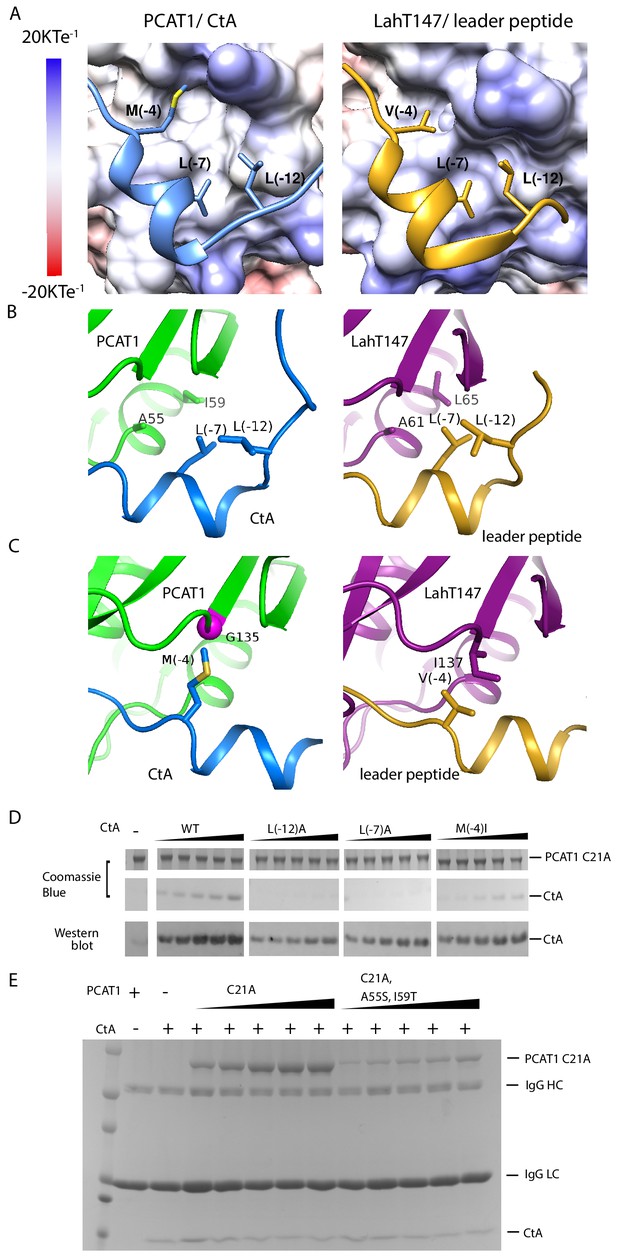
PCAT1 and LahT147 share common features in substrate recognition.
(A) Insertion of three hydrophobic residues onto a hydrophobic groove on the surface of the PEP domain, represented as electrostatic potential surfaces. CtA is shown in blue and the LahT147 substrate in gold. (B) A conserved hydrophobic knot. (C) Residue at position −4 of the substrate conferring specificity. (D) Pulldown of the wt or mutant CtA using PCAT1(C21A) conjugated resin. The CtA constructs contain a 3x Flag tag at their C-terminus. Western blot was performed using the anti-Flag M2 antibody. (E) Pulldown of PCAT1(C21A) using the M2 antibody resin against the Flag-tagged CtA. Mutation of hydrophobic residue A55 and I59 were introduced to PCAT1(C21A) background. IgG HC and IgG LC denote the heavy chain and the light chain of the M2 antibody, respectively.
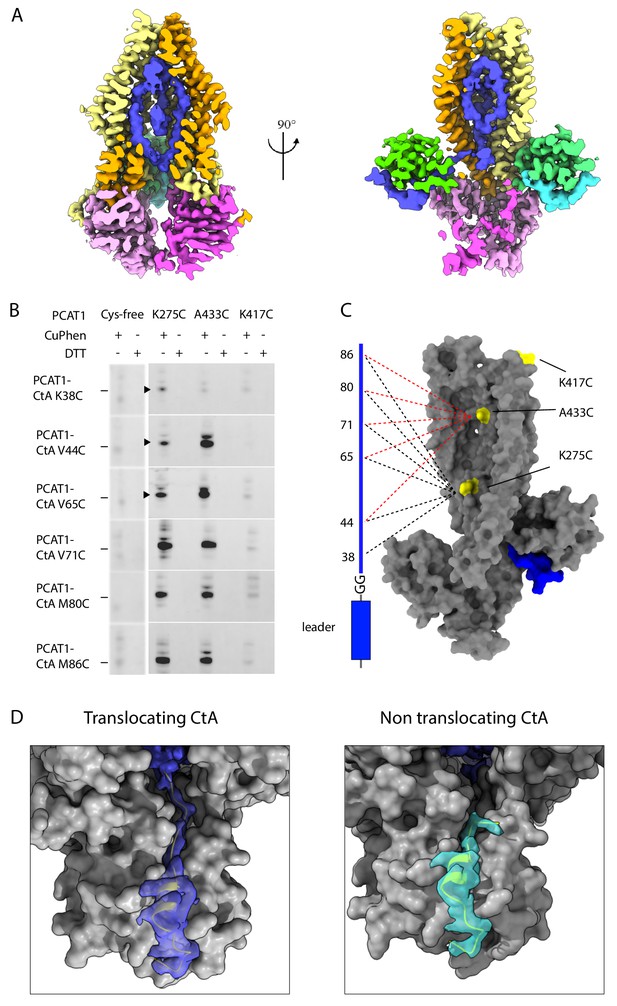
Positions of the two cargos.
(A) Two orthogonal views of the the translocation pathway. A composite cryo-EM map displays PCAT1 density contoured at 0.5 using Chimera, the CtA contoured at 0.24, and the density inside the translocation pathway contoured at 0.15. The cryo-EM density inside the TM cavity (shown in blue) likely corresponds to the C-terminal region of one CtA molecule. (B) Disulfide crosslinking experiments between PCAT1 and CtA. Crosslinked PCAT1-CtA products were detected by Western blot using an anti-HA antibody against HA-tagged CtA. Three bands (indicated by arrows) were excised from the SDS-PAGE and analyzed by mass spectrometry. In all three cases, peptide fragments with the correct disulfide bond were identified. (C) Summary of the crosslinking results. A PCAT1 monomer is shown as grey surface and CtA is represented by a cartoon. The dotted lines illustrate the crosslinked pairs (black, 275C crosslink pairs; red, 433C crosslinked pairs). (D) Comparison of the cryo-EM density at the two catalytic sites. The density is displayed as surface. Dark blue, the translocating CtA; light blue, the non-translocating CtA. The maps were contoured at 0.6 using Chimera. The TMD and PEP are shown as grey surfaces and the NBDs are omitted for clarity. The leader peptides are shown as ribbons.
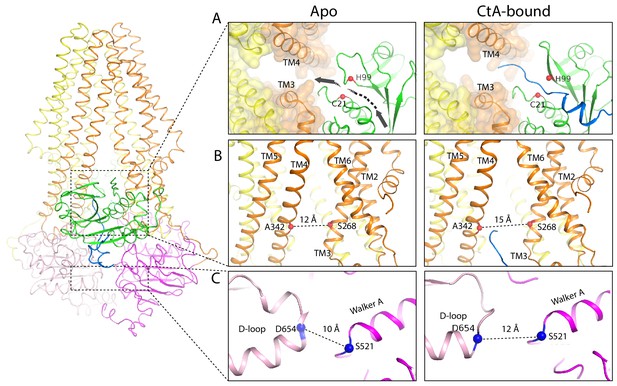
Conformational changes upon CtA binding.
(A,B) Widening of the lateral opening of the TM pathway. The arrow in the apo structure delineates the CtA binding groove. The catalytic residues C21 and H99 are labeled. The Cα distances between residues S268 and A342 that line the lateral opening are shown. (C) The NBDs are further apart in the CtA-bound structure. The Cα distances between S521 in the walker A motif and D654 in the D-loop of the other NBD are indicated.
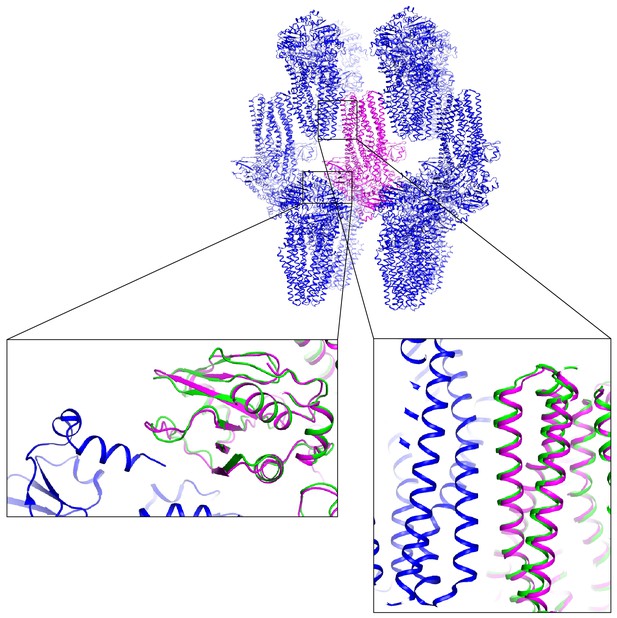
Crystal packing of apo PCAT1.
The crystal lattice of apo PCAT1 (PDBcode 4ry2, blue and magenta). In zoom-in views, the cryo-EM structure of PCAT1-CtA (green model) was superimposed on apo PCAT1, showing that regions making crystal contacts undergo little conformational changes upon CtA binding.
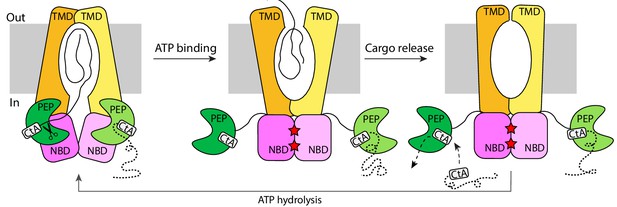
The alternating-access mechanism of PCAT1.
CtA is recruited and cleaved in the inward-facing conformation. ATP binding stabilizes the outward-facing conformation in which the PEP domains are disengaged. After cargo release, TMDs isomerize to form an occluded cavity. ATP hydrolysis resets PCAT1 back to the inward-facing conformation, allowing PEP to dock into the TMD-NBD interface.
Tables
Mass measurements from native MS analysis of PCAT1 samples.
| Sample | In UDM | In C8E4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected mass (Da) | Measured Mass ± SDa (Da) | Mass error (%)b | Measured Mass ± SDa (Da) | Mass error (%)b | |||||||
| wt PCAT1 dimer | 162,176 | 162,175 | ± | 1 | 0.0008 | - | 162,171 | ± | 1 | 0.003 | - |
| CtA | 10,207.9 | 10,206.5 | ± | 0.5 | 0.014 | 10,206.5 | ± | 0.3 | 0.014 | ||
| - | - | - | % Abun- dancec | - | - | % Abun- dancec | |||||
| C21A-PCAT1 dimer + 1 CtA | 172,320 | 172,337 | ± | 4 | 0.010 | 25 ± 2 | 172,341 | ± | 3 | 0.012 | 16 ± 2 |
| C21A-PCAT1 dimer + 2 CtA | 182,528 | 182,549 | ± | 3 | 0.012 | 75 ± 2 | 182,558 | ± | 6 | 0.017 | 84 ± 2 |
-
a From the average and S.D. of the mass values calculated from each charge state peak within a charge-state distribution (n ≥ 4 charge states).
b Mass accuracy measured by the relative difference between measured and expected masses divided by the expected mass.
-
c Based on relative peak intensities for the peak series corresponding to the 182 kDa and 172 kDa complexes within the same MS spectrum and corrected for gas-phase dissociation (see Figure 1—figure supplement 2 for more details) (n ≥ 3 conditions).
Summary of Cryo EM data and structure refinement statistics of PCAT1-CtA complex.
| Data collection | |
|---|---|
| Microscope | Titan Krios, 300 keV (FEI) |
| Detector | K2 summit direct electron detector (Gatan) |
| Energy Filter | 10 eV (Gatan) |
| Pixel size (Å) | 1.09 |
| number of movies | 3478 |
| Frames/movie | 60 |
| Total exposure time (s) | 12 |
| Exposure time per frame (s) | 0.2 |
| Total exposure (e/Å2) | 80.8 |
| Defocus range (µM) | 0.8 to 2.2 |
| Final reconstruction | |
| Number of particles | 133698 |
| B-factor correction (Å2) | −80 |
| RMS deviations | |
| bond length (Å) | 0.0028 |
| bond angles (°) | 1.2870 |
| Ramachandran | |
| favored (%) | 94.95 |
| Allowed (%) | 5.05 |
| Outlier (%) | 0.00 |
| Rotamer | |
| favored (%) | 87.30 |
| Allowed (%) | 9.73 |
| poor (%) | 2.97 |
| Reagent type | Designation | Source | Identifier | Additional information |
|---|---|---|---|---|
| Strain (Escherichia coli) | BL21 (DE3)-RIL | Agilent | #230280 | |
| Chemical compound | n-Dodecyl-β-D-Maltopyranoside, Sol-Grade | Anatrace | D310S | |
| Chemical compound | n-Undecyl-β-D-Maltopyranoside, Anagrade | Anatrace | U300 | |
| Chemical compound | C8E4 | Anatrace | T350 | |
| Chemical compound | Phenanthroline | Sigma-Alrich | 131377 | |
| Commercial assay or kit | M2 agarose affinity gel | Sigma-Alrich | A4596 | |
| Commercial assay or kit | Superdex 200 increase | GE Lifesciences | 28990944 | |
| Commercial assay or kit | Glutathione Sepharose 4B resin | GE Healthcare | 17075604 | |
| Commercial assay or kit | anti-HA mouse monoclonal antibody | Invitrogen RRID:AB_10978021 | 26183 | 1:10000 |
| Commercial assay or kit | goat anti-mouse Alexa Fluor 680 | Invitrogen RRID:AB_2535723 | A21057 | 1:1000 |
| Commercial assay or kit | M2 anti-flag mouse antibody | Sigma-Alrich | F 1804–50 UG | 1:5000 |
| Other | R1.2–1.3 400 mesh Au holey carbon grids | Quantifoil | 1210627 | |
| Software, algorithm | Leginon | RRID:SCR_016731 | http://emg.nysbc.org/redmine/projects/leginon/wiki/Leginon_Homepage | |
| Software, algorithm | Unblur | DOI:10.7554/eLife.06980 | http://grigoriefflab.janelia.org/unblur | |
| Software, algorithm | CryoSPARC | DOI: 10.1038/nmeth.4169 RRID:SCR_016501 | https://cryosparc.com/ | |
| Software, algorithm | CTFFIND4 | DOI: 10.1016/j.jsb.2015.08.008 RRID:SCR_016732 | http://grigoriefflab.janelia.org/ctf | |
| Software, algorithm | RELION | DOI: 10.7554/eLife.18722 DOI:10.7554/eLife.41266 RRID:SCR_016274 | http://www2.mrc-lmb.cam.ac.uk/relion | |
| Software, algorithm | gCTF | DOI: 10.1016/j.jsb.2015.11.003 RRID:SCR_016500 | https://www.mrc-lmb.cam.ac.uk/kzhang/ | |
| Software, algorithm | Gautomatch | https://www.mrc-lmb.cam.ac.uk/kzhang/ | ||
| Software, algorithm | alignparts_lmbfgs | DOI: 10.1016/j.jsb.2015.08.007 | https://sites.google.com/site/rubinsteingroup/direct-detector-align_lmbfgs | |
| Software, algorithm | FrealignX | DOI: 10.1016/bs.mie.2016.04.013 RRID:SCR_016733 | http://grigoriefflab.janelia.org/frealign | |
| Software, algorithm | COOT | DOI:10.1107/S0907444910007493 RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | PHENIX | DOI: 10.1107/S0907444909052925 RRID:SCR_014224 | https://www.phenix-online.org | |
| Software, algorithm | MolProbity | DOI: 10.1107/S0907444909042073;10.1093/nar/gkm216 RRID:SCR_014226 | http://molprobity.biochem.duke.edu | |
| Software, algorithm | Chimera | DOI: 10.1002/jcc.20084 RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera | |
| Software, algorithm | ChimeraX | DOI: 10.1002/pro.3235 RRID:SCR_015872 | https://www.https://www.rbvi.ucsf.edu/chimerax/ | |
| Software, algorithm | Pymol | PyMOL RRID:SCR_000305 | http://www.pymol.org | |
| Software, algorithm | APBS | DOI: 10.1093/nar/gkm276 RRID:SCR_008387 | http://www.poissonboltzmann.org | |
| Software, algorithm | 3V web server | DOI: 10.1093/nar/gkq395 | http://3vee.molmovdb.org/ | |
| Software, algorithm | UniDec v 3.2 | DOI: 10.1021/acs.analchem.5b00140 | https://github.com/michaelmarty/UniDec |





