State-dependent brainstem ensemble dynamics and their interactions with hippocampus across sleep states
Figures

Population activity in the brainstem across the sleep-wake cycle.
(A) A diagram of the experimental approach, showing the insertion of a silicon probe for extracellular recording in the brainstem and a screw for cortical EEG recording. Pupil dilation and EMGs were also monitored in a head-fixed condition. (B) An example of multiple electrophysiological readings across three behavioral states, including local field potentials (LFPs) in the brainstem (locally subtracted LFP signals), brainstem single unit activities (SUAs), cortical electroencephalograms (EEGs), electromyograms (EMGs) and normalized pupil diameter. REM, rapid eye movement sleep; NREM, non-REM sleep; AW, wakefulness. (C, E) Power spectrum density of cortical EEGs (C) and brainstem LFPs (E) across three behavioral states. Spectra were computed during every 4 s window. Errors indicate SEM. (D, F) Distribution of EMG signals (root mean square) (D) and normalized pupil diameter (F) across three behavioral states. Pupil diameter was normalized as z-score.
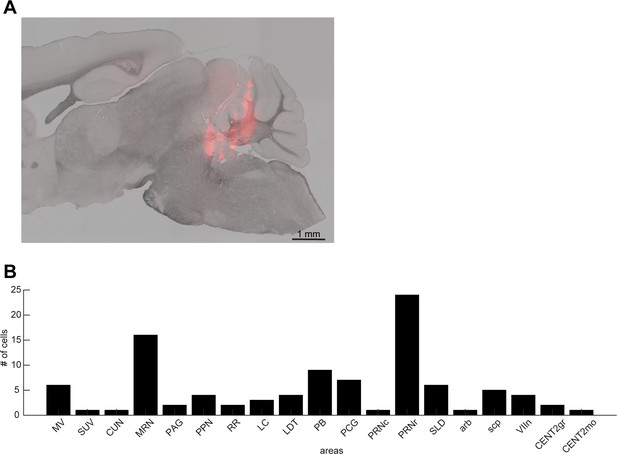
Histological analysis.
(A) A sagittal section showing the track of a silicon probe. (B) The number of neurons recorded from brainstem nuclei in this study. MV, medial vestibular nucleus; SUV, superior vestibular nucleus; CUN, cuneiform nucleus; MRN, midbrain reticular nucleus; PAG, periaqueductal gray; PPN, pedunculopontine nucleus; RR, retrorubral area; LC, locus ceruleus; LDT, laterodorsal tegmental nucleus; PB, parabrachial nucleus; PCG, pontine central gray; PRNc, pontine reticular nucleus, caudal part; PRNr, pontine reticular nucleus, rostral part; SLD, sublaterodorsal nucleus; arb, arbor vitae; scp, superior cerebellar peduncles; VIIn, facial nerve; CENT2gr, lobule II, granular layer; CENT2mo, lobule II, molecular layer.
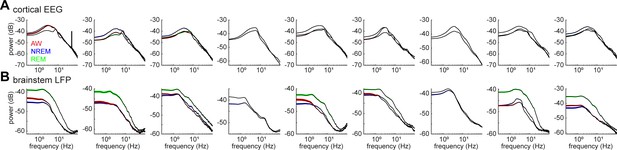
Power spectrum density of cortical EEGs (A) and brainstem LFPs (B) across all recording sessions.
The spectrum was computed in every 4 s window in each state.
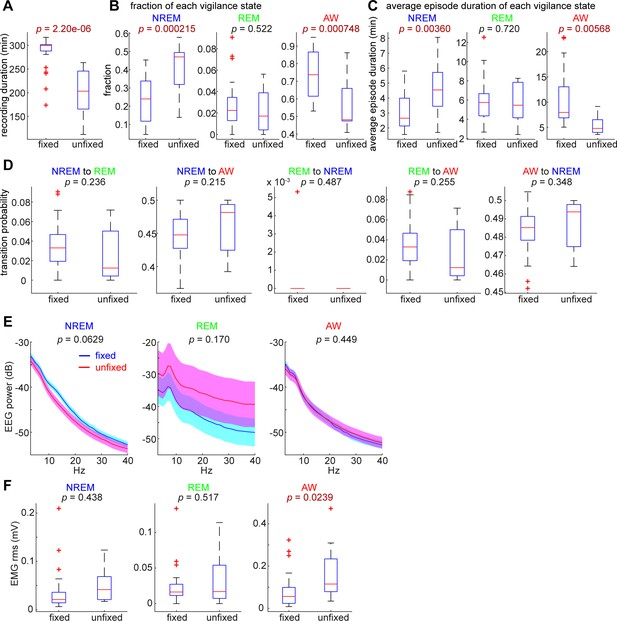
Comparisons of sleep architecture, cortical EEGs and EMGs between head-fixed (n = 24) and unfixed conditions (n = 12).
(A) The total duration of recording was significantly different (t-test). (B) The fraction of NREM sleep and awake (AW) states were significantly different. (C) The average duration of NREM sleep and AW states were significantly different. (D) There was no significant difference in state transition probabilities. (E) EEG spectral power was not significantly different between conditions across states. (F) The root-mean-square (rms) of EMGs during the AW state was significantly different.
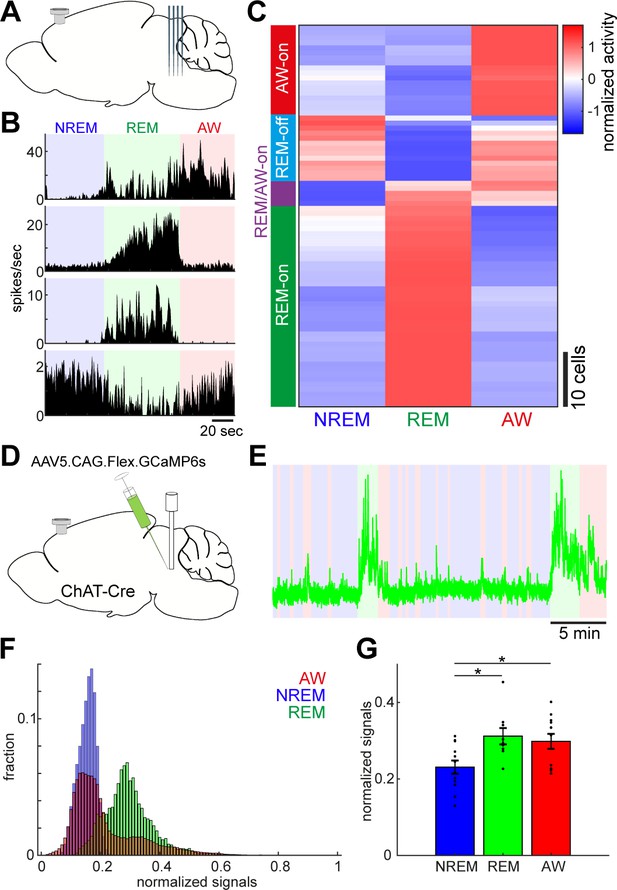
Diverse and cell-type-specific state-dependent firing in brainstem neurons.
(A) A diagram of the experimental approach, showing a silicon probe and a cortical EEG electrode. (B) Four examples of simultaneously recorded neurons. (C) Classification of functional classes. Firing rates across the three behavioral states were normalized as z-score for individual cells and hierarchical clustering was then applied. (D) A diagram of the experimental approach for fiber photometry-based Ca2+ imaging from pontine cholinergic neural populations in a freely behaving condition. (E) An example of fluorescent signals across sleep-wake cycles. Fluorescent signals (470 nm) were normalized by off-peak (405 nm) signals. Red, wakefulness; blue, NREM sleep; green, REM sleep. (F) Distributions of fluorescent signals across the three behavioral states. (G) Group statistics of average fluorescent signals from 12 recordings from four mice (F2,32 = 5.12, p=0.012, one-way ANOVA). *, p<0.05 with post-hoc Tukey’s honest significant difference criterion.
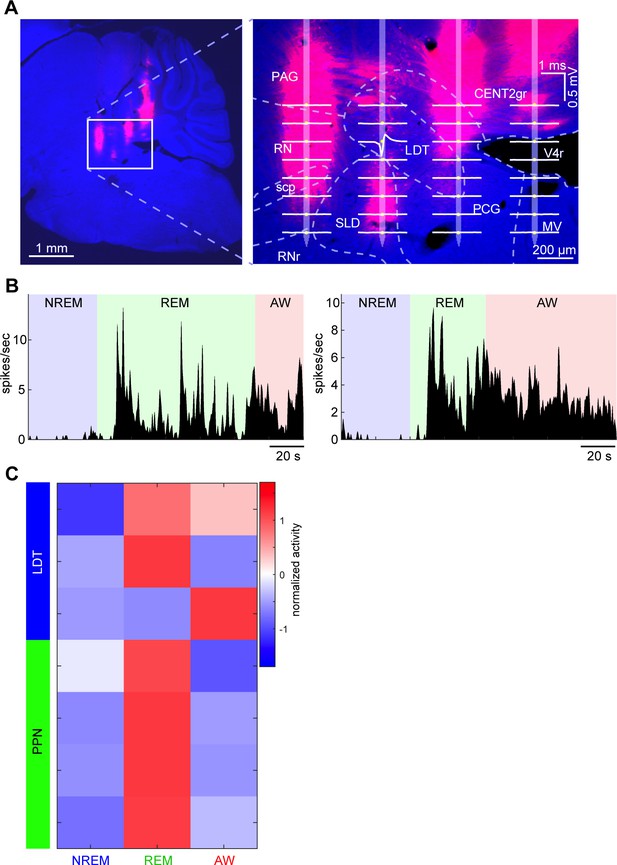
Neurons recorded from pontine cholinergic nuclei and their state dependency.
(A) A Nissl-stained sagittal section with DiI-labeled probe tracks (see also Figure 1—figure supplement 1A). Right, an enlarged image with an overlay of an estimated probe track and channel positions. Average waveforms of a LDT neuron (based on randomly picked 1000 spikes) were also overlaid. LDT, laterodorsal tegmental nucleus. (B) State-dependent firing of the LDT neuron shown in (A). The histograms were smoothed with a 1 s Gaussian kernel. (C) A summary of all neurons recorded from PPT and LDT. Firing rates across three behavioral states were normalized as z-score. All neurons were classified as either REM-on, AW-on or REM/AW-on neurons. The LDT neuron in (A) and (B) is the top one in this plot.
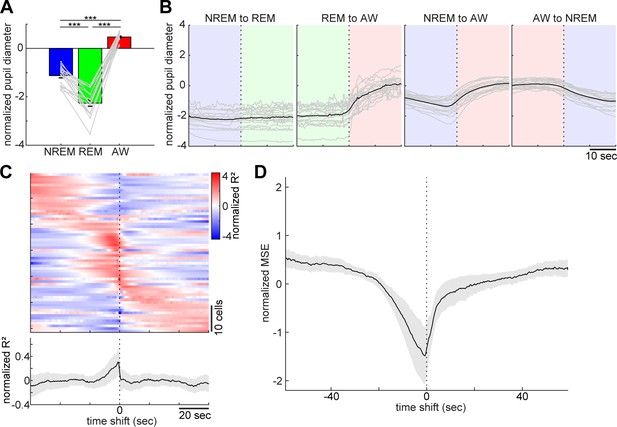
Pupil dilation across the sleep-wake cycle and prediction of pupil dilation by activity of brainstem populations.
(A) Mean normalized (z-scored) pupil diameter across the sleep-wake cycle (n = 18, F2, 53 = 220.33, p<0.0001. one-way ANOVA). ***, p<0.0001, with post-hoc Tukey’s honest significant difference criterion. (B) Pupil dilation during sleep-wake state transitions (n = 18). (C) Linear regression analysis to predict pupil diameter with individual neuronal activity. (Top) Normalized (z-scored) cross-validated R2 values are color-coded across neurons. (Bottom) The average of normalized cross-validated R2 values. Error, SEM. (D) Multiple linear regression analysis to predict pupil diameter according to simultaneously recorded neuronal activity. The average of normalized (z-scored) root mean square errors across datasets (n = 6). Error, SEM.
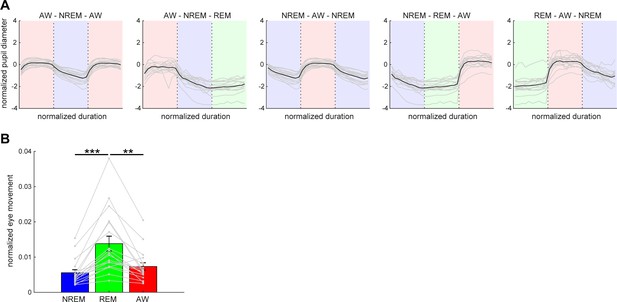
Pupil dilation and eye movement across vigilance states.
(A) Normalized (z-scored) pupil diameter across state transitions. The duration of each state episode was normalized. Pupil diameter gradually decreased before REM sleep and stayed fully constricted during REM sleep. (B) The average of eye movement across states. The effect of states on eye movement was significant (F2,53 = 8.93, p<0.001, one-way ANOVA with post-hoc Tukey’s honest significant difference criterion). **, p<0.01; ***p<0.001.
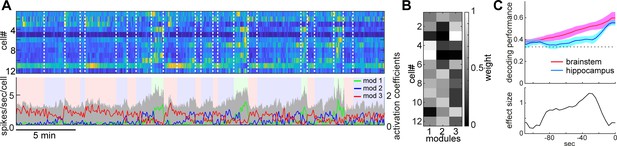
State-dependent brainstem population dynamics and their ability to predict vigilance states.
(A) Simultaneously recorded brainstem neurons across states and modules extracted with non-negative matrix factorization (NMF). (Top) Firing profiles of brainstem neurons. Firing rate was normalized by the maximum firing rate and the normalized values were color-coded. Dotted lines indicate state transitions. (Bottom) Average population activity (gray) and activation coefficients for each module derived by NMF. Background colors indicate behavioral states (red, AW; blue, NREM; green, REM). (B) Weights across neurons for each module. (C) Decoding of behavioral states from population activity. (Top) Decoding performance of brainstem and hippocampal CA1 neural populations for behavioral states as a function of time-shift. Error, SEM. (Bottom) Effect size as a function of time-shift.
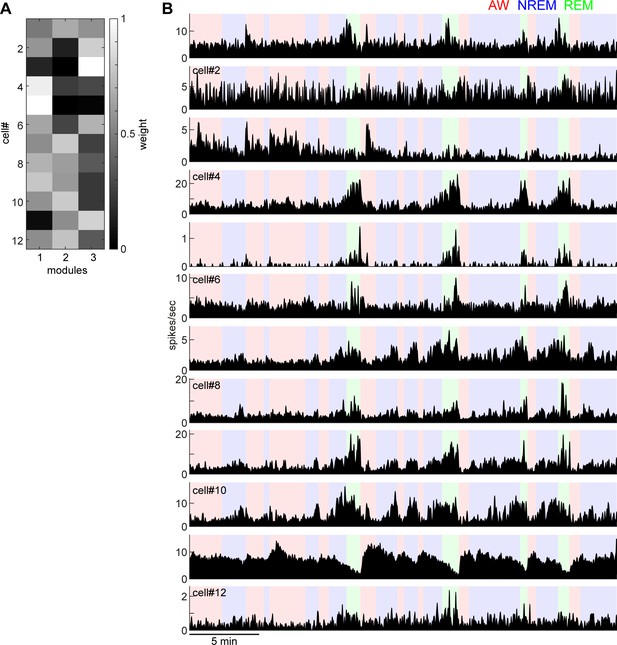
Weights of each module derived from non-negative matrix factorization (A) and individual firing profiles (B).
Neurons that have similar state-dependent firing patterns tend to have similar weights in each module. For example, cell#4 and cell#5 fire more during REM sleep and had higher weights in module 1.
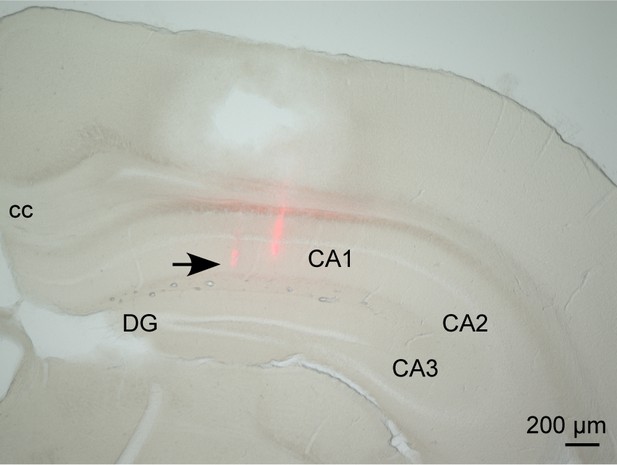
Histological analysis of dorsal hippocampal recording.
The track of a silicon probe was confirmed in the CA1 region of the dorsal hippocampus.
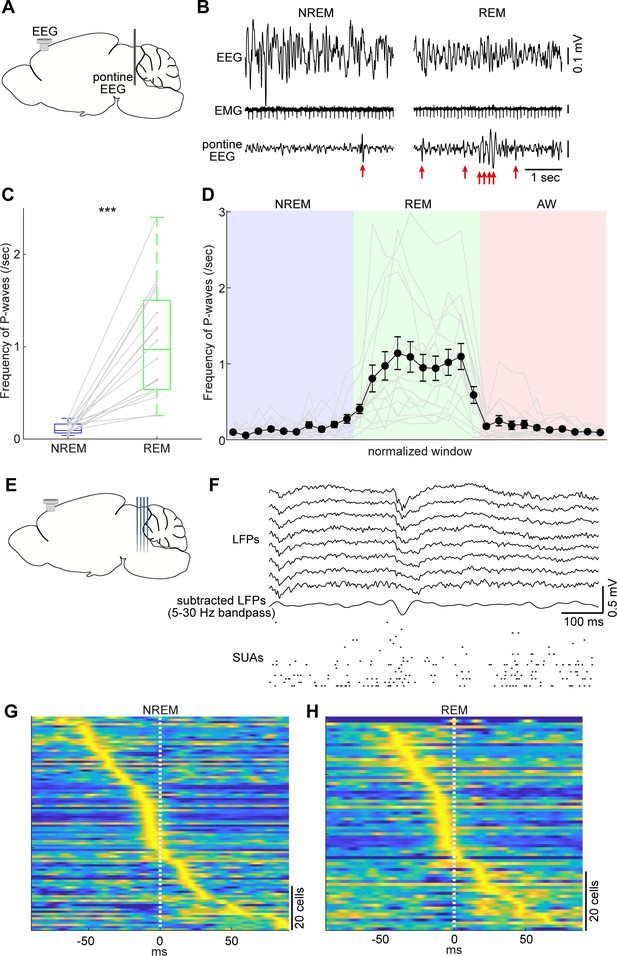
Pontine waves (P-waves) in the mouse.
(A) A diagram of the experimental approach, showing a bipolar electrode in the pons and a cortical EEG electrode. (B) Examples of P-waves during NREM (left) and REM sleep (right), showing cortical EEG and EMG traces. (C) Frequency of P-waves during NREM and REM sleep (n = 16). ***, p<0.0001, two-tailed t-test. (D) Temporal evolution of P-wave frequency. The duration of each state episode was normalized to one. Error, SEM. (E) A diagram of the experimental approach, showing a multi-shank silicon probe in the brainstem and a cortical EEG electrode. (F) An example of a P-wave, showing subtracted LFPs from a probe shank, and multiple single unit activity. (G, H) Pooled peri-event time histograms of brainstem single units relative to P-wave timing during NREM (G) and REM sleep (H). Time zero is the timing of P-waves (trough time). Each peri-event time histogram is color-coded by normalizing the maximum firing rate for each cell. The order of single units was sorted by the peak timing in each state.
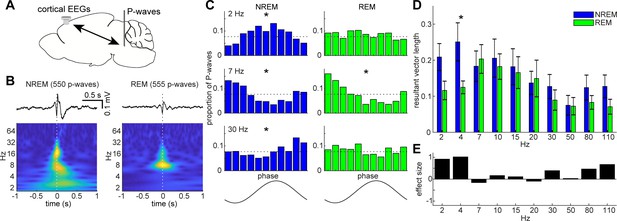
State-dependent interactions between P-waves and cortical oscillations.
(A) A diagram of an experimental approach showing a bipolar electrode in the pons and a cortical EEG electrode. (B) Examples of averaged event-triggered cortical EEGs and scalograms during NREM (left) and REM sleep (right). Time zero is the timing of P-waves (trough time). (C) Examples of phase-histograms. Cortical EEGs were filtered at certain frequency bands and the proportion of P-waves elicited in each phase bin was calculated. *, p<0. 01, Rayleigh test. (D) Phase modulations across frequency bands of cortical EEGs. Resultant vector length represents the mean measurement of circular spread of the phase relationship between cortical oscillations and P-waves. The effect of sleep states on resultant vector length was significant (F1,179 = 4.90, p<0.05, two-way ANOVA). *, p<0.05 (post-hoc Tukey’s hones significant difference test). Error, SEM. (E) Effect size of states across frequency bands.
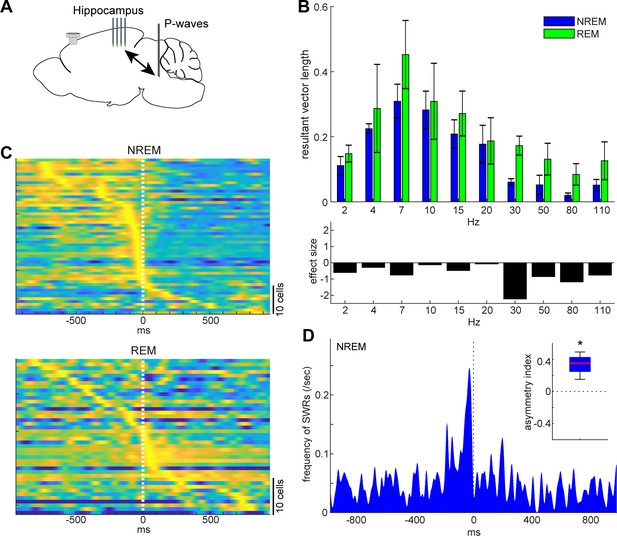
State-dependent interactions between P-waves and hippocampal activity.
(A) A diagram of the experimental approach showing a multi-shank silicon probe in the hippocampus CA1, a bipolar electrode in the pons and a cortical EEG electrode. (B) Phase modulations across frequency bands of hippocampal LFPs (top) and effect size of states (bottom). The effect of sleep states on resultant vector length was significant (F1,79 = 4.00, p<0.05, two-way ANOVA). Error, SEM. (C) Pooled peri-event time histograms of CA1 single units relative to P-wave timing in NREM (top) and REM sleep (bottom). Time zero is the timing of P-waves (trough time). Each peri-event time histogram is color-coded by normalizing the maximum firing rate for each cell. The order of single units was sorted by the peak timing in each state. (D) Frequency of hippocampal sharp wave-ripples (SWRs) relative to P-wave timing in NREM sleep. Time zero is the timing of P-waves (trough time). After binning at 1 ms resolution, the histogram was smoothed with a 50 ms Gaussian kernel. Inset, asymmetry index provides the normalized difference in the number of SWRs before and after P-waves in a 100 ms window. Larger positive values mean that more SWRs occurred before the P-waves. *, p<0.05 (t-test, n = 4).
Additional files
-
Supplementary file 1
Summary of experimental animals and recordings involved in this study.
BS-Si, silicon probe recording in the brainstem. HP-Si, silicon probe recording in the hippocampus. BS-EEG, EEG recording in the brainstem. FP, fiber photometry. Exc1, the animal was excluded because of electrode mispositioning. Exc2, the animal was excluded because of lack of histological data. Exc3, the animal was excluded because of eye closure during recording.
- https://cdn.elifesciences.org/articles/52244/elife-52244-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52244/elife-52244-transrepform-v2.docx





