Cell-specific non-canonical amino acid labelling identifies changes in the de novo proteome during memory formation
Figures
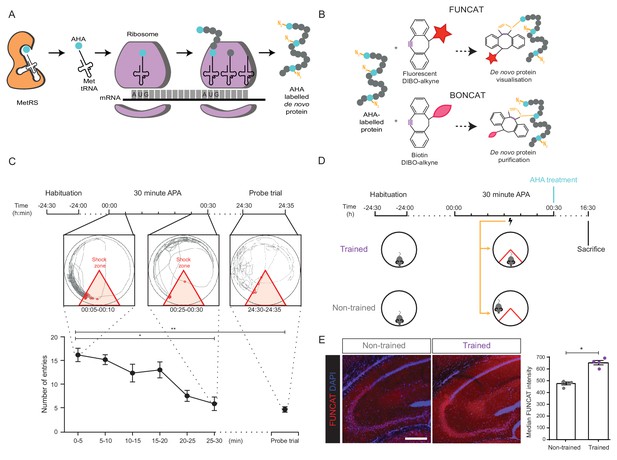
Long-term memory formation following training using the 30 min APA paradigm results in increased hippocampal de novo protein synthesis.
(A) AHA is recognised by mouse methionine tRNA synthetase, MetRS (Mars1), and labels de novo synthesised proteins at amino-terminal and internal methionine residues using the endogenous translational machinery. (B) NCAA-labelled proteins can be covalently bonded to various tags through reaction of the azide group (orange) of the NCAA with the alkyne group (purple) of the tag. This enables NCAA-labelled proteins to either be visualised using fluorescent non-canonical amino acid tagging (FUNCAT) or to be purified using bio-orthogonal non-canonical amino acid tagging (BONCAT). (C) The 30 min APA paradigm results in spatial long-term memory formation. Mice trained over 30 min learned to avoid a designated shock zone (red), with significantly fewer entries into the shock zone being recorded between 25–30 min compared to between 0–5 min. In a 5 min probe trial held 24 hr after training, mice continued to avoid entering the shock zone, even in the absence of shocks, indicative of the formation of spatial LTM (n = 6 mice, one-way ANOVA, Dunnett’s MCT, *p≤0.05, **p≤0.01). (D) Scheme of 30 min APA task for trained and non-trained mice. Trained mice received foot shocks upon entry into the designated shock zone, while non-trained mice received foot shocks at the same time as their trained partner and were therefore unable to undergo spatial LTM formation. Upon completion of the 30 min APA, mice were administered AHA and were perfused 16 hr later without undergoing a probe trial. (E) A significant increase in protein synthesis was observed in the hippocampus of trained compared to non-trained mice using FUNCAT (n = 4 mice, three sections per mouse, Student’s paired t-test, *p≤0.05). Scale bar = 400 µm.
-
Figure 1—source data 1
WT 30 minute APA behavioural data.
- https://cdn.elifesciences.org/articles/52990/elife-52990-fig1-data1-v1.xlsx
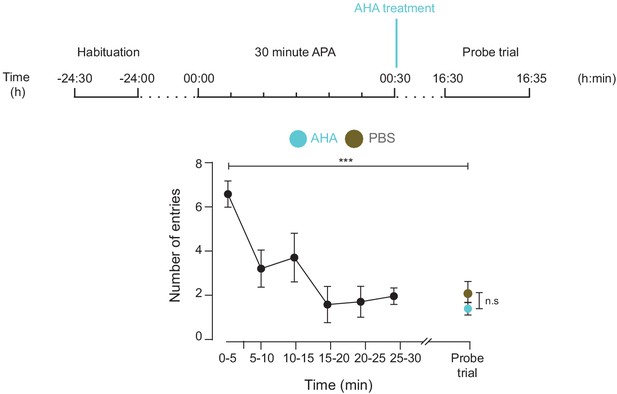
AHA treatment does not inhibit spatial LTM formation.
Both mice treated with either AHA or PBS immediately after training using the 30 min APA test could recall the location of the shock zone in a probe trial conducted 16 hr after training (n = 4 mice, two way ANOVA, Dunnett’s MCT, ***p≤0.001). There was no significant difference in performance in this probe trial between the two groups (two-way ANOVA, Sidak’s MCT, n.s. = non significant, p=0.85).
-
Figure 1—figure supplement 1—source data 1
AHA treated 30 minute APA behavioural data.
- https://cdn.elifesciences.org/articles/52990/elife-52990-fig1-figsupp1-data1-v1.xlsx
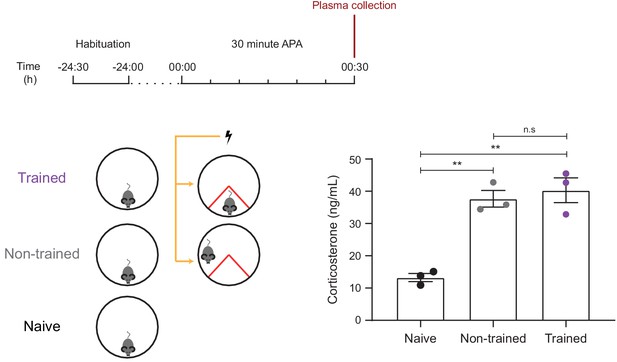
Trained and non-trained mice show similar levels of stress, as measured by plasma corticosterone levels.
Both trained and non-trained mice show increased plasma levels of corticosterone (a marker of increased stress) compared to a naïve controls which exposed to the 30 min APA arena without receiving foot shocks (n = 3 mice, one-way ANOVA, Tukey’s MCT, **p≤0.01). There was no significant difference in plasma corticosterone levels between trained and non-trained mice (n.s. = non significant, p=0.78).
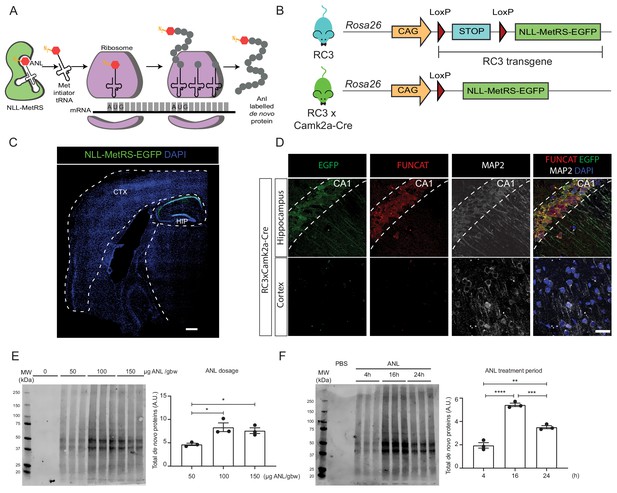
Rosa26 Cre Click-chemistry (RC3) mice enable cell type-specific labelling of newly synthesised proteins.
(A) Incorporation of ANL requires the expression of NLL-MetRS, which allows this NCAA to be incorporated at the amino-terminus as proteins are being synthesized. (B) In RC3 mice, the expression of NLL-MetRS-EGFP is prevented by an upstream FLOX-STOP cassette. In RC3xCamk2a-Cre mice, Cre-recombinase is expressed in the hippocampus, resulting in the excision of the FLOX-STOP cassette from the RC3 transgene. This enables expression of NLL-MetRS-EGFP restricted to hippocampal neurons. (C) Immunohistochemical analysis confirmed Cre-specificity of NLL-MetRS-EGFP expression in RC3xCamk2a-Cre mice, with the EGFP signal being confined to neurons in the CA1, CA2, CA2 and dentate gyrus regions of the hippocampus. Scale bar = 400 µm (D) FUNCAT staining confirms that ANL integration was restricted to hippocampal neurons in RC3xCamk2a-Cre mice. Scale bar = 40 µm. (E) BONCAT-WB analysis of RC3xCamk2a-Cre mice reveals maximal ANL labelling when mice were administered 100 µg ANL/gbw via i.p. injection (n = 3 mice, one way ANOVA, Tukey’s multiple comparison test, *p≤0.05). (F) BONCAT-WB analysis of RC3xCamk2a-Cre mice reveals maximal ANL labelling approximately 16 hr post injection (n = 3 mice, one way ANOVA, Tukey’s multiple comparison test **p≤0.01, ***p≤0.001, ****p≤0.0001).
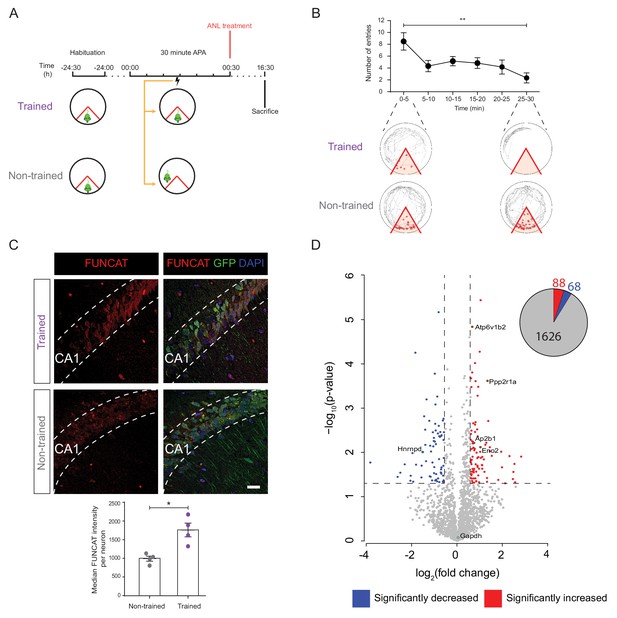
De novo proteomic analysis reveals altered hippocampal synthesis of selected proteins during spatial LTM formation.
(A) Schematic of the 30 min APA task for trained and non-trained (yoked) RC3xCamk2-Cre mice. Trained mice received foot shocks upon entry into the designated shock zone, while non-trained mice were paired by receiving foot shocks at the same time as their trained partner. Upon completion of the 30 min APA task, mice were administered ANL and then perfused 16 hr later. (B) RC3xCamk2a-Cre mice trained in the 30 min APA task reduced the number of entries (red circles) into the shock zone (red) over the 30 min training period (n = 4 mice, one-way ANOVA, Dunnett’s MCT, **p≤0.01). (C) Following spatial LTM formation, total protein synthesis was significantly increased in the hippocampal neurons of RC3xCamk2a-Cre mice. This was reflected by the increased FUNCAT signal observed in trained compared to non-trained mice (n = 4 mice, 30 neurons per mouse, Student’s paired t-test, *p≤0.05). Scale bar = 40 µm. (D) Volcano plot representing the relative abundance of de novo synthesised proteins in the hippocampus of trained and non-trained RC3xCam2ka-Cre mice. In total, 1782 proteins were quantified in four trained and non-trained mice each, using BONCAT-SWATH-MS. Proteins which were significantly increased in synthesised in trained mice (fold-change ≥1.5, p≤0.05) are shown in red, whereas proteins which exhibited significantly decreased synthesis (fold-change ≤0.66, p≤0.05) are shown in blue (n = 4 mice, Student’s t-test). Subsequently validated proteins are encircled in green (see Figure 5).
-
Figure 3—source data 1
ANL treated 30 minute APA behavioural data for protemic analysis.
- https://cdn.elifesciences.org/articles/52990/elife-52990-fig3-data1-v1.xlsx
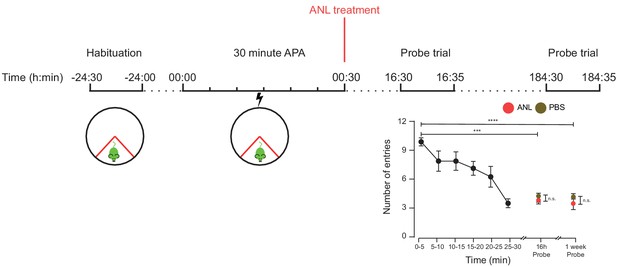
RC3xCamk2a-Cre mice treated with ANL and trained in the 30 min APA task were able to recall the location of the shock zone a week after training.
RC3xCamk2a-Cre treated with either ANL or PBS immediately after training showed a reduced number of shock zone entries in probe trials held 16 hr and one week following training (n = 4 mice, two way ANOVA, Dunnett’s MCT, ***p≤0.001, ****p≤0.0001). There was no significant difference in probe trial performance between ANL- and PBS-treated mice (two-way ANOVA, Sidak’s MCT, n.s. = non significant, p=0.99).
-
Figure 3—figure supplement 1—source data 1
ANL treated 30 minute APA behavioural data.
- https://cdn.elifesciences.org/articles/52990/elife-52990-fig3-figsupp1-data1-v1.xlsx
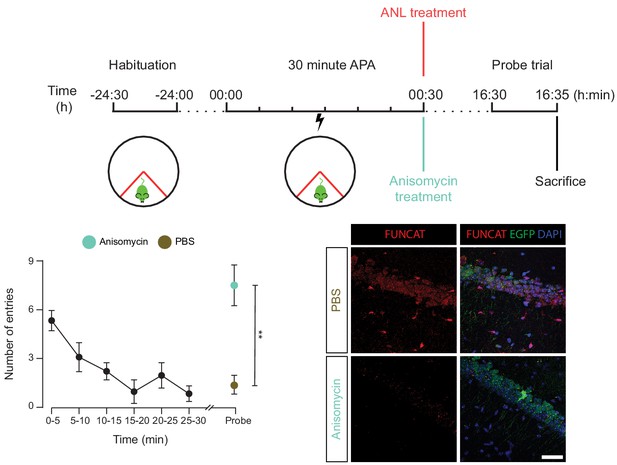
The spatial long-term memory formation observed in the 30 min APA task is dependent upon protein synthesis.
RC3xCamk2a-Cre mice were trained with the 30 min APA task, then administered ANL and then either the protein synthesis inhibitor anisomycin, or PBS. Mice were subsequently tested for their ability to form spatial LTMs using a probe trial held 16 hr following training. Unlike the PBS-treated control, anisomycin-treated mice did not show a reduced number of shock zone entries in the probe trail (n = 4 mice, two way ANOVA, Dunnett’s MCT, n.s. = non significant, p=0.51), and had significantly more shock zone entries than the PBS control (two-way ANOVA, Sidak’s MCT, **p≤0.01), indicating that anisomycin-treatment blocked spatial LTM formation. Anisomcyin-treatment was confirmed to block hippocampal protein synthesis using FUNCAT staining.
-
Figure 3—figure supplement 2—source data 1
Anisomycin treated 30 minute APA behavioural data.
- https://cdn.elifesciences.org/articles/52990/elife-52990-fig3-figsupp2-data1-v1.xlsx
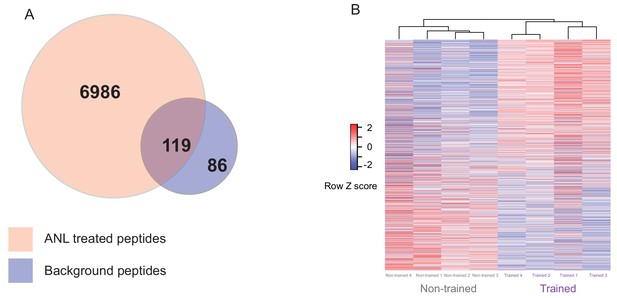
Proteomic analysis of Anl labelled proteins from trained and non-trained RC3xCamk2a-Cre mice.
(A) The overlap of peptides purified by BONCAT from ANL-treated RC3xCamk2a mice compared to background. Using 2D LC MS/MS, 7105 unique peptides were identified from ANL-treated trained and non-trained RC3xCamk2a-Cre mice following BONCAT purification. In order to control for background purification, we removed 119 peptides which had been identified in BONCAT purification from PBS-treated negative control animals. The remaining 6986 peptides were then used to form a custom peptide library for SWATH-MS analysis. (B) Heatmap of the relative abundance in individual samples of the 1782 de novo synthesised proteins from trained and non-trained mice. Euclidian cluster analysis revealed that trained RC3xCamk2a-Cre mice cluster separately from their non-trained RC3xCamk2a-Cre pairs.
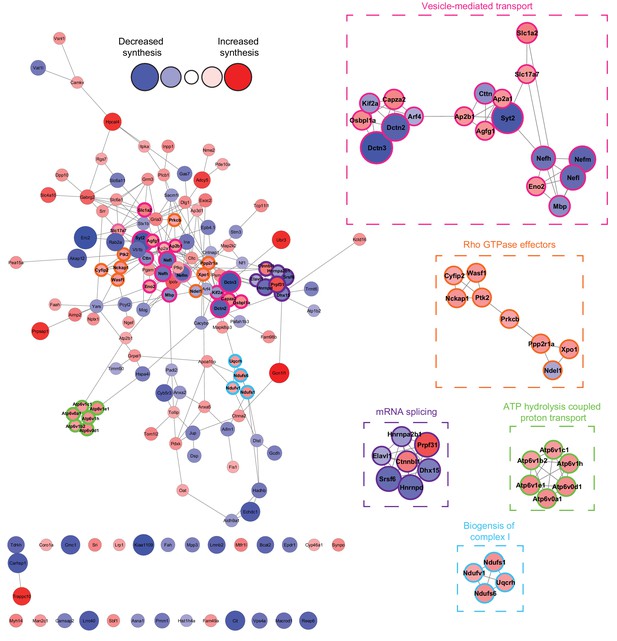
Network analysis reveals that the synthesis of distinct clusters of proteins is significantly altered during spatial LTM formation.
Network analysis using the STRING database reveals that of the 156 proteins identified to be altered in synthesis during spatial LTM formation (|fold-change| ≥ 1.5, p≤0.05), 125 (≈80%) showed evidence of interaction (STRING score cut off ≥0.4) with at least one other significantly regulated protein, forming a highly interconnected network. Within this network, there was a median of 4 interactions per node. MCODE cluster analysis revealed the presence of 5 distinct clusters which were associated with mRNA splicing, ATP hydrolysis-coupled proton transport, vesicle-mediated transport, biogenesis of mitochondrial complex I, and Rho GTPase effectors. Proteins in clusters are depicted by a coloured border and are magnified in the inserts. The distance between each node is representative of the STRING score. Proteins which did not display interactions are not shown. The absolute fold-change is represented by the node size, and the directionality of the fold-change by the node colour.
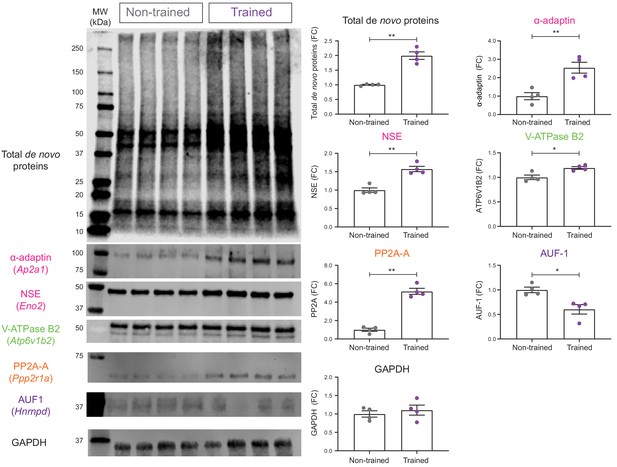
Validation of de novo proteomic comparison of trained vs non-trained.
RC3xCamk2a-Cre mice using BONCAT western blotting BONCAT-WB confirms that in trained mice, there is increased hippocampal synthesis of α- α-adaptin (Ap2a1), neuron specific enolase (NSE: Eno2), V-ATPase subunit B2 (V-ATPase B2: Atp6v1b2), and the α isoform of the structural subunit of protein phosphatase 2A (PP2A-A: Ppp2r1a) compared to non-trained controls. Synthesis of the ARE binding protein ARE/poly(U)-binding/degradation factor 1 (AUF-1: Hnrnpd) was also confirmed to be decreased during spatial LTM formation. The synthesis of the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was unchanged in the hippocampal neurons of trained RC3xCamk2a-Cre mice compared to non-trained controls (p=0.48), together validating the SWATH analysis (n = 4 mice, Student’s paired t-test, *p≤0.05, **p≤0.01, ***p≤0.001).
Complete images of the western blots shown in Figure 5.
Additional files
-
Supplementary file 1
List of peptides identified by 2D-LC MS/MS from BONCAT purified from ANL-treated RC3xCamk2a-Cre mice and PBS-treated negative controls.
- https://cdn.elifesciences.org/articles/52990/elife-52990-supp1-v1.xlsx
-
Supplementary file 2
SWATH-MS data from quantification of BONCAT purified proteins from trained and non-trained RC3xCamk2a-Cre.
For each of the 1782 proteins quantified, the name, Uniprot identifier, gene name, fold change in trained compared to non-trained mice, p-value, Benjamini and Hochberg adjusted p-value, number of peptides sequences used for quantified, and the normalised protein peak area are given. Proteins contained within clusters identified by MCODE analysis are highlighted.
- https://cdn.elifesciences.org/articles/52990/elife-52990-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/52990/elife-52990-transrepform-v1.pdf





