Development of a confinable gene drive system in the human disease vector Aedes aegypti
Figures
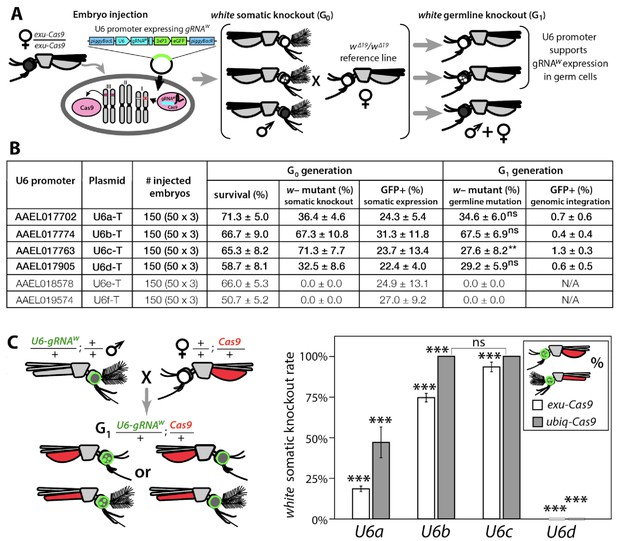
Functional identification of polymerase III promoters in Ae. aegypti.
(A) exu-Cas9 embryos were injected with one of 6 piggyBac plasmids, each utilizing a different U6 promoter expressing a guide RNA targeting the white eye pigmentation gene, gRNAw (Figure 1—figure supplement 2). The frequency of somatic white eye phenotypes in the resulting G0 progeny was used to assess promoter efficiency and to confirm gRNA target site accessibility. Germline mutagenesis rates were assessed by crossing G0 to white loss-of-function (w∆19/w∆19) lines to determine the white eye phenotype frequency in G1 progeny. (B). Two types of w– knockouts were observed: complete white eyes and mosaic white eyes. Out of 6 tested U6 promoters, four U6 promoters (U6a, U6b, U6c, and U6d) induced white knockout phenotypes. Statistical differences between germline and somatic mutation rates were estimated by equal variance t-test. (C) Transgenic males harboring piggyBac-integrated U6-gRNAW were outcrossed to either exu-Cas9 or ubiq-Cas9 females (left panel), and eye phenotypes were scored in G1 trans-heterozygous progeny (right panel, Supplementary file 1). Statistical differences in mutation knockout rates were estimated by equal variance t-test. (P ≥ 0.05ns, p<0.05*, p<0.01**, and p<0.001***).
-
Figure 1—source data 1
The metadata of the Aedes aegypti whole-genome sequences.
- https://cdn.elifesciences.org/articles/51701/elife-51701-fig1-data1-v1.xlsx
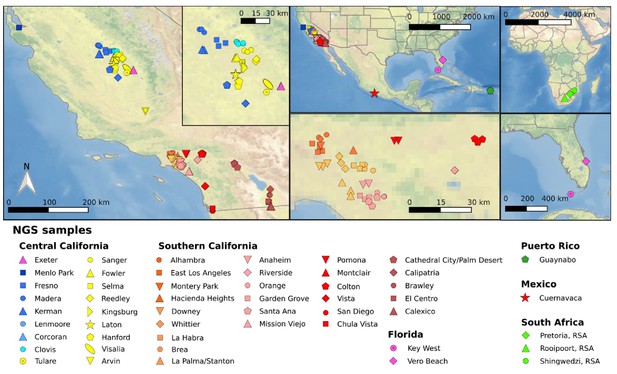
Sample locations of mosquitoes utilized for whole genome sequencing.
Left map depicts the location of all samples from California (CA) with the inset enlarging the Fresno/Clovis area. Top center shows a North American map including CA, Florida, and Cuernavaca, Mexico. Bottom central shows southern California near Greater Los Angeles area. Top right shows an African map with three locations in South African sample origin. Bottom right shows a Florida map with sampling locations. Each symbol represents a single individual mosquito. These plots were generated using CleanTOPO2 basemap (Patterson, 2008).
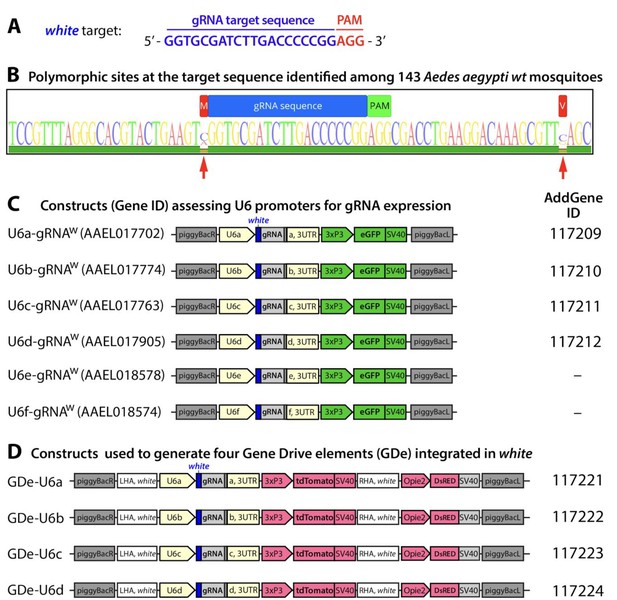
Guide RNA (gRNA) sequence and gene constructs developed in the study.
Nucleotide sequence for the gRNAw genomic target sequence in exon 3 of white gene with the PAM sequence indicated in red (A). Conservation of the RNAw genomic target sequence identified among 143 Ae. aegypti mosquitoes sampled from the field (B). Within the target sequence (AaegL5. 1:107,955,440–107,955,462, Vectorbase.org) (Dudchenko et al., 2017)(Matthews et al., 2018) no polymorphism sites were detected. The nearest polymorphic sites detected (red arrows) were −1 bp upstream (107,955,439) and +22 bp downstream (107,955,484). Schematic maps of six constructs utilized to functionally test U6 promoter activity in Ae. aegypti (C). Schematic maps of the four split drive elements (GDe) developed in this study. Plasmids generated in the study were deposited at Addgene.org and the corresponding ID numbers are listed.
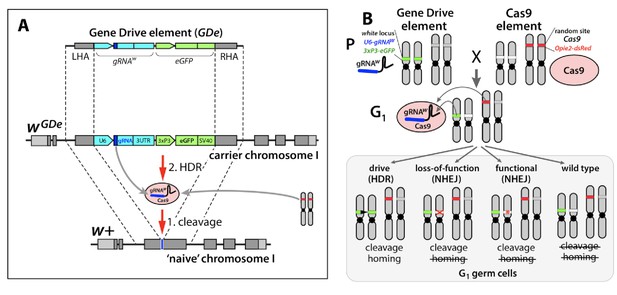
Design of the split-drive, a confined and high-threshold population replacement system.
To confine a gene drive’s propensity to spread, we separated the complete CRISPR/Cas9 gene drive system into two components: a Gene Drive element (GDe) comprising a gRNA and a fluorescent marker both flanked by Left and Right Homology Arms (LHA and RHA); and Cas9 endonuclease (Cas9). GDe integrated in a target gene, here it was white locus, can spread from the carrier chromosome into a naive chromosome in the presence of Cas9 transgene randomly integrated at a separate locus (A). First, Cas9/gRNA complex cuts the target site (cleavage); when GDe can be copied from the carrier chromosome into a naive chromosome via Homology Directed Repair (HDR). Each element is inactive on its own and can be maintained as a homozygous parental line (P). The cross between the homozygous lines results in 100% trans-heterozygous G1 progeny that carry both elements (B). gRNA expressed by GDe directs cleavage at white locus by Cas9, which can be repaired in three different ways: via HDR using GDe as a repair template and result in homing of GDe, and via Non-Homology End Joining (NHEJ) and lead to a loss-of-function mutant allele or an in-frame functional mutant allele. NHEJ-induced mutated alleles carry insertions/deletions (indels) at the cut site and become resistant to the same Cas9/gRNA system.
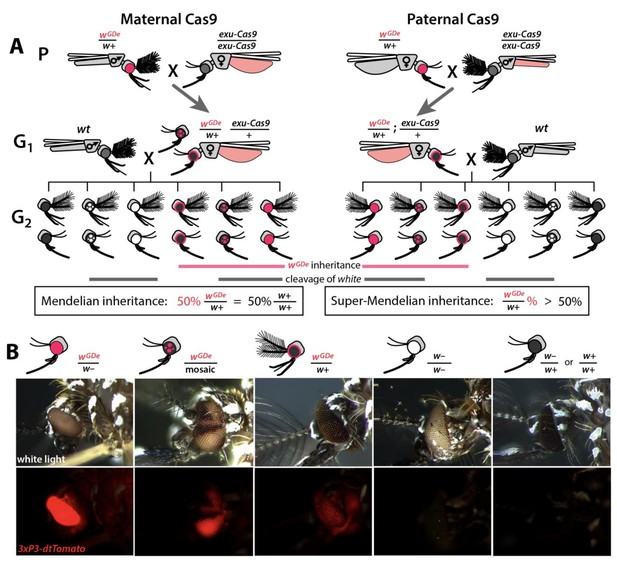
Assessing super-Mendelian inheritance of split drives.
(A) Crossing scheme of the wGDe and exu-Cas9 parent strains (P) to generate trans-heterozygotes (G1), and the outcrossing of their progeny to wild-type (wt) mosquitoes (G2). tdTomato eye and dsRed abdominal expression were the wGDe and exu-Cas9 transgene inheritance markers, respectively. wGDe transmission and white loss-of-function mutation rates were estimated among G2 progeny of trans-heterozygous female and wt male crosses. Super-Mendelian inheritance of wGDe occurred when the transmission rate of wGDe in G2 progeny was >50%, as expected by standard Mendelian inheritance. (B) Examples of G2 progeny eye phenotypes and corresponding genotypes.
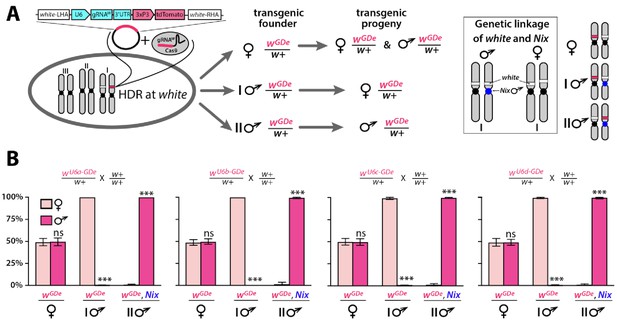
Sex biased inheritance of the GDe is due to linkage of white and Nix, a male determining gene in Ae. aegypti.
Schematic of site-specific integration and sex-linked inheritance of Gene Drive element (GDe) (A). To direct site-specific integration of GDe inside white locus (white stripe on chromosome I) via homology directed repair (HDR), wild type (wt) Ae. aegypti embryos were injected with a Cas9/gRNAw complex and a plasmid carrying GDe flanked by homology regions complementary to left and right genomic regions at the white cut site. GDe contains two genes: U6-gRNAw to direct a site-specific cleavage and the 3xP3-tdTomato transgenesis marker (red stripe). Female mosquitoes carrying wGDe/w+ passed wGDe randomly to both genders, while transgenic males transmitted wGDe either to females (type I) or males (type II) nearly exclusively. Close genetic linkage of white and Nix genes causes tight sex-linked inheritance of wGDe via male founders (B). Nix is located near the centromere of chromosome I (Dudchenko et al., 2017; Matthews et al., 2018) and consequently, type I male (♂) founders have wGDe inserted on the chromosome copy without Nix and therefore pass wGDe exclusively to female progeny. On the other hand, type II male founders have wGDe integrated on the chromosome with Nix, and thus they transfer wGDe and Nix together exclusively to male progeny (box). Bars show average ± SD estimated for 20 data points. Statistical significance between gender frequencies was estimated by an equal variance t test. (P ≥ 0.05ns and p<0.001***).
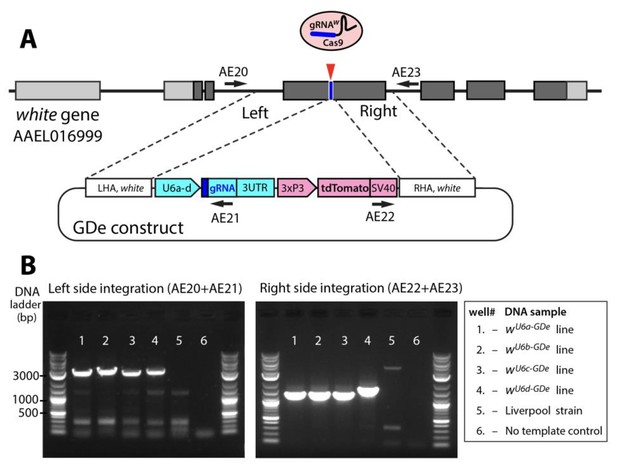
Site-specific integration of Gene Drive element (GDe).
Schematic showing positions of Left and Right Homology Arms (LHA and RHA) of GDe relative to genomic white sequence. The cut site is presented over the genetic structure of white, with darker gray boxes representing exonic coding sequences. GDe constructs were integrated at the cut site via homology directed repair (HDR) relying on the construct’s LHA and RHA to white. Arrows depict primers (AE20, AE21, AE22 and AE23) used for PCR and their location relative to genomic and constructs sequences. Two gel images showing specific PCR amplicons for each side of integration (B). The same DNA samples, listed in the box to the left, were used to PCR of both fragments. To confirm PCR specificity, amplicons were Sanger sequenced from both ends.
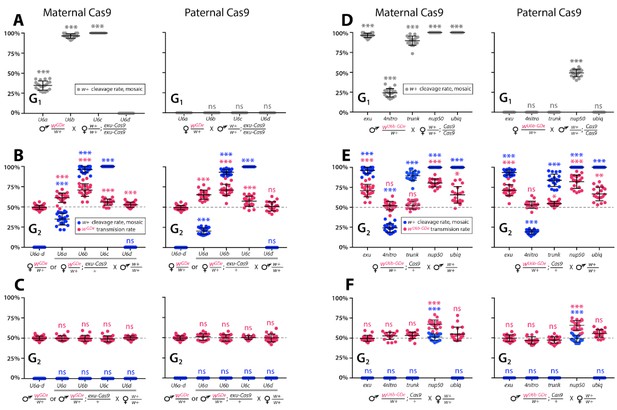
Timing and expression of drive components affect cleavage and transmission rates.
We bidirectionally crossed trans-heterozygous mosquitoes that contained both components of a gene drive: the gene drive element (GDe) and the Cas9 transgene (wGDe/w+; Cas9/+). (Figure 2A). (A–C) Four wGDe/w+ lines, each with a different gRNAw U6 promoter, were crossed to the exu-Cas9 strain to compare cleavage and homing activity. (A) Maternally deposited Cas9 protein induced white cleavage in G1 trans-heterozygotes harboring wU6a-GDe, wU6b-GDe, and wU6c-GDe but not wU6d-GDe. (B) In comparison to wGDe/w+, trans-heterozygous wGDe/w+; exu-Cas9/+ females, (C) but not males, exhibited super-Mendelian transmission of wGDe. The wU6b-GDe/w+; exu-Cas9/+ females transmitted wU6b-GDe to 70.9 ± 7.8% of G2 progeny. (D–F) Five lines expressing Cas9 from different promoters were crossed to wU6b-GDe/w+. (D) Maternally deposited Cas9 resulted in white cleavage in G1 trans-heterozygotes. (E) Three out of five tested Cas9 lines, exu-Cas9, nup50-Cas9, and ubiq-Cas9, induced super-Mendelian transmission of wGDe by trans-heterozygous females. The wU6b-GDe/w+; nup50-Cas9/+ females transmitted wU6b-GDe to 80.5 ± 5.0% of G2 progeny. (F) All trans-heterozygous males transmitted wGDe following a regular Mendelian inheritance except for the wU6b-GDe/w+; nup50-Cas9/+ males that induced white cleavage in 51.0 ± 3.9% and transmitted the wU6b-GDe allele to 66.9 ± 5.4% of G2 progeny. Point plots show the average ± standard deviation (SD) over 20 data points. Grey dotted line indicates standard Mendelian inheritance rates. Statistical significance was estimated using an equal variance t-test. (P ≥ 0.05ns, p<0.05*, p<0.01**, and p<0.001***).
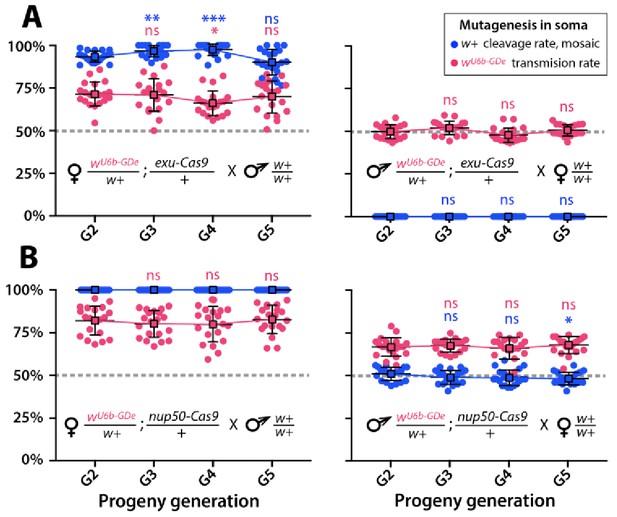
Split-drive over multiple generations.
Frequencies of white cleavage and wU6b-GDe transmission were plotted over multiple generations. The wU6b-GDe/w+; exu-Cas9/+ and wU6b-GDe/w+; nup50-Cas9/+ trans-heterozygous females, or males were outcrossed to wt each generation and both transmission and cleavage rates were scored. Average transmission rates at consecutive generations were compared to the corresponding rates at G2. Point plots show the average ± SD over 20 data points. Statistical significance was estimated using a t test with equal variance. (P ≥ 0.05ns, p<0.05*, and p<0.001***).

Sequences of de novo resistance alleles at white locus (wR).
Mosquitoes carrying resistance alleles (wR) were identified among progeny of genetic crosses between wU6b-gRNA/w+; nup50-Cas9/+ trans-heterozygous mosquitoes with wt (Supplementary file 6) and were further crossed to the Ae. aegypti loss-of-function w∆19/w∆19 line, to place novel knockout alleles (wR) into the recessive genetic background. To sample wR alleles from w∆19/wR heterozygous mosquitoes, PCR amplicons amplified from an individual mosquitoes were cloned into a plasmid, and a seven clones were Sanger sequenced in both directions and aligned against the w+ and w∆19 alleles in SnapGene 4.2. Genders and genotypes of analyzed mosquitoes as well as the generation when they were recovered are provided on the left side. Numbers of bases deleted is indicated below the corresponding deletion. Notably, while the deletions of 3 and 18 bases re-established the reading frame for white gene, they still displayed the loss-of-function phenotype.
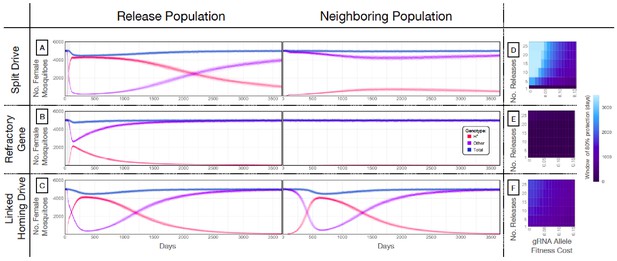
Mathematical model predictions for best performing split drive.
Model predictions for releases of Ae. aegypti mosquitoes homozygous for (A) the split drive system, (B) a disease-refractory gene, or (C) a homing drive system in which the components of the split drive system are linked at the same locus. Parameters correspond to those for the best performing split drive system (wU6b-GDe/w+; nup50-Cas9/+) (Supplementary file 7b). Releases are carried out in a population with an equilibrium size of 10,000 adults and a 1% per mosquito per generation migration rate with a neighboring population of the same equilibrium size. Model predictions were computed using 100 realizations of the stochastic implementation of the MGDrivE simulation framework (Sánchez et al., 2018). Weekly releases of 10,000 homozygous split drive males or the disease-refractory gene were simulated over a 10 week period, while a single release was simulated for the linked homing drive system. Total female population size (‘total’, dark blue), adult females with at least one copy of the disease-refractory allele (‘H*”, red), and disease-susceptible adult females without the disease-refractory allele (‘other’, purple) were plotted for each group. Notably, the split drive system is: i) largely confined to its release population, ii) reversible, and iii) present at a high frequency (>85% of adult females having at least one copy) for over three years. The split drive system outperforms inundative adult male release of the disease-refractory gene in population disease refractoriness and outperforms the confinability of a linked homing drive system. In the right column, heatmaps are shown for (D) the split drive system, (E) inundative releases of a disease-refractory gene, and (F) a linked homing drive system, and depict the window of protection in days that the proportion of mosquitoes in the release population with at least one copy of the disease-refractory allele exceeds 80%. The fitness cost (reduction in mean adult lifespan) associated with gRNA/refractory allele homozygotes is varied along the x-axis, and the number of weekly releases along the y-axis. Notably, for the split drive system, the window of protection exceeds three years following 10 or more weekly releases for gRNA/refractory allele fitness costs of 10% in homozygotes.
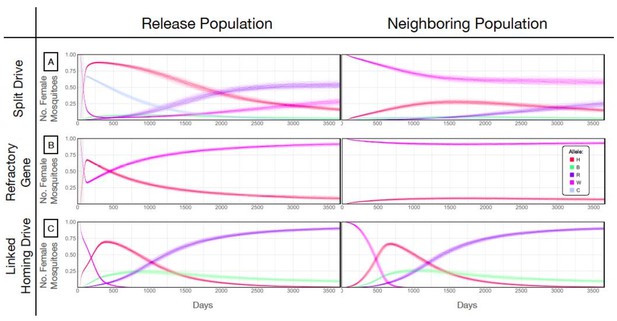
Allele frequencies for best performing split-drive construct.
Model predictions for releases of Ae. aegypti mosquitoes homozygous for the split drive system (A), a disease-refractory gene (B), or a homing drive system in which the components of the split drive system are linked at the same locus (C). Parameters correspond to those for the best performing split drive system (wU6b-GDe/w+; nup50-Cas9/+) (Supplementary file 7b). Releases are carried out in a population with an equilibrium size of 10,000 adults that exchanges migrants with a neighboring population of the same equilibrium size at a rate of 1% per mosquito per generation. Model predictions were computed using 100 realizations of the stochastic implementation of the MGDrivE simulation framework (Sánchez et al., 2018). Weekly releases of 10,000 males homozygous for the split drive system or disease-refractory gene were carried out over a 10 week period, while a single release was carried out for the linked homing drive system. Results are plotted for female allele frequencies. Red denotes the gRNA/disease-refractory allele (H), pink denotes the wild-type allele at this locus (W), and dark blue and green represent in-frame/cost-free and out-of-frame/costly resistant alleles (R and B). Light blue represents the allele frequency of Cas9 at the second locus for the split drive system (C). Notably, the H allele reaches a frequency of ~30% in the neighboring population for the split drive releases, before being eliminated by virtue of a fitness cost. At the release site, the Cas9 allele is reduced to a population frequency of ~10% within four years of the final release, leading to a progressive decline of the gRNA/refractory allele in both populations. Reversibility can be accelerated through dilution of transgenes by releases of wild-type males.
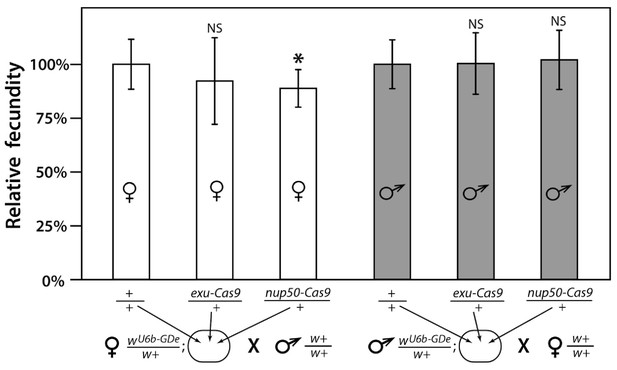
Fecundity of trans-heterozygous females only slightly reduced in mathematical models.
Progeny numbers from 20 individual crosses for each parent combination crossed to wt (w+/w+) mosquitoes of the opposite sex were used to estimate fecundity. To transform progeny numbers into fecundity percentages, each progeny number was normalized to the average progeny number in the corresponding control group, heterozygous wU6b-GDe/w+ females or males crossed to w+/w+ mosquitoes. nup50-Cas9 caused a significant reduction in fecundity, from 100.0 ± 11.6% to 88.8% ± 8.7%. Average fecundity percentages were compared to that in the corresponding control group. Bars show averages ± SD estimated from 20 data points. Statistical significance was estimated by an equal variance t-test. (p<0.02*).
Additional files
-
Supplementary file 1
Frequency of white somatic knockout in G1 gRNA[w]/+; Cas9/+ trans-heterozygotes depends on U6 promoter driving gRNA[w] expression.
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp1-v1.xlsx
-
Supplementary file 2
Genetic linkage of Gene Drive element (GDe) integrated at white locus and Nix (male dominant locus), raw data.
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp2-v1.xlsx
-
Supplementary file 3
Split drive cleavage and transmission rate.
(a) Eye phenotypes in G1 progeny from ♂wGDe/w+ X ♀ exu-Cas9/exu-Cas9. (b) Eye phenotypes in G1 progeny from ♀wGDe/w+ X ♂exu-Cas9/exu-Cas9. (c) Eye phenotypes in G2 progeny from trans-heterozygous ♀wGDe/w+; exu-Cas9/+ with Maternal Cas9 X ♂w+/w+ (wt). (d) Eye phenotypes in G2 progeny from trans-heterozygous ♂wGDe/w+; exu-Cas9/+ with Maternal Cas9 X ♀w+/w+ (wt). (e) Eye phenotypes in G2 progeny from trans-heterozygous ♀wGDe/w+; exu-Cas9/+ with Paternal Cas9 X ♂w+/w+ (wt). (f) Eye phenotypes in G2 progeny from trans-heterozygous ♂wGDe/w+; exu-Cas9/+ with Paternal Cas9 X ♀w+/w+ (wt).
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp3-v1.xlsx
-
Supplementary file 4
Cleavage and homing efficiency variability between Cas9 lines.
(a) Eye phenotypes in G1 progeny from ♂wU6b-GDe/w+ X ♀Cas9/Cas9 (b) Eye phenotypes in G1 progeny from ♀wU6b-GDe/w+ X ♂Cas9/Cas9. (c) Eye phenotypes in G2 progeny from ♀wU6b-GDe/w+; Cas9/+ with Maternal Cas9 X ♂w+/w+ (wt). (d) Eye phenotypes in G2 progeny from ♂wU6b-GDe/w+; Cas9/+ with Maternal Cas9 X ♀w+/w+ (wt). (e) Eye phenotypes in G2 progeny from ♀wU6b-GDe/w+; Cas9/+ with Paternal Cas9 X ♂w+/w+ (wt). (f) Eye phenotypes in G2 progeny from ♂wU6b-GDe/w+; Cas9/+ with Paternal Cas9 X ♀w+/w+ (wt).
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp4-v1.xlsx
-
Supplementary file 5
Multi-generation split drive stability.
(a) Eye phenotypes in G3, G4, G5 progeny from ♀wU6b-GDe/w+; exu-Cas9/+ X ♂w+/w+ (wt). (b) Eye phenotypes in G3, G4, G5 progeny from ♀wU6b-GDe/w+; nup50-Cas9/+ X ♂w+/w+ (wt). (c) Eye phenotypes in G3, G4, G5 progeny from ♂wU6b-GDe/w+; exu-Cas9/+ X ♀w+/w+ (wt). (d) Eye phenotypes in G3, G4, G5 progeny from ♂wU6b-GDe/w+; nup50-Cas9/+ X ♀w+/w+ (wt).
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp5-v1.xlsx
-
Supplementary file 6
Eye phenotypes in progeny from the w- mosquitoes sampled at G2, G3, G4, and G5 (w[U6b-GDe]/w+; nup50-Cas9/+ X w+/w+) and w∆19/w∆19 reference mosquitos.
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp6-v1.xlsx
-
Supplementary file 7
Transgene and model fitness parameters.
(a) Effect of transgenes on fitness. Comparisons of several fitness parameters between wildtype (WT) and transgenic lines expressing gRNAs targeting the w gene (wU6b-GDe) only, Cas9 only (nup50-Cas9) or both gRNAs targeting the w gene and Cas9 (wU6b-GDe/nup50-Cas9). The transgenic and WT mosquitoes only differed significantly in one fitness parameter, female fecundity, which suggests that the transgenic strains do not have a major impact on mosquito fitness. The survivorship of transgenic versus WT mosquitoes is also shown. (b) Parameter values used in Ae. aegypti population model. (Carvalho et al., 2015; Christophers, 1960; Focks et al., 1993; Horsfall, 1972; Otero et al., 2006; Simoy et al., 2015; Taylor et al., 2001).
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp7-v1.xlsx
-
Supplementary file 8
Methods supporting information.
(a) Key resources table (b) Primer sequences used in this study.
- https://cdn.elifesciences.org/articles/51701/elife-51701-supp8-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51701/elife-51701-transrepform-v1.pdf





