The genetic basis of aneuploidy tolerance in wild yeast
Figures
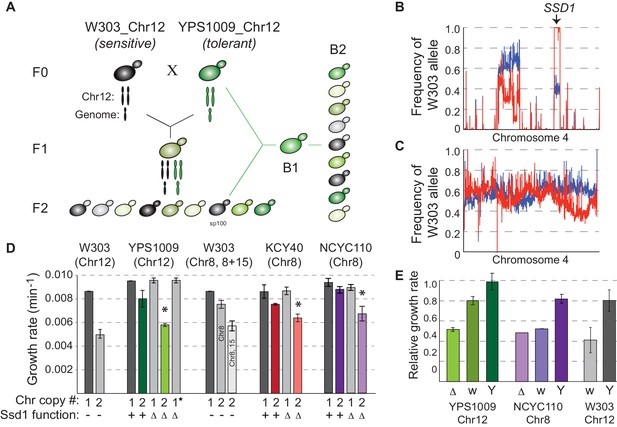
SSD1 is required for aneuploidy tolerance.
(A) Mapping schema, see Materials and methods. (B) W303 allele frequency across Chr4 in the pool of aneuploidy-sensitive (red) versus -tolerant (blue) B2 segregants or C) small (red) versus large (blue) colony pools from the euploid-control cross. (D) Average and standard deviation of growth rates for denoted strains with amplified chromosomes, indicated above. Number of chromosomes per haploid genome, SSD1 status (Δ, deletion; –, ssd1W303), and star indicating euploid revertant are indicated below. Asterisk, p<0.005, T-test comparing aneuploids with and without SSD1. (E) Average and standard deviation of growth of aneuploid ssd1- strains harboring empty vector (Δ), ssd1W303 (w) or SSD1YPS1009 (Y), relative to the isogenic aneuploid wild type with empty vector (or euploid cells with empty vector in the case of W303 ssd1W303 cells).
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/52063/elife-52063-fig1-data1-v2.zip
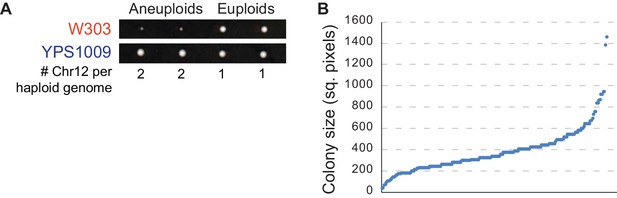
Aneuploidy tolerance varies in W303 and YPS1009 strains.
(A). Tetrads resulting from dissection of diploid W303_Chr12-3n or YPS1009_Chr12-3n, each trisomic for Chr12, onto solid YPD medium. Aneuploidy segregates 2:2 (confirmed by qPCR as outlined in Materials and methods), with extreme colony size differences evident in aneuploid versus euploid W303 (top) but not YPS1009 (bottom). (B) Colony sizes of haploid, Chr12 aneuploid F2 segregants from the hYPS1009_Chr12 x W303_Chr12 cross. Cells are sorted by colony size.
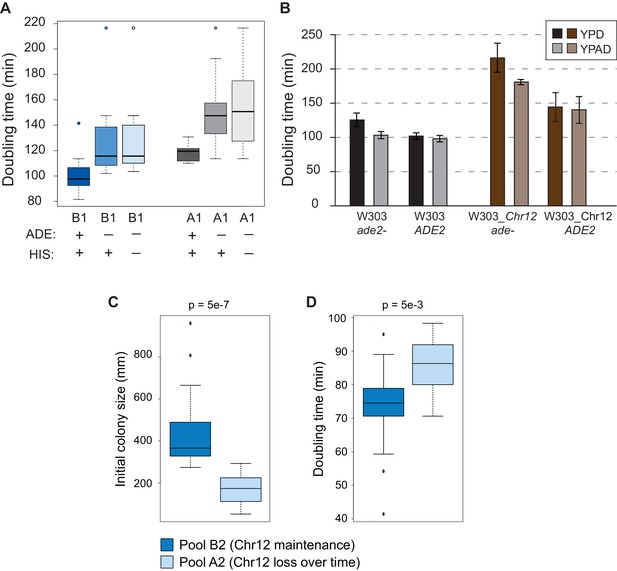
Auxotrophies influence aneuploidy tolerance.
W303 harbors multiple auxotrophies that likely influence cell growth and potentially aneuploidy tolerance. We therefore phenotyped aneuploid F2s from the cross for auxotrophies including adenine (ade-), histidine (his-), uracil, leucine and tryptophan, by plating cells on synthetic medium lacking each nutrient. (A) Doubling-time distributions of aneuploidy-tolerant (B1 pool) and aneuploidy-sensitive (A1 pool) F2s in liquid YPD medium. Pools were defined based on propensity of each culture to lose Chr12 after ~20 generations of growth. Auxotrophies were determined at the time of tetrad dissection by replica plating onto drop-out media. Ade- cells in Pool B1 that maintained Chr12 after passaging showed slower growth in rich medium compare to Ade+ cells, with little additional effect if cells were also auxotrophic for histidine (his-), whereas the aneuploidy sensitive pool showed a wider range of growth rates. (B) To test if adenine auxotrophy influences the trait, we supplemented adenine to the media or reintroduced the missing ADE2 gene. Figure 1—figure supplement 2B shows the average and standard deviation of doubling times for euploid (gray scale) and aneuploid (brown scale) W303 with or without the ADE2 gene integrated into the native genomic locus, as cells were grown in rich medium (YPD) or rich medium supplemented with additional adenine (YPAD). Reintroducing the ADE2 gene into euploid W303 improved growth (i.e. decreased doubling time) to the same effect as adenine supplementation. However, reintroducing ADE2 in the W303_Chr12 aneuploid produced a much more dramatic effect than supplementation, decreasing doubling time by nearly 30%. Thus, adenine prototrophy specifically improves the growth rate of aneuploid W303. Propensity of the culture to lose Chr12 is highly correlated with growth rate in backcrossed spores. Spores from the backcross (see Figure 1A) were partitioned into aneuploidy-tolerant pool B2 and aneuploidy-sensitive pool A2 based only on the propensity of the culture to lose Chr12 after passaging. This phenotype was highly correlated with initial colony size after dissection (C) and doubling time (D), even though colony size and growth rate were not considered in the pooling. In both cases, the aneuploidy-sensitive strains showed considerably reduced growth (p-value above plots, Welch’s T-test).

Multipool output for initial cross.
As shown in Figure 1B but for each of the 16 yeast chromosomes from the initial cross.
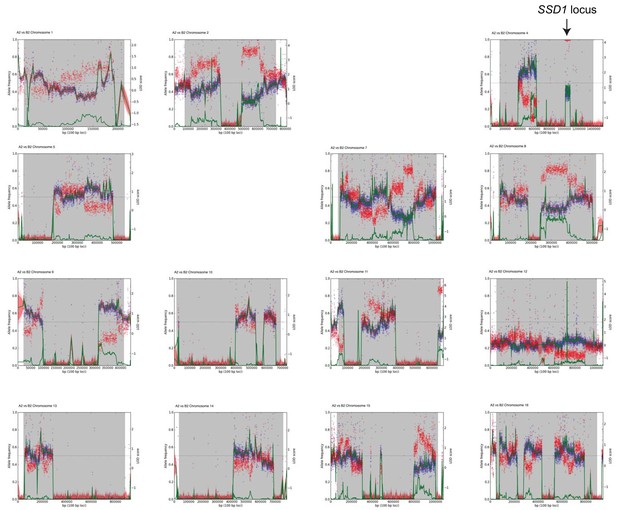
Multipool output for the backcross.
As shown in Figure 1B but for each of the 16 yeast chromosomes from the YPS1009_Chr12 backcross.
LOD traces are shown in green. Missing plots for Chr three and Chr six result from failed Multipool runs, presumably due to low recombination rates in the backcross. We focused on peaks enriched for W303 alleles in both the initial cross (Figure 1—figure supplement 3) and backcross that were not significant in the euploid cross scoring growth rate.
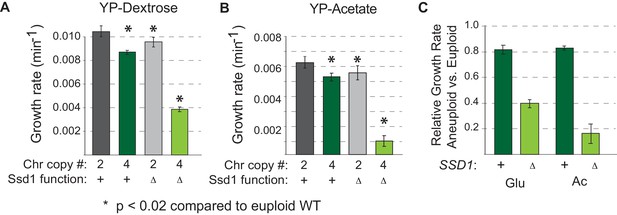
Ssd1 is required for aneuploidy tolerance in diploid YPS1009.
As shown in Figure 1D. for diploid YPS1009 with two or four copies of Chr12, with and without SSD1 according to the key, for cells grown on A) dextrose or B) acetate as a sole carbon source. (C) Growth rates in each aneuploid relative to the paired euploid is shown for direct comparison to haploid strains in Figure 4A. The relative growth rate of diploid cells tetrasomic for Chr12 was more severely affected by SSD1 deletion, especially on acetate where cells grew extremely slowly. The latter result is consistent with our published results in which we were unable to make diploid W303 aneuploids that could maintain respiration, whereas haploid W303 with extra chromosomes was defective but showed some respiratory capability (Hose et al., 2015). Interestingly, unlike in haploids, the diploid YPS1009 euploid strain showed a subtle but statistically significant (*=p < 0.02, paired T-test) growth reduction compared to wild-type, euploid cells, implicating Ssd1 in base-ploidy specific effects.
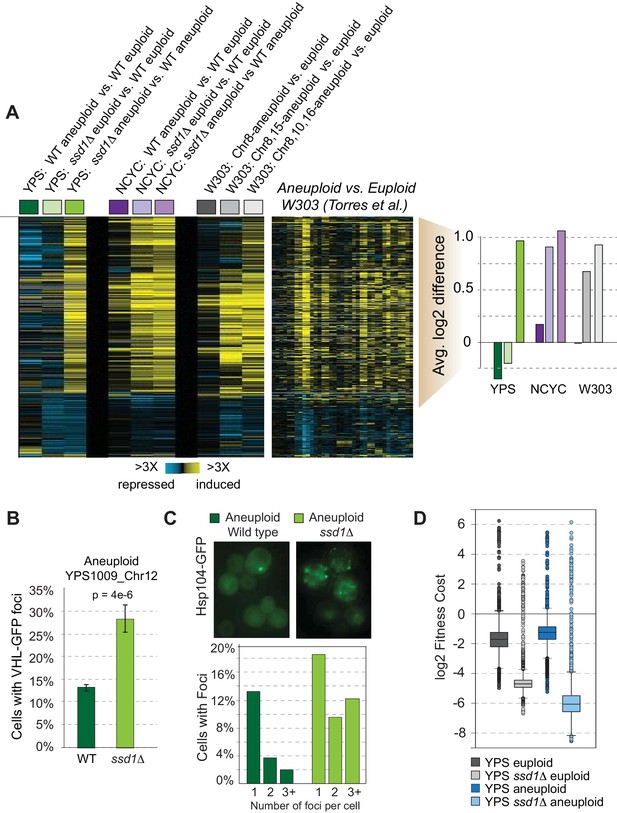
SSD1 deletion induces aneuploidy signatures.
(A) Replicate-averaged log2 expression differences for strain comparisons (columns) across 861 genes (rows) differentially expressed in mutant versus wild-type aneuploids, see text. Strains include haploid YPS1009 disomic for Chr12, diploid NCYC110 tetrasomic for Chr 8, or haploid W303 derivatives with different chromosome amplifications. Corresponding data from Torres et al. and average log2 differences in expression of induced genes are shown, where colors indicate strain labels from left. (B–C) Quantification of B) VHL-GFP foci and C) Hsp104-GFP in aneuploid strains. Data represent average and standard error of the mean (SEM) across biological triplicates, p from Fisher’s exact test. (D) Distribution of replicate-averaged fitness costs from high-copy plasmid over-expression in each strain (see Materials and methods).
-
Figure 2—source data 1
Transcriptome data shown in Figure 2A.
- https://cdn.elifesciences.org/articles/52063/elife-52063-fig2-data1-v2.txt
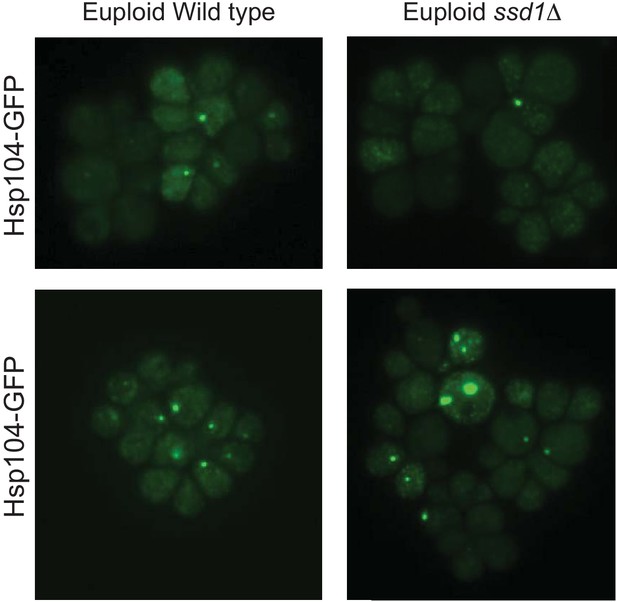
Hsp104-GFP foci in euploid cells.
Representative images of euploid YPS1009 wild type and ssd1Δ cells. For the most part, the euploid mutant looked very similar to the wild-type strain in terms of Hsp104-GFP foci number and quality, with only occasional cells harboring >1 Hsp104-GFP focus (bottom example). Thus, defects in proteostasis are occasionally seen in the euploid mutant but exacerbated by Chr12 duplication in YPS1009_Chr12 ssd1Δ cells.
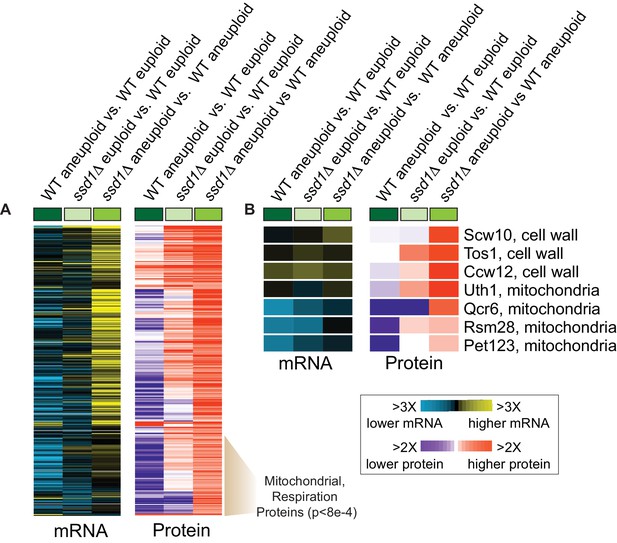
Ssd1 affects the proteome in aneuploid YPS1009 cells.
(A) Replicate-averaged log2(fold difference) in abundance across 301 significant proteins (FDR < 0.05) and their corresponding mRNAs (rows) for denoted comparisons (columns), where colors represent the magnitude of change according to the key. The indicated cluster is enriched for mitochondrial proteins and respiration factors (hypergeometric test). (B) Representative Ssd1-bound transcripts from (A).
-
Figure 3—source data 1
source data for Figure 3.
- https://cdn.elifesciences.org/articles/52063/elife-52063-fig3-data1-v2.txt
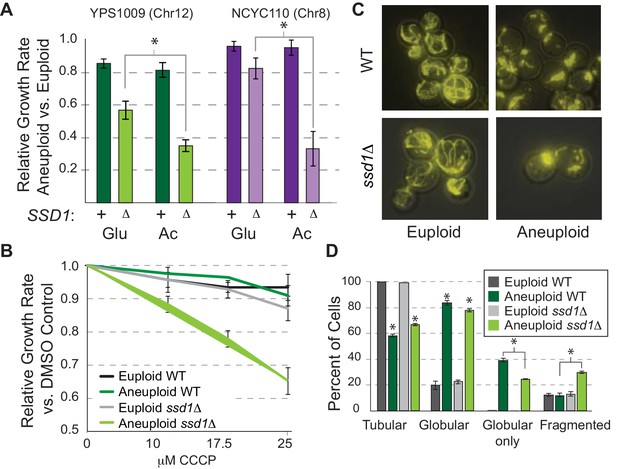
Ssd1 affects mitochondrial function and morphology.
(A) Average and standard deviation of growth rates for denoted aneuploids versus euploids ± SSD1 in glucose (Glu) or acetate (Ac). Asterisk, p<2e-4, replicate-paired T-test. (B) Average growth rates across CCCP doses. (C) Representative images of rhodamine-B stained mitochondria and D) quantified morphologies for cells with any tubular, any globular, only globular, or fragmented mitochondria (average and SEM, see Materials and methods). p<0.0001, Fisher’s exact test.
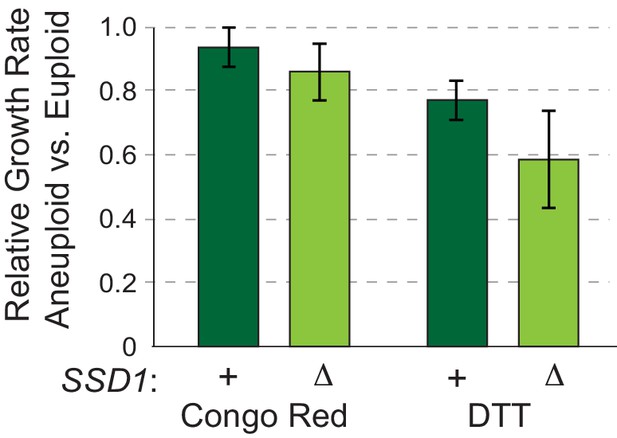
No synergistic defects between aneuploidy and cell-wall or ER stress.
Average and standard deviation of growth rates in aneuploid relative to euploid cells in the presence of 0.02 mg/mL Congo red or 2.5 mM dithiolthreitol (DTT). As shown in Figure 4A. Aneuploid YPS1009_Chr12 ssd1Δ cells are not significantly more sensitive than wild-type cells to cell-wall stress inflicted by Congo red or ER stress via DTT (p>0.05, replicate-paired T-test). The lack of synergistic sensitivity to these drugs indicates that the synergistic sensitivity of the mutant to aneuploidy plus mitochondrial stress, non-fermentable acetate, and NTC (see main text) is not due to overall stress sensitivity of the aneuploid mutant but rather implicates a specific defect in mitochondrial function and proteostasis in aneuploid ssd1Δ cells.
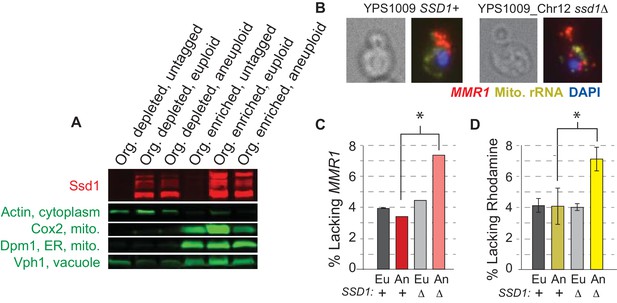
Ssd1 affects mRNA localization.
(A) Representative Western blot showing Ssd1-GFP and markers of mitochondria, ER, and vacuole as detected in organelle-depleted and organelle-enriched fractions from SSD1-GFP or untagged-Ssd1 strains (see Methods). Ssd1 fragments migrating below the expected top band emerge during the fractionation incubations. (B) Representative smFISH showing bud-localized MMR1 transcript in wild-type and ssd1Δ aneuploids. (C) Quantification of percent buds lacking MMR1 from smFISH (see Materials and methods). (D) Percent of cells lacking Rhodamine staining. Histograms represent average and SEM across biological triplicates; *, p<0.02, Fisher’s exact test.
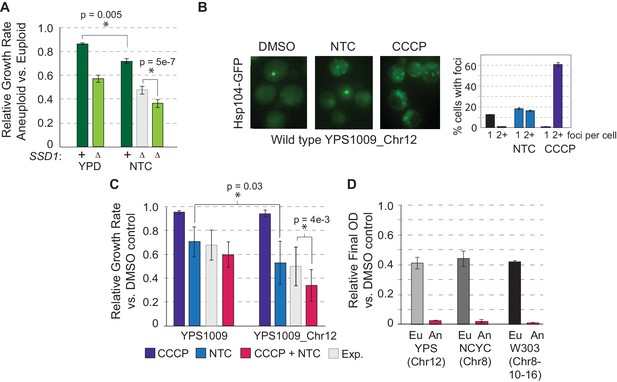
Protein misfolding and mitochondrial dysfunction sensitize aneuploids.
(A) Average and standard deviation of relative growth rates in rich YPD medium or with 1 ug/mL NTC. The expected (Exp, light gray) additive effect was calculated based on the fold-drop in growth rate of NTC-treated euploid cells applied to the wild-type aneuploid growth rate in the absence of NTC. (B) Representative Hsp104-GFP foci triggered by 1 ug/mL NTC or 25 uM CCCP and quantification in YPS1009_Chr12 (average and SEM). (C) Average relative growth rate over three generations for indicated treatments or additive expectation (Exp, paired T-test). (D) Relative final optical density after overnight CCCP + NTC treatment in euploid (Eu) and aneuploid (An) strains (see Materials and methods).
-
Figure 6—source data 1
Clustal Omega alignment of YPS1009 and seven other strains with truncated alleles.
Fasta files were recapitulated by mapping SNPs from the published vcf file8 onto the S288c sequence and performing a multiple alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Allelic differences are highlighted in yellow below. Only regions with polymorphisms are shown.
- https://cdn.elifesciences.org/articles/52063/elife-52063-fig6-data1-v2.pdf
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifier | Additional information |
|---|---|---|---|---|
| Gene (kanr) | kanr | Yeast Knockout Collection; Horizon Discovery | kanMX | |
| Gene (Klebsiella pneumoniae) | hph | pAG26; Goldstein AL, McCusker JH | hphMX | |
| Gene (Streptomyces noursei) | nat1 | pPKI | natMX | |
| Gtrain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG | this study | AGY731 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat alpha Euploid, hoΔ::HYG | this study | AGY732 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG | this study | AGY735 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat alpha Disome12, hoΔ::HYG | this study | AGY736 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, ssd1Δ::KAN | this study | AGY1444 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, ssd1Δ::KAN | this study | AGY1445 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, ssd1-Δ2 (KANMX removed) | this study | AGY1503 | Haploid, marker rescued for plasmid expression, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, ssd1-Δ2 (KANMX removed) | this study | AGY1517 | Haploid, marker rescued for plasmid expression, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, SSD1-GFP-SSD1YPS1009-terminator-NATMX | this study | AGY1446 | Haploid, GFP tagged Ssd1, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, SSD1-GFP-SSD1YPS1009-terminator-NATMX | this study | AGY1447 | Haploid, GFP tagged Ssd1, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, his3Δ::KAN | this study | AGY1504 | Haploid, his3 deletion enabling HIS3 selection, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, his3Δ::KAN | this study | AGY1505 | Haploid, his3 deletion enabling HIS3 selection, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, his3Δ::KAN, ssd1Δ::KAN | this study | AGY1506 | Haploid, his3 deletion enabling HIS3 selection, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, his3Δ::KAN, ssd1Δ::KAN | this study | AGY1507 | Haploid, his3 deletion enabling HIS3 selection, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, his3Δ::KAN, PET123-GFP-ADH1terminator-HIS3M × 6 | this study | AGY1513 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, his3Δ::KAN, PET123-GFP-ADH1terminator-HIS3M × 6 | this study | AGY1514 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, his3Δ::KAN, HSP104-GFP-ADH1terminator-HIS3M × 6 | this study | AGY1518 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, his3Δ::KAN, HSP104-GFP-ADH1terminator-HIS3M × 6/HSP104 | this study | AGY1519 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009 Mat a Euploid, hoΔ::HYG, his3Δ::KAN, ssd1Δ::KAN, HSP104-GFP-ADH1terminator-HIS3M × 6 | this study | AGY1520 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, his3Δ::KAN, ssd1Δ::KAN, HSP104-GFP-ADH1terminator-HIS3M × 6/HSP104 | this study | AGY1521 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-YPS1009_Chr12.2n Euploid | Hose et al. | AGY613 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-YPS1009_Chr12.4n Aneuploid | Hose et al. | AGY614 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-YPS1009_Chr12.2n Euploid, ssd1Δ::KAN/ssd1Δ::KAN | this study | AGY1560 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-YPS1009_Chr12.4n Aneuploid, ssd1Δ::KAN/ssd1Δ::KAN | this study | AGY1561 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303 Mat a Euploid ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16Δ::KAN | this study | AGY1387 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr12 Mat a Disome12 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16Δ::KAN/ade16Δ::HYG | this study | AGY768 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303 Mat a Euploid ADE2+ his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16Δ::KAN | this study | AGY1388 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr12 Mat a Disome12 ADE2+ his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16Δ::KAN/ade16Δ::HYG | this study | AGY1389 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303 Mat a Euploid ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ | this study | AGY103 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr8 Mat a Disome8 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ | this study | AGY1495 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr8-15 Mat a Disome8,15 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ | this study | AGY1496 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr8-10-16 Mat a Disome8,10,16 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ | this study | AGY1497 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009xW303 (sp100) Mat alpha Disome12 trp1-1 ade16Δ::KAN HYG+ | this study | AGY1548 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110 Euploid | Hose et al. | AGY729 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid | Hose et al. | AGY703 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110 Euploid, ssd1Δ::KAN/ssd1Δ::KAN | this study | AGY1493 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid, ssd1Δ::KAN/ssd1Δ::KAN | this study | AGY1494 | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | KCY40 (or VC580) Euploid, hoΔ::MFAprom-HYGMX-NATMX | Hose et al. | AGY806 | Haploid |
| Strain (Saccharomyces cerevisiae) | KCY40 (or VC580) Disome8, hoΔ::MFAprom-HYGMX-NATMX | Hose et al. | AGY1105 | Haploid |
| Strain (Saccharomyces cerevisiae) | KCY40 (or VC580) Euploid, hoΔ::MFAprom-HYGMX-NATMX, ssd1Δ::KAN | this study | AGY1385 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | KCY40 (or VC580) Disome8, hoΔ::MFAprom-HYGMX-NATMX, ssd1Δ::KAN | this study | AGY1386 | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG + pPKI | this study | AGY735 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, ssd1Δ::KAN + pPKI | this study | ABY1445 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, ssd1Δ::KAN + pJH1-SSD1-W303 | this study | ABY1445 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | YPS1009_Chr12 Mat a Disome12, hoΔ::HYG, ssd1Δ::KAN + pJH1-SSD1-YPS1009 | this study | ABY1445 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid + pJH1 | this study | AGY703 tranformed with plasmid | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid, ssd1Δ::KAN/ssd1Δ::KAN + pJH1 | this study | AGY1494 transformed with plasmid | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid, ssd1Δ::KAN/ssd1Δ::KAN + pJH1-SSD1-W303 | this study | AGY1494 transformed with plasmid | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | d-NCYC110_Chr8-4n Aneuploid, ssd1Δ::KAN/ssd1Δ::KAN + pJH1-SSD1-YPS1009 | this study | AGY1494 transformed with plasmid | Diploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303 Mat a Euploid ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade1::HIS3, lys2::KAN | Torres et al. | AGY487 | Haploid |
| Strain (Saccharomyces cerevisiae) | W303_Chr12 Mat a Disome12 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16::HIS3 ade16::KAN | Torres et al. | AGY488 | Haploid |
| Strain (Saccharomyces cerevisiae) | W303 Mat a Euploid ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade1::HIS3, lys2::KAN + pJH1 | this study | AGY487 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr12 Mat a Disome12 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16::HIS3 ade16::KAN + pJH1 | this study | AGY488 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Strain (Saccharomyces cerevisiae) | W303_Chr12 Mat a Disome12 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 Gal+ ade16::HIS3 ade16::KAN + pJH1-SSD1-YPS1009 | this study | AGY488 transformed with plasmid | Haploid, available on request from the Gasch Lab |
| Antibody | Rabbit polyclonal Anti-GFP | Abcam | Abcam catalog #ab290 | Rabbit polyclonal; 1:2000 |
| Antibody | Mouse monoclonal Anti-Actin | Thermo Fisher Scientific | Thermo Fisher Scientific catalog #MA1-744 | Mouse monoclonal; 1:1000 |
| Antibody | Mouse monoclonal Anti-COX2 | Abcam | Abcam catalog #ab110271 | Mouse monoclonal; 1:500 |
| Antibody | Mouse monoclonal Anti-DPM1 | Abcam | Abcam catalog #ab113686 | Mouse monoclonal; 1:250 |
| Antibody | Mouse monoclonal Anti-VPH1 | Abcam | Abcam catalog #ab113683 | Mouse monoclonal; 1:1000 |
| Recombinant DNA reagent | pXIPHOS | GenBank accession MG897154 | PAM sgRNA sequence (GAATCGAATG CAACCGGCGC) that targeted KanMX | Higgins et al., Wrobel et al. |
| Recombinant DNA reagent | pPKI | this study | AGB185 | CEN plasmid with the natMX selection marker. |
| Recombinant DNA reagent | pJH1 | this study | AGB090 | CEN plasmid derived from pKI that has natMX selection marker. pJH is equivalent to pKI except for a fragment of unexpressed DNA that was removed during generation. |
| Recombinant DNA reagent | pJH1-SSD1-YPS1009 | this study | ORF + 1000 bp upstream and 337 bp downstream of SSD1 from YPS1009 genomic DNA. Plasmid has natMX selection marker | |
| Recombinant DNA reagent | pJH1-SSD1-W303 | this study | ORF + 1000 bp upstream and 337 bp downstream of SSD1 from aW303 genomic DNA. Plasmid has natMX selection marker | |
| Recombinant DNA reagent | Molecular Barcoded Yeast (MoBY) v2.0 ORF Library | other | obtained from Great Lakes Bioenergy Research Center (GLBRC) | Ho, CH. et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotech. 27 (Holland and Cleveland, 2012), 369–377 (2009). |
| Sequence-based reagent | MMR1 FISH probes | Stellaris | designed against MMR1 mRNA | |
| Sequence-based reagent | Mitochondrial rRNA FISH probes | Stellaris | designed against 15 s and 21 s rRNA | |
| Peptide, recombinant protein | von Hippel-Lindau (VHL) tumor suppressor | Kaganovich et al. | Addgene catalog #21053 | Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008 Aug 28. 454 (7208):1088–95. |
| Peptide, recombinant protein | Aequorea victoria GFP (S65T) | Huh et al. | Huh W, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, and O'Shea EK (2003) Global Analysis of Protein Localization in Budding Yeast Nature 425:686–691. | |
| Commercial assay or kit | Mitochondrial Yeast Isolation Kit | Abcam | Abcam catalog #ab178779 | |
| Commercial assay or kit | Illumina TruSeq Total RNA Stranded | Illumina | Illumina catalog #20020597; previously RS-122–2203 | |
| Commercial assay or kit | NEBNext Ultra DNA Library Prep Kit for Illumina | New England Biolabs | NEB catalog #E7370L | |
| Commercial assay or kit | Yeast Mitochondrial Stain Sampler Kit | Thermo Fisher Scientific | Thermo Fisher Scientific catalog #Y7530 | |
| Chemical compound, drug | Nourseothricin-dihydrogen sulfate(clonNAT) | Werner BioAgents | Werner BioAgents catalog #5.005.000 | |
| Chemical compound, drug | 4',6-Diamidino-2-phenylindole, dihydrochloride (DAPI) | Thermo Fisher Scientific | Thermo Fisher Scientific catalog #PI62247 | |
| Chemical compound, drug | Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) | Sigma-Aldrich | Sigma catalog #C2759 | |
| Chemical compound, drug | Radicicol, Humicola fuscoatra | A.G. Scientific | A.G. Scientific catalog #R-1130 | |
| Chemical compound, drug | GFP-Trap Magnetic Agarose | Chromotek | Chromotek catalog #gtma-20 |
Additional files
-
Supplementary file 1
Zipped file with MULTIPOOL output files, as described in the MULTIPOOL manual, comparing pools A1 versus B1, A2 versus B2, and D versus F as described in Materials and methods.
Plots represent the W303 allele frequency in the aneuploidy-sensitive or small-colony pools (red) versus the aneuploidy-tolerant or larger-colony pools (blue).
- https://cdn.elifesciences.org/articles/52063/elife-52063-supp1-v2.zip
-
Supplementary file 2
Compiled information and data.
- https://cdn.elifesciences.org/articles/52063/elife-52063-supp2-v2.xlsx
-
Supplementary file 3
Normalized absolute protein abundance for each sample, see Materials and methods.
- https://cdn.elifesciences.org/articles/52063/elife-52063-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52063/elife-52063-transrepform-v2.pdf





