Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides
Figures
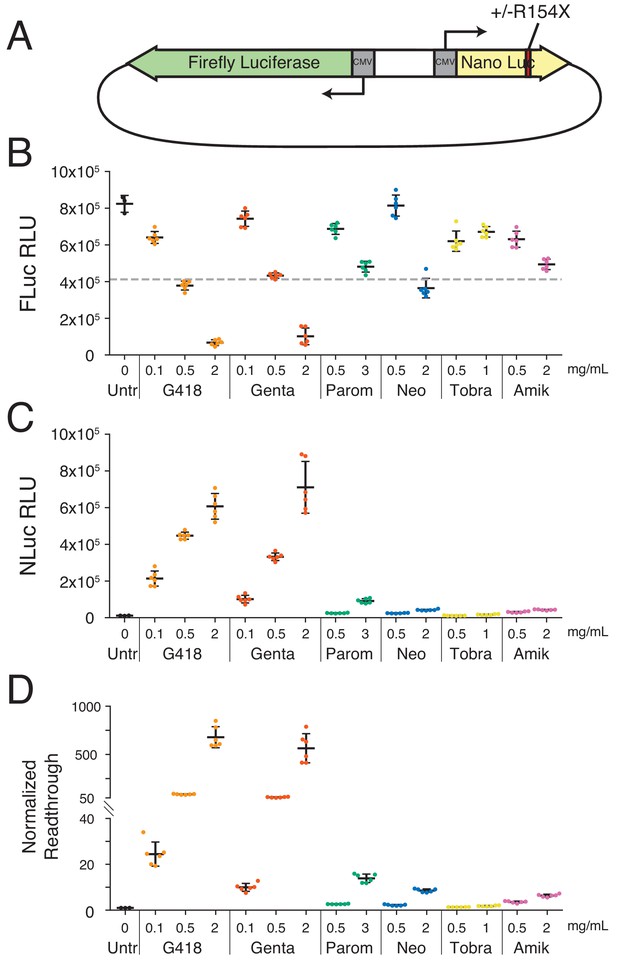
Aminoglycosides stimulate readthrough of a PTC in luciferase reporter assays.
(A) Schematic of reporter constructs used in this study. Full-length firefly luciferase (FLuc) is expressed from the reverse strand and nano luciferase (NLuc) encoding either an arginine, or a UGA stop codon at amino acid 154 is expressed from the forward strand. (B-D) HEK293T cells transiently transfected with the PTC-containing reporter after 24 hr of AG treatment. (B) Activity of FLuc, (C) or NLuc measured in Relative Light Units (RLUs) as well as (D) normalized readthrough calculated as the NLuc to FLuc ratio for each individual well and normalized to the mean NLuc to FLuc ratio in untreated cells. Mean RLUs or readthrough values are shown with error bars indicating the standard deviations of each measurement. Normalized readthrough in (D) is plotted on two separate axes. Varying concentrations were tested for G418 (0.1, 0.5, 2.0 mg/mL), gentamicin (0.1, 0.5, 2.0 mg/mL), paromomycin (0.5, 3.0 mg/mL), neomycin (0.5, 2.0 mg/mL), tobramycin (0.5, 1.0 mg/mL), and amikacin (0.5, 2.0 mg/mL). Six replicates (separate wells) were collected for each drug treatment condition in this experiment, and at least three separate experiments were performed with all compounds. The dashed line in (B) corresponds to a 50% reduction in FLuc signal relative to untreated cells.
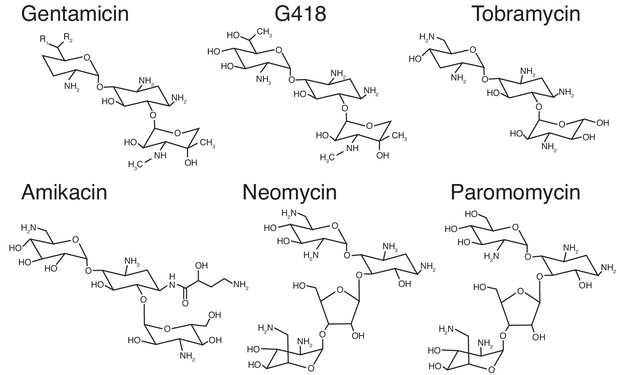
Aminoglycosides tested in this study.
Molecular structures of aminoglycosides tested in this study are depicted here. Gentamicin is a heterogeneous mix of compounds with sites of substitutions indicated by R groups.
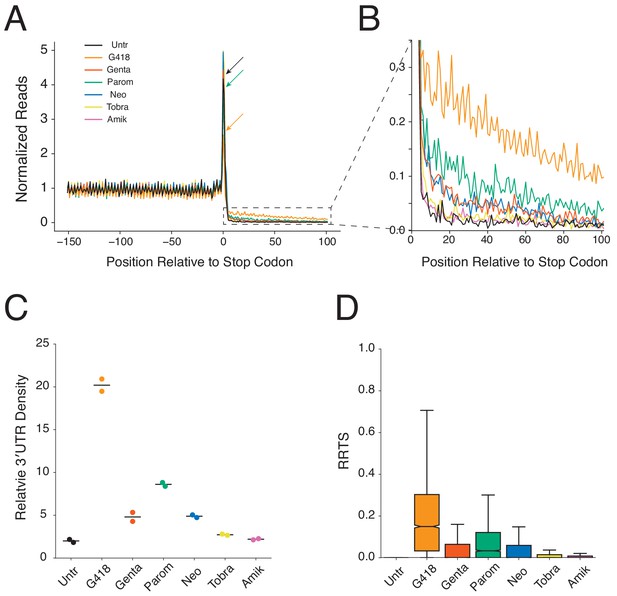
Aminoglycosides stimulate genome-wide stop codon readthrough.
(A) Average gene plot showing normalized ribosome densities relative to the distance, in nucleotides, from the stop codon at position 0. Ribosome densities from untreated cells (black), or cells treated for 24 hr with G418 (orange, 0.5 mg/mL), gentamicin (red, 0.5 mg/mL), paromomycin (green, 3 mg/mL), neomycin (blue, 2 mg/mL), tobramycin (yellow, 1 mg/mL), and amikacin (pink, 2 mg/mL) cells are overlaid. Arrows demonstrate the height of peaks at stop codons for Untr, G418, and paromomycin to facilitate comparison. (B) Magnified view of the 3′UTR showing increased densities of ribosomes in this region for AG treated cells. (C) Densities of ribosomes in 3′UTRs (from +5 to +100) are plotted relative to densities of ribosomes in the coding sequence (positions −147 to −16) for each AG. Each replicate is displayed, along with the mean value. (D) RRTS values are displayed for all genes in pooled replicates using box and whisker plots. Median values are represented with the notch indicating the 95% confidence interval, and whiskers representing 1.5 times the interquartile range. Outliers are not shown.
-
Figure 2—source data 1
Source data from ribosome profiling analysis used in Figure 2.
- https://cdn.elifesciences.org/articles/52611/elife-52611-fig2-data1-v2.xlsx
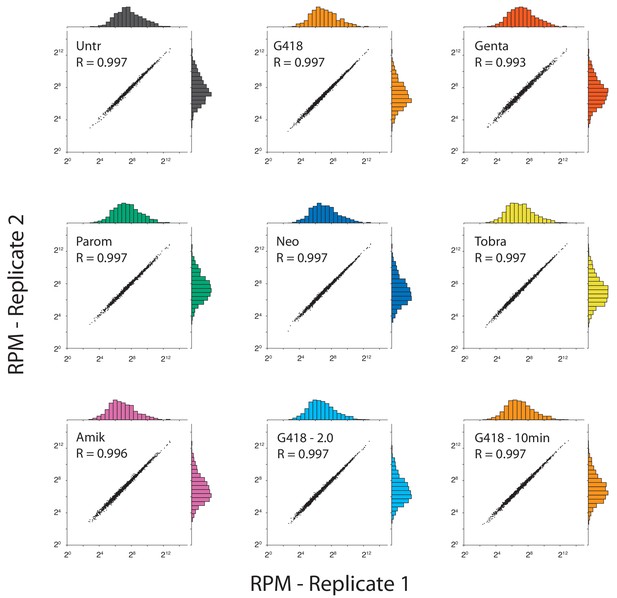
Ribosome profiling sample replicates are well correlated.
CDS densities are shown comparing replicates for each AG treatment. Densities were sorted into histograms with 20 bins each, and plotted on the corresponding axis for each sample. In addition to the samples analyzed in Figure 2, a higher concentration of G418 (2.0 mg/mL, light blue) and a 10-minute G418 treatment (0.5 mg/mL, orange) were also tested. Pearson correlations of log-transformed CDS densities show very strong correlations (R > 0.99) as expected.
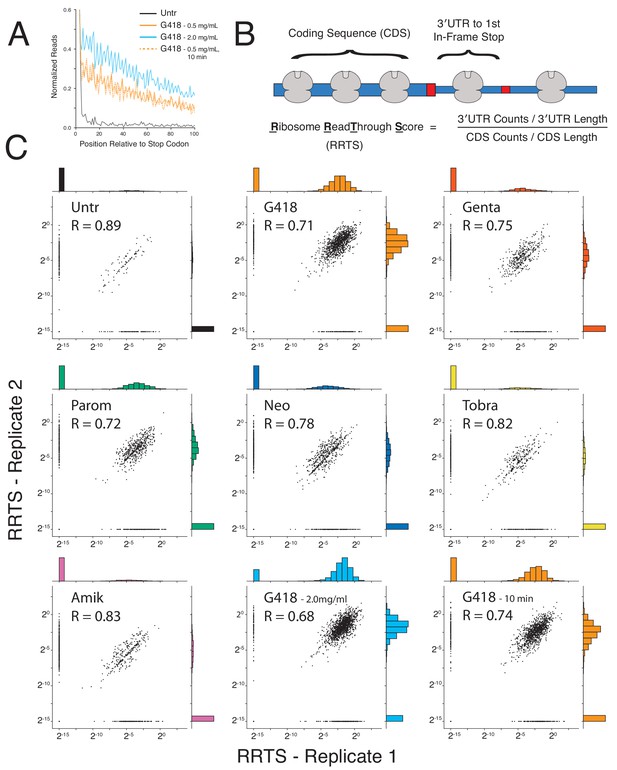
RRTS values are well correlated among replicates.
(A) Average gene plot 3′UTR regions as in Figure 2B of untreated cells (black) or cells treated with G418 (24 hr – 0.5 mg/mL – orange solid line, 24 hr – 2.0 mg/mL – cyan, and 10 min – 0.5 mg/mL – orange dashed line). (B) Ribosome ReadThrough Scores were calculated for all genes by computing the density of ribosomes in the 3′UTR between the NTC and the first in-frame stop codon and dividing this value by the density of ribosomes in the coding sequence. (C) Scatterplots showing log-transformed RRTS values were compared between replicates. Transcripts with RRTS values of zero were assigned the arbitrarily small value of 2−15 and not included when calculating correlations. Histograms were generated for each sample by sorting RRTS values into 20 bins. Pearson correlations reveal strong correlation of this metric between biological replicates (R > 0.7).
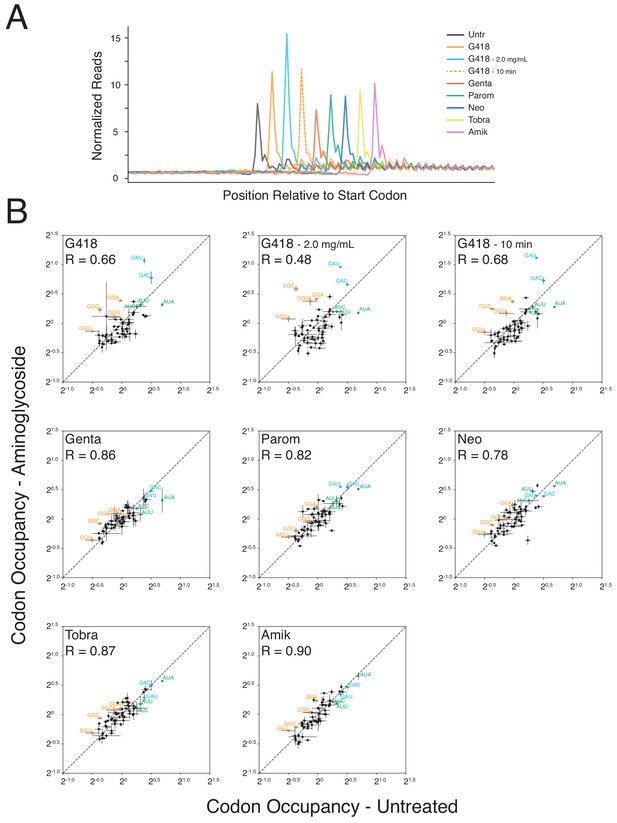
Aminoglycosides perturb translation initiation and elongation.
(A) Average gene plot showing normalized ribosome densities relative to the distance of the start codon for untreated cells (black), or cells treated with G418 (orange - 0.5 mg/mL, cyan - 2.0 mg/mL, dashed orange – 0.5 mg/mL - 10 min), gentamicin (red - 0.5 mg/mL), paromomycin (green - 3 mg/mL), neomycin (blue - 2 mg/mL), tobramycin (yellow - 1 mg/mL), and amikacin (pink - 2 mg/mL). All cells were treated for 24 hr with the exception of the 10-minute G418 treatment. Each trace is shifted to the right by six nucleotides (two codons) to facilitate comparison of start codon peaks. (B) Codon occupancies are plotted for aminoglycoside treatments (y-axis) compared to untreated cells (x-axis). The average occupancy between two biological replicates is plotted for all 61 sense codons with error bars indicating the standard deviation of these measurements. Glycine (orange), aspartic acid (blue) and isoleucine (green) codons are highlighted, and Pearson correlations are displayed for each treatment.
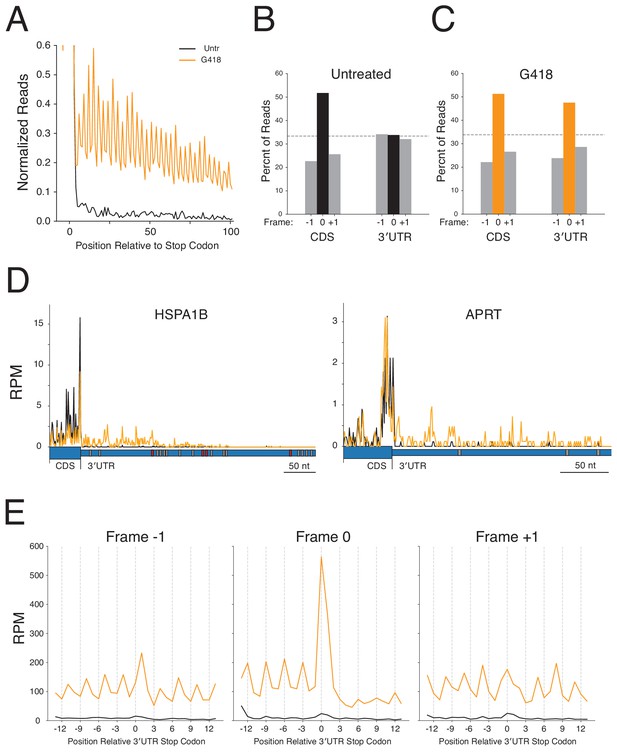
3′UTR ribosomes in G418-treated cells derive from stop codon readthrough.
(A) Average gene plot showing increased density of ribosomes in 3′UTRs in G418-treated cells (orange) relative to untreated cells (black). Reading frame is analyzed for (B) Untreated and (C) G418-treated cells showing the percent of ribosomes in a given frame in the CDS and 3′UTR. (D) Gene models of HSPA1B (left) and APRT (right) showing translation of the 3′UTRs of these genes. G418-treated cells (orange lines) are overlaid onto untreated cells (black lines). The wider blue bar below the plot indicates the CDS and the narrow blue bar represents the 3′UTR. In-frame 3′TCs are colored in red, while out-of-frame 3′TCs are colored in gray. (E) Average gene plots show total ribosome density in the region surrounding the first in-frame 3′TC when found in frame −1 (left), frame 0 (center), or frame +1 (right). Transcripts with additional 3′TCs in this window were excluded for this analysis.
-
Figure 3—source data 1
Source data from ribosome profiling analysis used in Figure 3 and Figure 4.
- https://cdn.elifesciences.org/articles/52611/elife-52611-fig3-data1-v2.xlsx
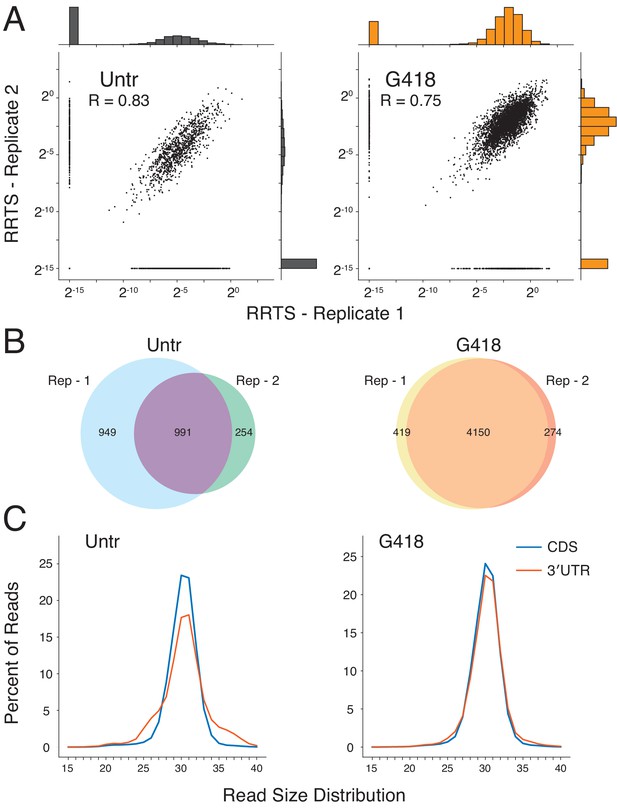
Defining features of RPFs in deeper sequencing libraries.
(A) RRTS correlations were compared between biological replicates from ribosome profiling libraries with greater sequencing depth. Pearson correlations indicate strong correlation between replicates (R > 0.7). (B) Venn diagrams indicate the total number of transcripts with non-zero RRTS values for both replicates of untreated (left) and G418-treated (right) cells and the reproducibility of readthrough detection between replicates. (C) Read size distributions showing strong agreement of read lengths between the CDS (blue) and the 3′UTR (red).
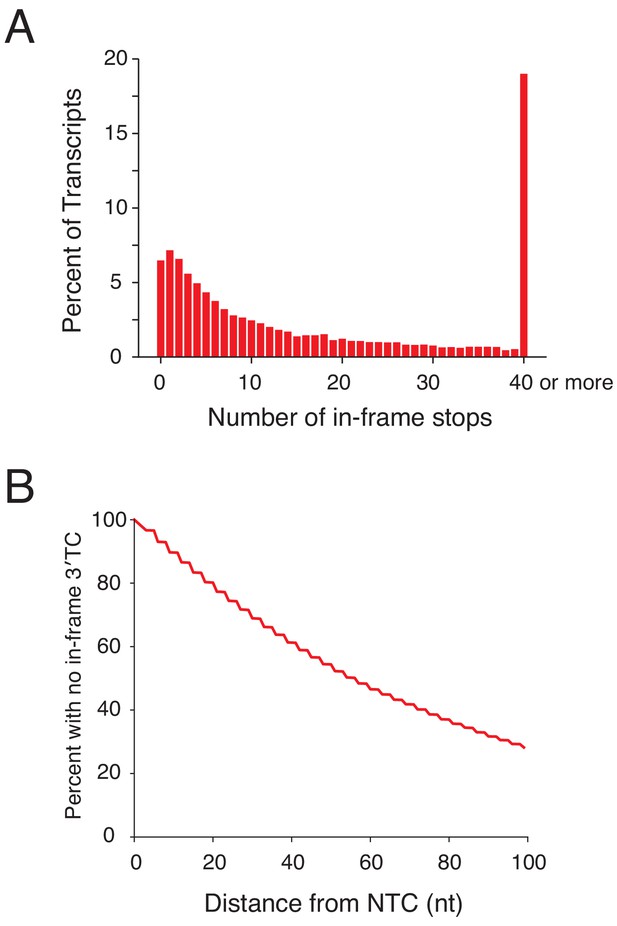
Analysis of in-frame stop codons in 3′UTRs genome-wide.
(A) All transcripts were sorted by the number of in-frame 3′TCs present in 3′UTRs. Approximately 7% of transcripts have no in frame termination codon. Transcripts with greater than 40 in-frame 3′TCs were pooled into the final bin. (B) A survival curve demonstrating the percent of transcripts that have not yet encountered an in-frame 3′TC, as a function of distance from the NTC in nucleotides.
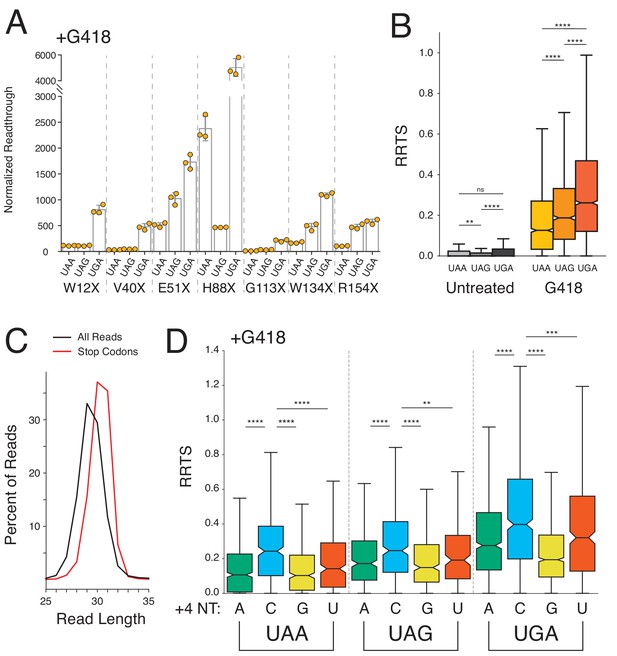
Stop codon identity influences readthrough probability genome-wide.
(A) Luciferase activity in HEK293T cells treated with 0.5 mg/mL G418 for 24 hr was measured for seven different PTCs (W12X, V40X, E51X, H88X, G113X, W134X, and R154X) in NLuc testing all three possible stop codons at each position. Normalized readthrough values - NLuc/FLuc ratios normalized to the lowest NLuc/FLuc ratio in untreated cells (Figure 4—figure supplement 1A, V40X-UAG), are plotted for each stop codon. Experiments were performed in triplicate with error bars representing one standard deviation. (B) Box and whisker plot showing RRTS values of transcripts sorted by NTC identity. Two-sided Mann-Whitney U tests were performed to test for significant differences between groups of transcripts. (C) Read size distributions comparing lengths of reads at stop codons (red) to all reads (black) in untreated cells. (D) Distribution of RRTS values, in G418-treated cells, comparing the effect of the 4 nt termination codon on readthrough. Within each 3 nt stop codon, presence of a C at the +4 position significantly increases RRTS values (One-sided Mann-Whitney U test, p<0.01 for all comparisons). Outliers are not shown.
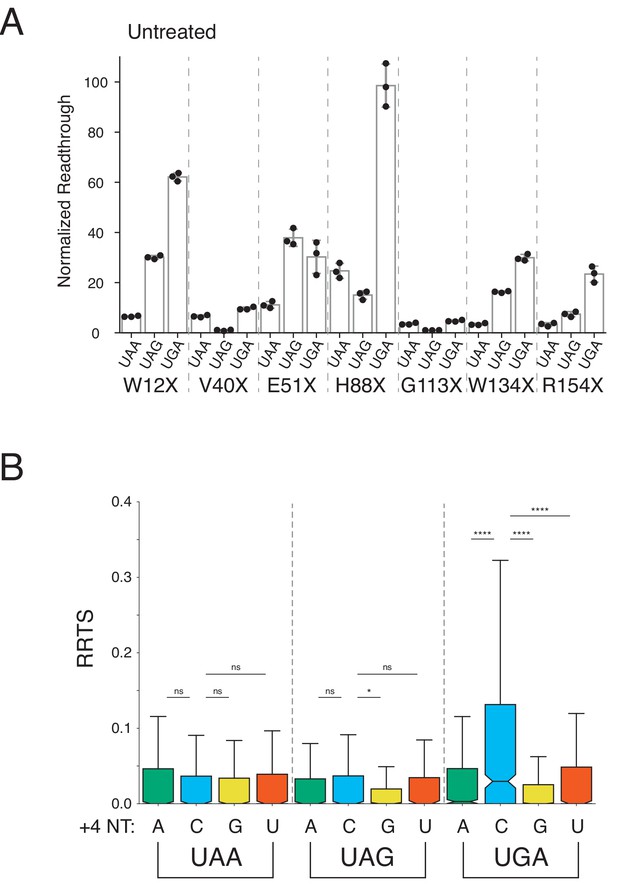
Stop codon identity influences readthrough in untreated cells.
(A) Luciferase activity in untreated HEK293T cells was measured for the same PTCs as in Figure 4A. NLuc/FLuc ratios are normalized to the stop codon with the lowest average signal, V40X-UAG. (B) RRTS values were calculated for untreated cells, as in Figure 4D. One-sided Mann-Whitney U tests were performed testing whether transcripts with a C at the +4 position had higher RRTS values. UGAC had significantly higher RRTS values than all other UGA codons. For other stop codons, the only significant difference was UAGC to UAGG. Outliers are not shown.
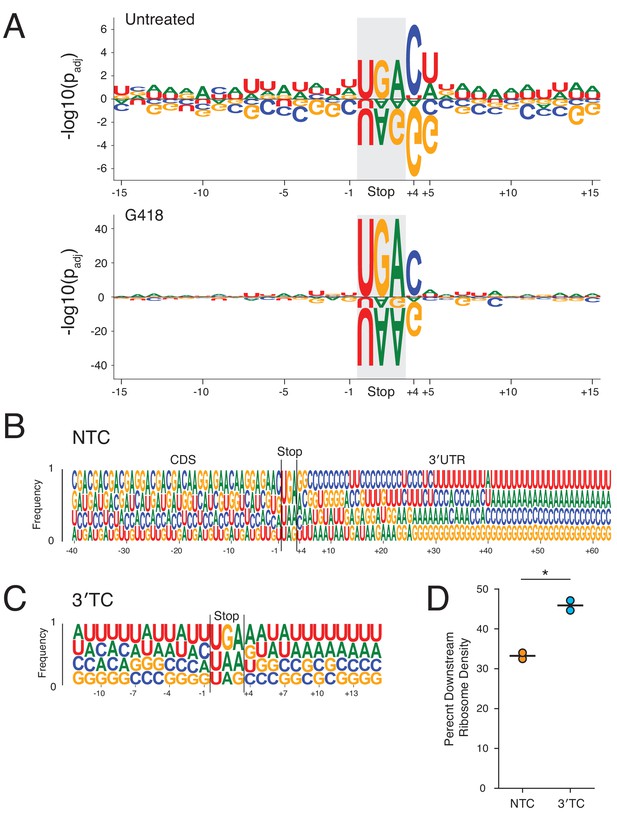
Surrounding sequence context influences stop codon readthrough genome-wide.
(A) Within a sequence window corresponding to the footprint of a translating ribosome at the NTC (15 nt upstream to 12 nt downstream), the likelihood of each nucleotide increasing or decreasing RRTS is plotted with positive values indicating more readthrough and negative values indicating less readthrough. Each nucleotide was tested using one-sided t-tests against all other nucleotides at each position for untreated (top) and G418-treated (bottom) cells. P values were adjusted using the Benjamini-Hochberg correction. Letters are scaled in proportion to the adjusted P value. (B) The frequencies of each nucleotide are plotted for all positions 40 nt upstream to 60 nt downstream of the stop codon. Nucleotides are plotted in order of increasing frequencies. (C) As in (B), nucleotide frequencies are plotted for first in-frame 3′TCs, 12 nt upstream to 12 nt downstream of the 3′TC. (D) Using normalized ribosome densities in a window 30 nt upstream to 30 nt downstream of stop codons, ribosome density downstream of stop codons was calculated for NTCs and the first in-frame 3′TCs in G418-treated cells. A paired t-test was performed revealing significant differences between levels of SCR between 3′TCs and NTCs (p=0.02).
-
Figure 5—source data 1
Source data from ribosome profiling analysis used in Figure 5.
- https://cdn.elifesciences.org/articles/52611/elife-52611-fig5-data1-v2.xlsx
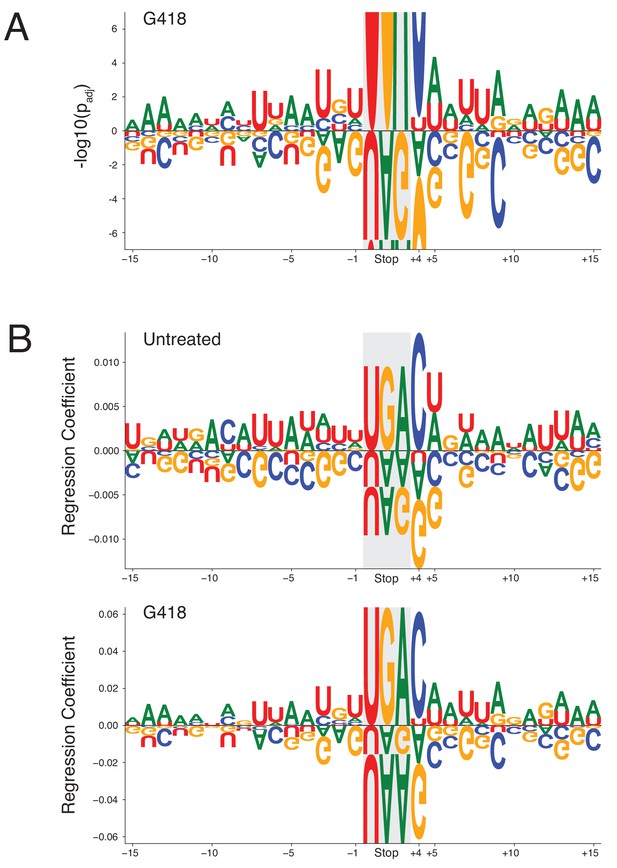
Linear regression analysis of stop codon readthrough.
(A) A magnified view of Figure 5A (bottom) is shown to facilitate comparison between untreated and G418-treated cells. (B) Ridge regressions were performed to calculate regression coefficients for each nucleotide in the same sequence window as in Figure 5A (15 nt upstream to 12 nt downstream of the NTC). The three nucleotides of the stop codon were encoded by a single position. Positive values indicate increased likelihood of readthrough while negative values indicate decreased likelihood of readthrough. Letters are scaled to the size of the regression coefficient. Regression coefficients were calculated for untreated (top) and G418-treated cells (bottom).
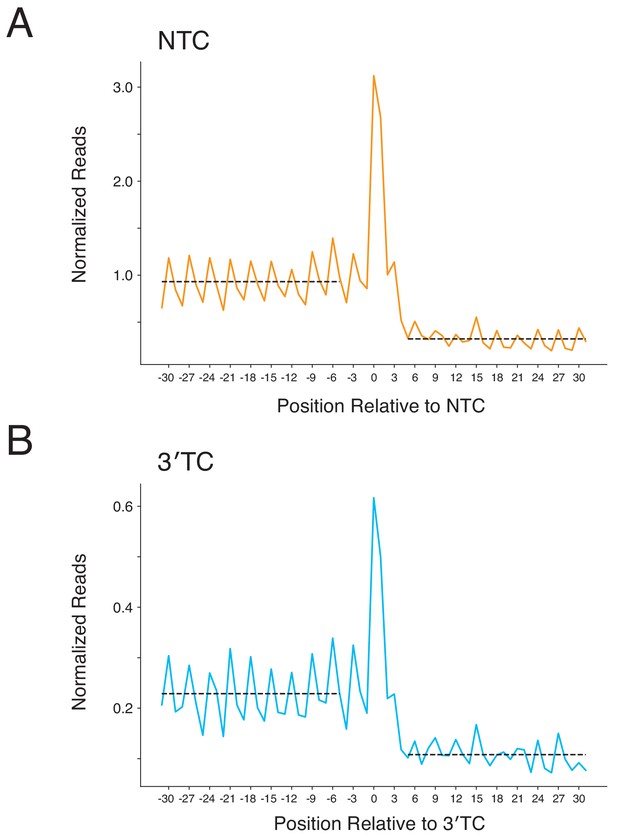
Metagene plots for readthrough at NTCs and 3′TCs.
(A–B) Normalized ribosome densities were calculated in a window 30 nt upstream to 30 nt downstream of NTCs (A) and 3′TCs (B). Average ribosome densities upstream and downstream of the stop codons were calculated, and are indicated by dashed lines for these regions. These densities were used for calculations in Figure 5D. The codons before and after the stop codon were excluded from this analysis.
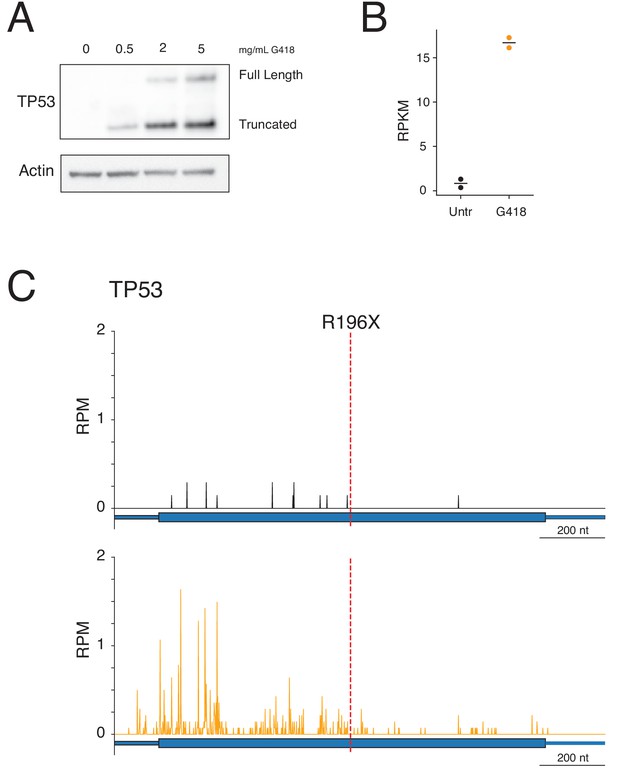
G418 stimulates readthrough of a PTC in TP53.
(A) Western blot showing TP53 protein levels (top) following 24-hour G418 treatment in Calu-6 cells harboring a nonsense mutation in TP53 at R196X. Full-length protein corresponds to the expected size of the readthrough product and the truncated band corresponds to translation termination at the PTC. (B) Quantification of mRNA levels in Calu-6 cells by RNA-seq in two biological replicates reveals increased mRNA abundance of TP53 following G418 treatment. (C-D) Ribosome profiling reads mapped to TP53 gene model for (C) untreated and (D) G418-treated cells. The red line at nucleotide 722 of the mRNA indicates the position of the R196X PTC.
-
Figure 6—source data 1
Source data from ribosome profiling and RNA-seq analysis used in Figure 6.
- https://cdn.elifesciences.org/articles/52611/elife-52611-fig6-data1-v2.xlsx
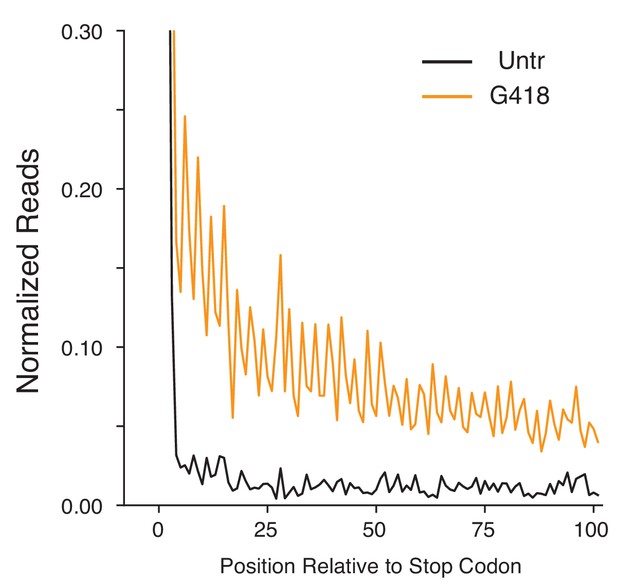
G418 stimulates genome-wide SCR in Calu-6 cells.
Average gene plot showing 3′UTR regions of Calu-6 cells as in Figure 3A. G418 (orange) increases ribosome density relative to untreated (black) cells.
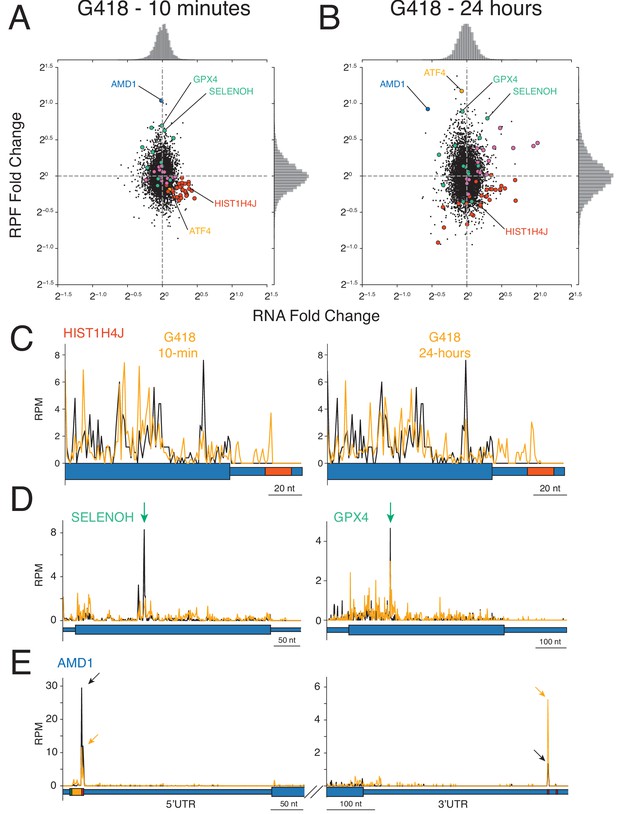
Impact of G418 on gene expression.
(A–B) Changes in mRNA abundance (x-axis) and RPF abundance (y-axis) were calculated for cells (A) treated with G418 (0.5 mg/mL) for 10 min or (B) treated with G418 (2.0 mg/mL) for 24 hr. Several noteworthy genes are highlighted including histone genes (red), selenoproteins (green), chaperone proteins and foldases (purple), ATF4 (orange), and AMD1 (blue). (C-E) Gene models show translation of several genes altered by G418 treatment (orange) relative to untreated (black) cells. Blue bars indicate the position along the mRNA with wide bars representing the CDS and narrow bars representing UTRs. (C) Translation of a representative histone gene HIST1H4J at the 10 min (left) and 24 hr (right) time points reveal that G418 induces translation into RNA hairpins of histone mRNAs indicated by the red box in the 3′UTR. (D) Translation of two selenoproteins, SELENOH (left) and GPX4 (right). Green arrows indicate UGA codons normally decoded by selenocysteine tRNAs. (E) Translation of AMD1 is altered by G418 treatment. Reduced peaks of ribosomes stalled in the 5′UTR at the stop codon of the uORF encoding the MAGDIS peptide (left). Arrows indicate the height of peaks at the stop codon for untreated (black) and G418-treated (orange) cells to facilitate comparison. The boxes depicting the start (green) and stop (red) codons and coding sequence (orange) of the uORF are enlarged. G418 treatment increases the peak of ribosomes found in the 3′UTR of AMD1 (right, compare orange arrow to black arrow). No in-frame 3′TCs are present between the NTC and stalling site.
-
Figure 7—source data 1
Source data from ribosome profiling and RNA-seq analysis used in Figure 7.
- https://cdn.elifesciences.org/articles/52611/elife-52611-fig7-data1-v2.xlsx
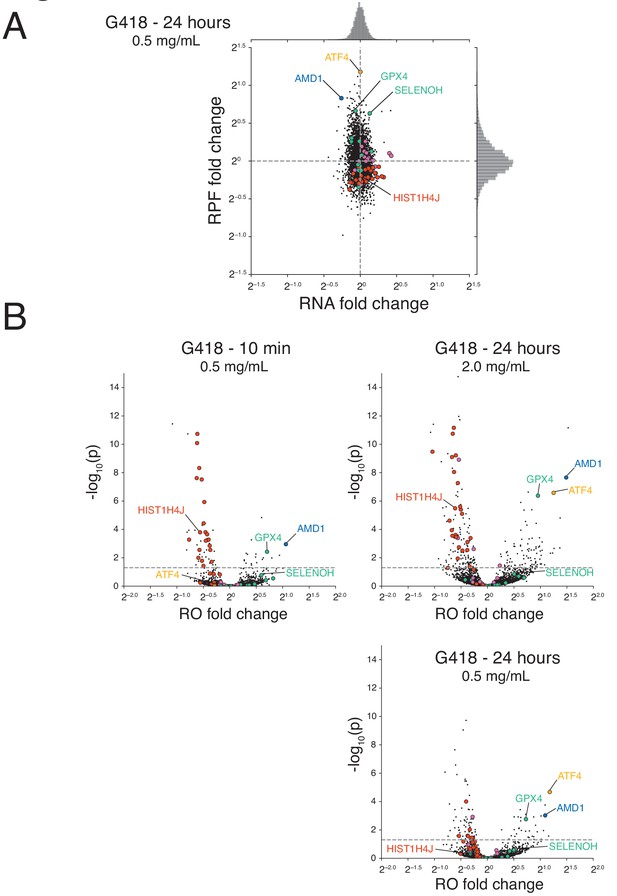
Statistical analysis indicating that G418 alters ribosome occupancy of a subset of genes.
(A) Changes in RNA and translation (RPF) levels are plotted for cells treated with 0.5 mg/mL G418 for 24 hr. As in Figure 5A and B, histones (red), selenoproteins (green), chaperone proteins and foldases (purple), ATF4 (orange), and AMD1 (blue) are highlighted. (B) Changes in ribosome occupancies (ROs) of G418-treated cells are plotted according to the magnitude of the fold change (x-axis) and the significance of the change (y-axis) calculated using Xtail. Changes are compared at the 10 min (0.5 mg/mL, left) time point and the 24 hr time point (2.0 mg/mL, upper right and 0.5 mg/mL, lower right).
Additional files
-
Supplementary file 1
Key resources table.
- https://cdn.elifesciences.org/articles/52611/elife-52611-supp1-v2.docx
-
Supplementary file 2
RefSeq Identifiers of sequences used for rRNA depletion.
- https://cdn.elifesciences.org/articles/52611/elife-52611-supp2-v2.docx
-
Supplementary file 3
Statistical test results.
- https://cdn.elifesciences.org/articles/52611/elife-52611-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52611/elife-52611-transrepform-v2.docx





