Phenotypic plasticity as a mechanism of cave colonization and adaptation
Figures
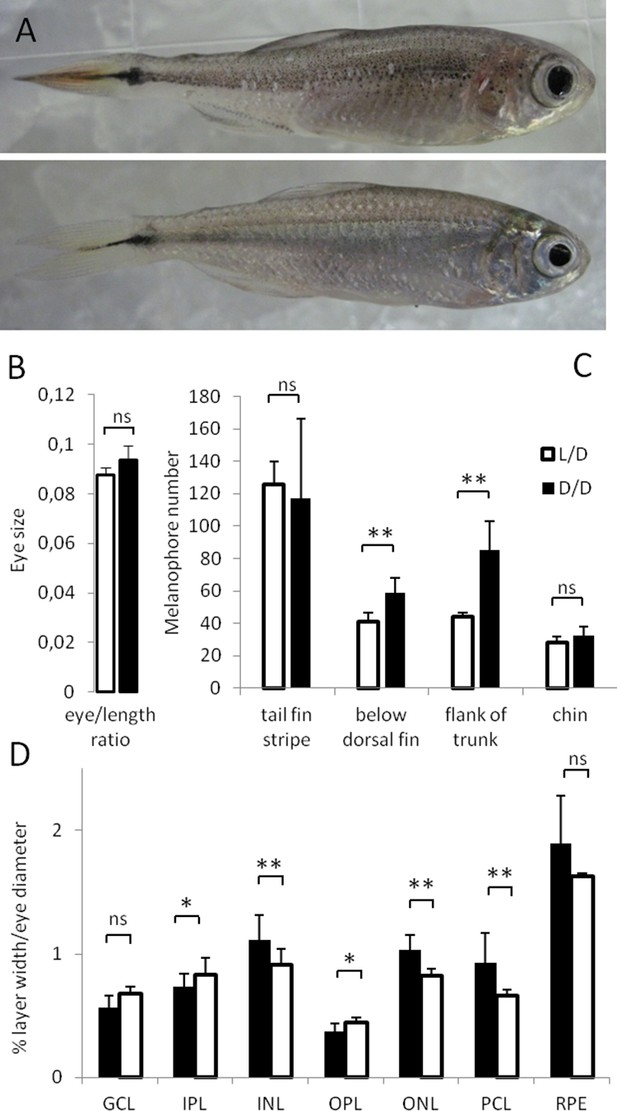
Morphological differences in Astyanax mexicanus surface fish maintained in different light regimes.
(A) Surface fish (SF) kept in constant dark (D/D; top frame) vs. light/dark (L/D; bottom frame) photoperiod for 1 year. (B) Eye size normalized by body length in D/D vs. L/D SF kept in the experimental conditions for 1 to 2 years. (N = 8) (C) Number of melanophores in 1 year-old D/D vs. L/D SF determined in four different body regions. (N = 5) (D) Thickness of retinal layers in D/D (N = 4) vs. L/D fish (N = 3): GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; PCL photoreceptor cell layer; RPE, retinal pigment epithelium measured as a ratio to eye diameter. (Error bars: SD; T-test Ns – not significant, *p<0.05, **p<0.001). Figure 1—source data 1 contains raw data and summary statistics.
-
Figure 1—source data 1
Raw data and summary statistics for Figure 1.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig1-data1-v1.xlsx
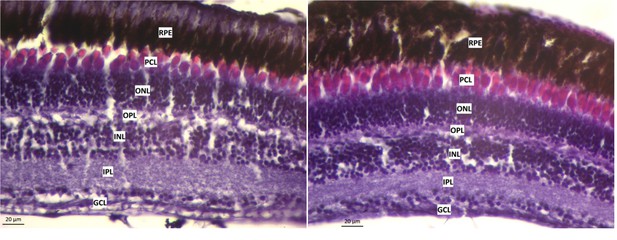
Cross section of retinal layers in surface fish reared in L/D (top) or D/D (bottom) conditions.
GCL - ganglion cell layer; IPL - inner plexiform layer; INL - inner nuclear layer; OPL - outer plexiform layer; ONL - outer nuclear layer; PCL - photoreceptor cell layer; RPE - retinal pigment epithelium. Scale bar 20 µm.
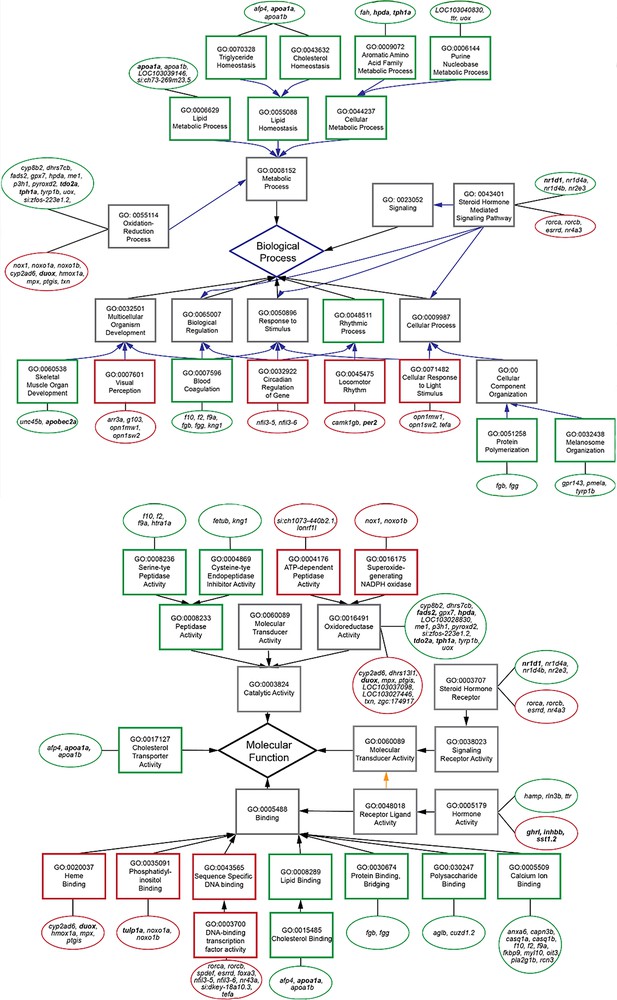
Subset of relevant, enriched GO terms (boxes) for biological processes (top) and molecular functions (bottom) of differentially expressed genes (circles) in the transcriptome.
Genes tested by RT-PCR are in bold. Red outlines are down-regulated terms and genes, and green outlines are up-regulated.
-
Figure 2—source data 1
List of differentially expressed genes between surface fish maintained in different light regimes.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig2-data1-v1.docx
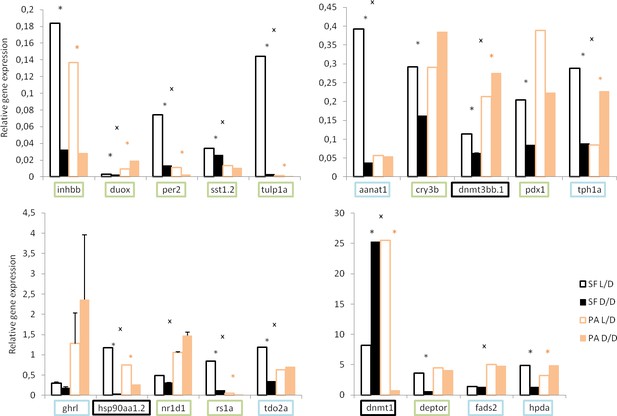
Normalized relative expression levels of genes in D/D and L/D SF and PA determined by RT-PCR.
(Error bars: SD; ANOVA with Bonferroni adjustments p<0.05 black * for SF D/D vs. SF L/D; pink * for PA D/D vs. PA L/D; X for SF vs. PA,). Genes that showed the same direction of change in transcriptome and rtPCR are in green, genes that do not show the latter changes in blue, and genes chosen based on previous work in Astyanax in black.
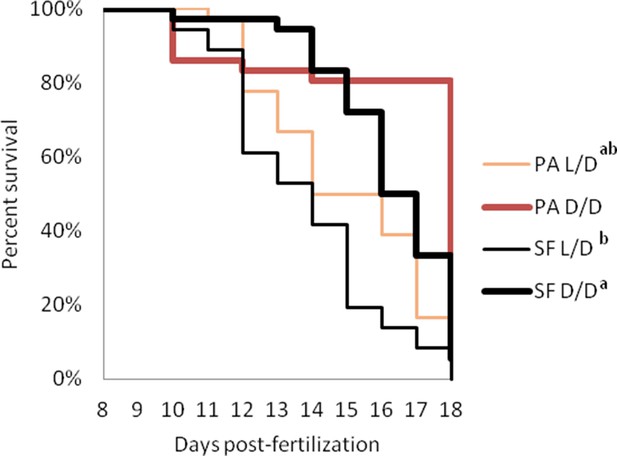
Survival curves of starvation resistance in Astyanax mexicanus surface fish (SF) and Pachón cavefish (PA) raised in complete darkness (D/D) or a normal photoperiod (L/D).
Graphs show the percent of surviving fish (from the initial 36) on each day. Groups of SF and PA larvae from each condition were starved starting at seven dpf. Vertical drops represent individuals lost at a given time point. Groups in the legend that share a superscript are not statistically different, p values calculated by Cox proportional hazards model followed by generalized linear hypothesis test. Figure 4—source data 1 contains raw data.
-
Figure 4—source data 1
Starvation survival of Astyanax mexicanus surface fish and Pachón cavefish raised in different light conditions.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig4-data1-v1.xlsx
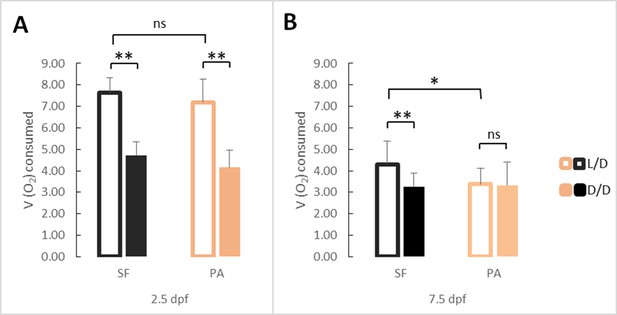
Average oxygen consumption at 2.5 (A) and 7.5 dpf (B) in SF and PA larvae kept in D/D versus L/D conditions.
At 2.5 dpf N = 18 (SF L/D), 19 (SF D/D), 18 (PA L/D), 25 (PA D/D), and at 7.5 dpf N = 20 (SF L/D), 24 (SF D/D), 20 (PA L/D) and 24 (PA D/D). (Error bars represent standard deviation, ns: not significant, *p<0.05, **p<0.01 as calculated by ANOVA and Tukey HSD Test.) Figure 5—source data 1 contains raw data and summary statistics.
-
Figure 5—source data 1
Raw data and statistics for Figure 5.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig5-data1-v1.xlsx
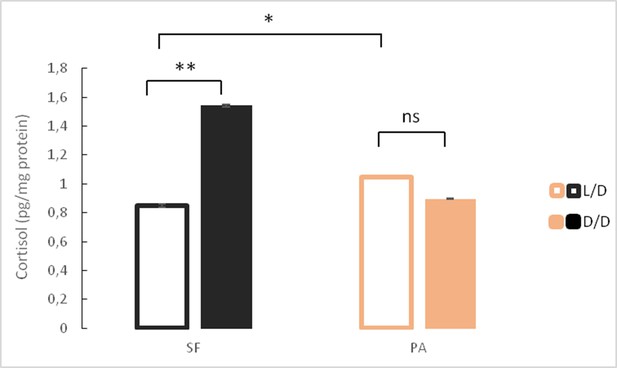
Mean cortisol levels in adult surface fish (SF) and Pachón cavefish (PA) kept in D/D or L/D conditions for 1.5 to 2 years (N = 4/group).
(Error bars represent SD in three technical replicates, ANOVA and Tukey HSD Test: Ns – not significant, *p<0.05, **p<0.01). Figure 6—source data 1 contains raw data and summary statistics.
-
Figure 6—source data 1
Raw data and summary statistics for Figure 6.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig6-data1-v1.xlsx
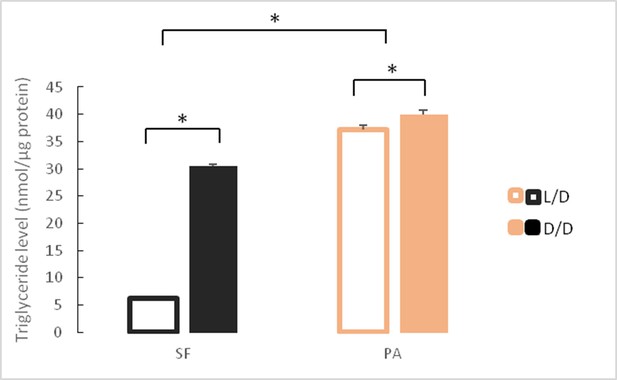
Mean triglyceride levels in SF and PA raised under D/D versus L/D conditions for approximately 1 year since < 24 hpf.
N = 3 fish/group (Error bars represent standard deviation. *p<0.01; ANOVA and Tukey HSD Test). Figure 7—source data 1 contains raw data and summary statistics.
-
Figure 7—source data 1
Raw data and summary statistics for Figure 7.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig7-data1-v1.xlsx
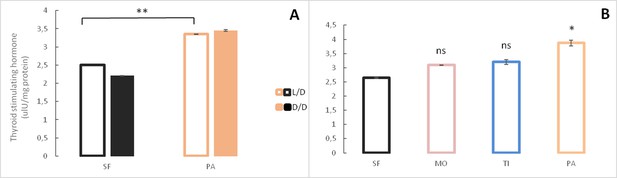
Levels of Thyroid stimulating hormone in Astyanax mexicanus under different experimental conditions and from different populations.
(A) Mean Thyroid stimulating hormone levels normalized by protein concentration in adult surface fish (SF) and Pachón cavefish (PA) kept in D/D or L/D conditions for 1.5 to 2 years. N = 3 (SF L/D), 4 (SF D/D), 3 (PA L/D), 3 (PA D/D). (B) Mean thyroid stimulating hormone levels in SF (N = 8) and three different CF populations: Pachón (PA) (N = 5), Tinaja (TI) (N = 3) and Molino (MO) (N = 4) caves. (Error bars represent SD in three technical replicates. N ranges from 3 to 8 fish/group; *p<0.05; **p<0.01 as calculated by ANOVA and Tukey HSD Test. In B ns or * denotes significance in comparison to SF.) Figure 8—source data 1 contains raw data and summary statistics.
-
Figure 8—source data 1
Raw data and summary statistics for Figure 8.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig8-data1-v1.xlsx
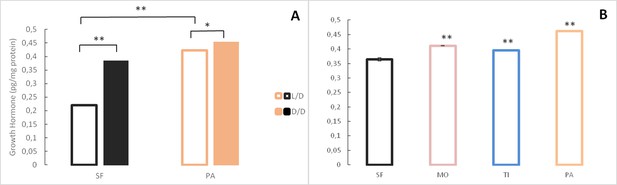
Levels of Growth hormone in Astyanax mexicanus under different experimental conditions and from different populations.
(A) Mean Growth hormone levels normalized by protein concentration in adult surface fish (SF) and Pachón (PA) cavefish kept in D/D or L/D conditions for 1.5 (SF) and 2 years (PA) since < 3 dpf. N = 3 (SF L/D), 4 (SF D/D), 3 (PA L/D), 3 (PA D/D). (B) Mean growth hormone levels in 3–4 month old SF and three different CF populations: PA, Tinaja (TI), and Molino (MO). (Error bars represent SD in three technical replicates. N = 3 to 8/group. *p<0.05; **p<0.01 as calculated by ANOVA and Tukey HSD Test. In B, **p<0.01 compared to SF). Figure 9—source data 1 contains raw data and summary statistics.
-
Figure 9—source data 1
Raw data and summary statistics for Figure 9.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig9-data1-v1.xlsx
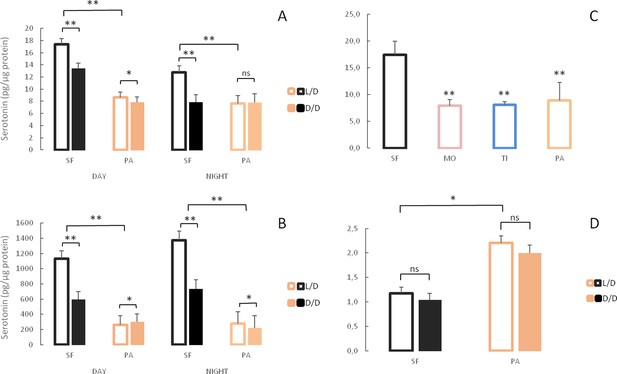
Serotonergic system changes in adults and larvae of light/dark (L/D)- and dark/dark (D/D)-reared surface fish and cavefish.
(A, B) Serotonin levels in adult brains (A) and bodies (B) of D/D and L/D reared surface fish (SF) and Pachón cavefish (PA) collected in the middle of the day (DAY) and the middle of the night (NIGHT). (Error bars represent the standard error of the means.) (C) Mean serotonin levels in brains of adult SF and three different CF populations: Molino (MO), Tinaja (TI) and PA. (D) Mean serotonin levels in pooled samples of 5 larvae aged seven dpf placed in the experiment within first few hours post fertilization. (Error bars SEM; ns – not significant, *p<0.05; **p<0.01 as calculated by ANOVA and post-hoc Tukey HSD Test. In C, **p<0.01 vs SF. The number of each fish type subjected to analysis ranged from 4 to 10 per group.) Figure 10—source data 1 contains raw data and summary statistics.
-
Figure 10—source data 1
Raw data and summary statistics for Figure 10.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig10-data1-v1.xlsx
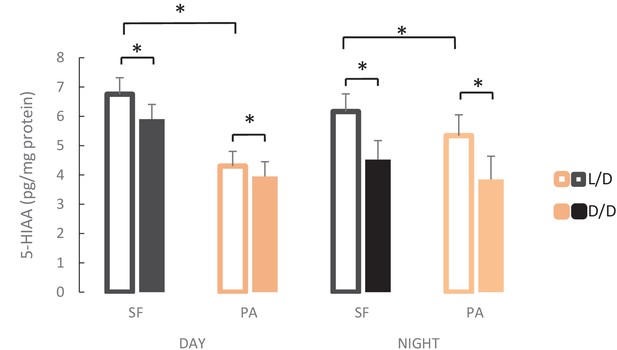
Levels of 5-Hydroxyindoleacetic acid (5-HIAA), the main metabolite of serotonin, in adult brains of L/D or D/D-reared surface fish (SF) and Pachón cavefish (PA) collected in the middle of the day (DAY) and the middle of the night (NIGHT).
Error bars represent the standard error of the means.
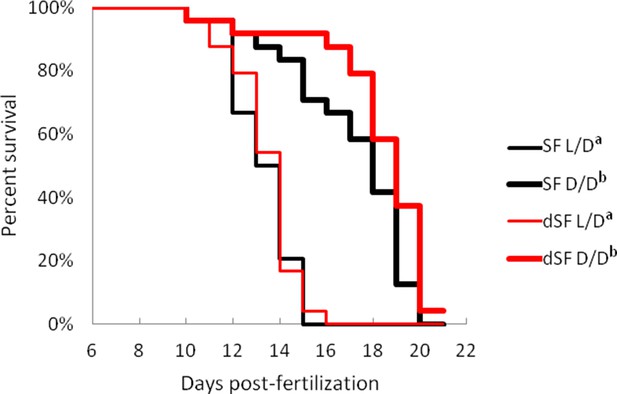
Survival curve of starvation resistance in G1 offspring of surface fish kept in normal light/dark photoperiod (SF) and G1 offspring of surface fish raised in total darkness for 2 years (dSF).
Graphs show the percent of surviving fish (from the initial 24) on each day. One group of larvae from each fish type (SF, dSF) and each lighting condition (D/D, L/D) was starved starting at seven dpf (a vs. b p<0.0001). Vertical drops represent individuals lost at a given time point, groups in the legend that share a superscript are not statistically different, p values calculated by Cox proportional hazards model followed by generalized linear hypothesis test. Figure 11—source data 1 contains raw data.
-
Figure 11—source data 1
Raw data for Figure 11.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig11-data1-v1.xlsx
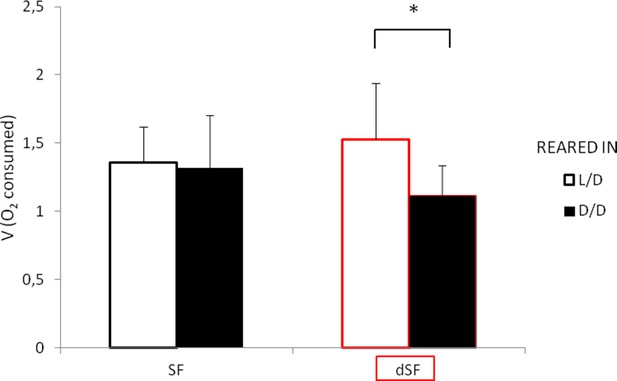
Average oxygen consumption of 11 dpf G1 offspring surface fish kept in the normal light/dark photoperiod (SF) and surface fish kept in total darkness for 2 years (dSF).
Each group of offspring was exposed to D/D or L/D conditions within first 24 hpf. (Error bars represent standard deviation; *p<0.05, as calculated by ANOVA and Tukey HSD Test.). Figure 12—source data 1 contains raw data and statistics.
-
Figure 12—source data 1
Raw data and statistics for Figure 12.
- https://cdn.elifesciences.org/articles/51830/elife-51830-fig12-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Biological sample (Astyanax mexicanus surface fish) | Surface fish, SF | Jeffery laboratory | ||
| Biological sample (Astyanax mexicanus Pachón cavefish) | Pachón, cavefish, CF, PA | Jeffery laboratory | ||
| Biological sample (Astyanax mexicanus Tinaja cavefish) | Tinaja, TI | Jeffery laboratory | ||
| Biological sample (Astyanax mexicanus Molino cavefish) | Molino, MO | Jeffery laboratory | ||
| Commercial assay or kit | TruSeq mRNA Library Prep Kit | Illumina | Cat# RS-122–2001 | |
| Commercial assay or kit | Cortisol ELISA Kit | Cayman Chemical | Cat#500360 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 | |
| Commercial assay or kit | Fish growth hormone(GH) ELISA Kit | Cusabio | Cat#CSB-E12121Fh | |
| Commercial assay or kit | Fish thyroid stimulating hormone(TSH) ELISA Kit | Cusabio | Cat#CSB-EQ02726Fl | |
| Commercial assay or kit | Triglyceride Quantification Assay Kit | Abcam | Cat#ab65336 | |
| Chemical compound, drug | Tricaine methanesulfonate | Western Chemical Inc | Cat#TRS1 | |
| Chemical compound, drug | Trizol | Invitrogen | Cat#15596026 | |
| Chemical compound, drug | Superscript III and IV Reverse Transcriptase | Invitrogen | Cat#18080044, 18090050 | |
| Chemical compound, drug | NP-40 | Abcam | Cat#ab142227 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij/ | RRID:SCR_003070 | |
| Software, algorithm | FASTQC | http://www.bioinformatics. babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 | |
| Software, algorithm | Bowtie2 v2.3.2 | http://bowtie-bio.sourceforge .net/bowtie2/index.shtml | RRID:SCR_005476 | |
| Software, algorithm | TopHat v2.1.1 | https://ccb.jhu.edu/ software/tophat/index.shtml | RRID:SCR_013035 | |
| Software, algorithm | Cufflinks v2.1.1 | http://cole-trapnell-lab. github.io/cufflinks/ | RRID:SCR_014597 | |
| Software, algorithm | R package ‘cummeRbund’ | https://bioconductor.org/packages/release/bioc/html/cummeRbund.html | ||
| Software, algorithm | R.Studio v1.0.136 | https://www.rstudio.com/products/rstudio/#Desktop | ||
| Software, algorithm | R v3.5.1 | https://cran.r-project.org/bin/windows/base/old/3.5.1/ | ||
| Software, algorithm | DAVID Bioinformatics Resources | https://david.ncifcrf.gov/ | RRID:SCR_001881 | |
| Software, algorithm | Reactome | https://reactome.org/ | RRID:SCR_003485 | |
| Software, algorithm | RefFinder | https://www.heartcure.com.au/for-researchers/ | RRID:SCR_000472 | |
| Software, algorithm | R version 3.5.3 | https://cran.r-project.org/src/base/R-3/ | ||
| Software, algorithm | R package ‘multcomp‘ | https://cran.r-project.org/web/ packages/multcomp/index.html | ||
| Software, algorithm | CSW32 data program | https://www.dataapex.com/products/csw32.php (product was discontinued) | ||
| Software, algorithm | SigmaStat version 3.5 | https://sigmastat.software. informer.com/3.5/ | RRID:SCR_010285 |
Additional files
-
Supplementary file 1
List of genes and primers used in RT-PCR experiments.
- https://cdn.elifesciences.org/articles/51830/elife-51830-supp1-v1.docx
-
Supplementary file 2
Summary statistics of Illumina output: the number reads, total base pairs, quality trimmed reads retained for each treatment, and the overall mapping rate from Tophat2 using Bowtie2.
- https://cdn.elifesciences.org/articles/51830/elife-51830-supp2-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/51830/elife-51830-transrepform-v1.docx





