Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite
Figures
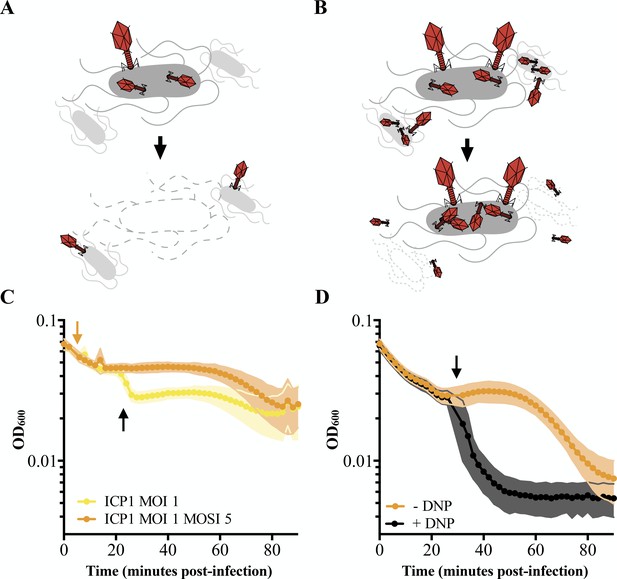
Characterizing lysis inhibition.
Schematic of T4 infection of E. coli. (A) At low multiplicities of infection (MOI), available E. coli are readily infected by T4 (red). Under these conditions, an infected cell is hijacked to produce progeny phage and lyses releasing the phages into the environment where they go on to infect neighboring cells. (B) At high MOIs, phage outnumber the hosts resulting in initial infection by phage followed by secondary superinfection (second phage infecting the same cell). Superinfection delays lysis through LIN, enabling the production of more virions and protecting progeny phage from an environment devoid of uninfected hosts. (C) ICP1 exhibits LIN at intermediate multiplicities of infection and upon superinfection. PLE (-) V. cholerae infected with ICP1 at MOI = 1 demonstrate a lysis event 20 minutes post-infection (black arrow) before the optical density (OD600) stabilizes. Superinfection (orange arrow) of a culture four minutes post-infection with ICP1 at MOI = 1 with ICP1 at a multiplicity of superinfection (MOSI) of 5 triggers lysis inhibition and stabilizes the OD600 before a lysis event can occur. Data from three biological replicates are shown. (D) ICP1 LIN is sensitive to chemical collapse. PLE (-) V. cholerae infected with ICP1 at MOI = 5 maintain OD600 for an extended period but the OD600 collapses when 2,4-dinitrophenol (DNP) is added (black arrow). Data from four biological replicates are shown. For all graphs, points show the average of replicates; shading shows the standard deviation.
-
Figure 1—source data 1
This spreadsheet contains the data used to create Figure 1C and D.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig1-data1-v1.xlsx
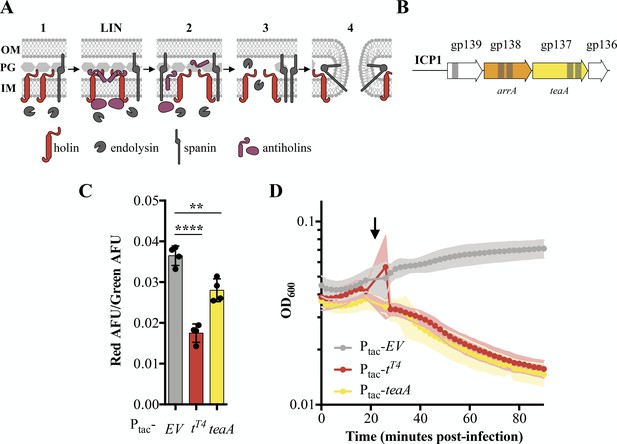
Identification and characterization of ICP1’s holin TeaA.
(A) Schematic of canonical T4 lysis in E. coli. Lysis occurs in four steps with a potential delay caused by lysis inhibition. Step one: Phage encoded proteins including holins, endolysins, and spanins accumulate in the cell. In the event of superinfection, antiholins halt the lysis program causing lysis inhibition (LIN). Step two: Holins are triggered collapsing the proton motive force and forming holes in the inner membrane (IM) releasing endolysins to the periplasm. Step three: The peptidoglycan (PG) is degraded by endolysins. Step four: Spanins fuse the inner and outer membrane (OM). (B) Transmembrane domains (grey bars) were predicted in three gene products (gp) in a conserved locus of ICP1. (C) Relative state of the proton motive force as measured by the ratio of red to green arbitrary fluorescence units (AFU) from DiOC2(3) after ectopic gene expression. Points represent individual replicates; bar height is the average; error bars display the standard deviation of samples. Significance was calculated via one-way ANOVA followed by Dunnett’s test. ****p≤0.0001; **p≤0.01. (D) Chemical triggering with 2,4-dinitrophenol (arrow) after ectopic gene expression as measured by optical density (OD600). Points show the average of four replicates; shading shows the standard deviation.
-
Figure 2—source data 1
Fluorescence Source Data.
This spreadsheet contains the data used to create Figures 2C and 4C.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig2-data1-v1.xlsx
-
Figure 2—source data 2
DNP Source Data.
This spreadsheet contains the data used to create Figures 2D and 4D.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig2-data2-v1.xlsx
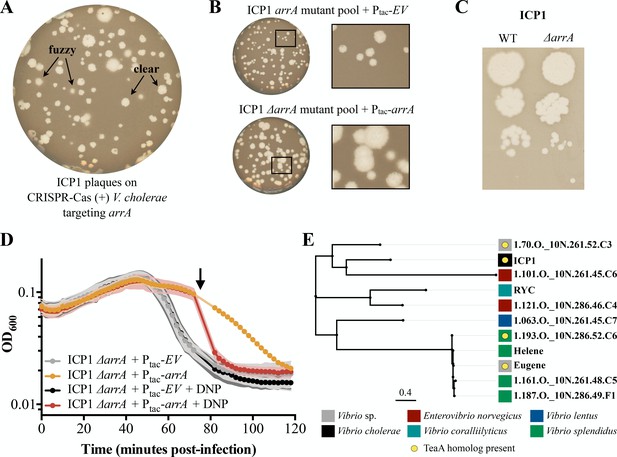
Antiholin ArrA identification and characterization.
(A) Initial plaquing of wild type ICP1 on CRISPR-Cas (+) V. cholerae targeting arrA yielded a mixture of plaque phenotypes. (B) Plaques with clear edges can be found in populations of phage that overcame targeting with various mutations in arrA when plaqued on V. cholerae harboring an empty vector (EV) control (Top). Providing ArrA in trans restores the fuzzy edge phenotypes within the mutant population. (C) Using a repair template we created a clean arrA deletion and found that ΔarrA ICP1 yield plaques with clear edges. (D) During infection, once lysis begins as observed through changes in optical density (OD600), ΔarrA ICP1 causes rapid lysis consistent with abolishment of lysis inhibition. When ArrA is expressed in trans the lysis inhibition phenotype is rescued: a small lysis event is visible 40 minutes post-infection after which the optical density is stabilized for about 30 minutes. Consistent with ArrA restoring lysis inhibition, the stabilization of optical density is sensitive to chemical disruption of the proton motive force by 2,4-dinitrophenol (DNP) (arrow). Points show the average of four or greater replicates; shading shows the standard deviation. (E) BLASTP was used to find proteins with 20% identity to ArrA over 75% of the query; these homologs are displayed in an unrooted tree displaying the name of the phage the protein is found in. Colored blocks show the identity of the host that each phage infects, and yellow circles denote that a protein with homology to TeaA is present in the phage. A table with extended information for each homolog is available in Supplementary file 3.
-
Figure 3—source data 1
This spreadsheet contains the data used to create Figure 3D.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig3-data1-v1.xlsx
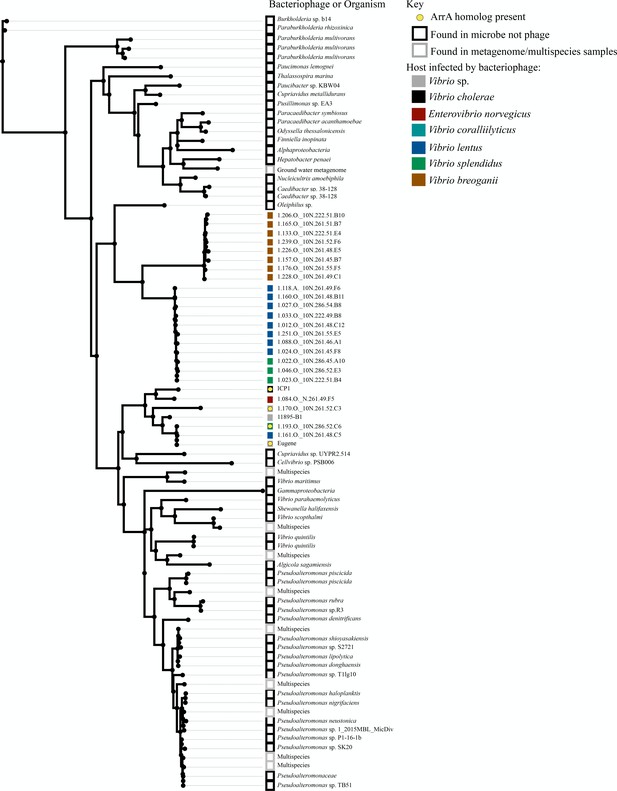
Proteins with similarity to TeaA.
This unrooted tree shows proteins identified with BLASTP that share 30% identity with TeaA over 85% of the query. The source of the sequence is shown in the label on the tree while colored blocks denote the host the phage infects or whether the sequence was found in a microbe or multispecies sequence entry and yellow circles denote that a protein with homology to ArrA is present in the phage.
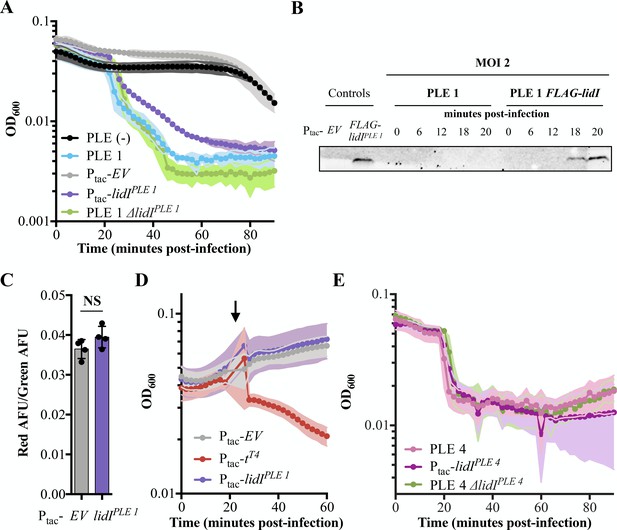
Accelerated lysis by PLE and LidI.
(A) The optical density (OD600) of ICP1 MOI = 5 infections was followed in PLE (-), PLE 1, and PLE 1 ΔlidI V. cholerae strains as well as V. cholerae strains containing induced empty vector (EV) and lidIPLE 1 constructs. Data points represent the average reading of n ≥ 3 replicates and shaded regions display the standard deviation of experiments. The LidI sequence is available in a PRALINE alignment shown in Figure 4—figure supplement 1. (B) Tagged LidIPLE 1 expressed in trans is readily observable by Western blot. Tagged LidIPLE 1 in the native PLE context is visible 18 to 20 minutes post infection with ICP1 at MOI = 2. Complete blot available in Figure 4—figure supplement 2. (C) Proton motive force as measured by the ratio of red to green arbitrary fluorescence units (AFU) from DiOC2(3) after ectopic gene expression. Points represent individual replicates; bar height is the average; error bars display the standard deviation of samples. NS signifies ‘no significance’ by two-tailed t-test. (D) Chemical holin triggering with 2,4-dinitrophenol (arrow) after ectopic gene expression as measured by OD600. Points show the average of four replicates; shading shows the standard deviation. (E) The OD600 of ICP1 at MOI = 5 infections was followed in PLE 4, and PLE 4 ΔlidI V. cholerae strains as well as V. cholerae containing the induced lidIPLE 4 construct. Data points represent the average reading of n ≥ 5 replicates and shaded regions display the standard deviation of experiments.
-
Figure 4—source data 1
This spreadsheet contains the data used to create Figure 4A and E.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig4-data1-v1.xlsx
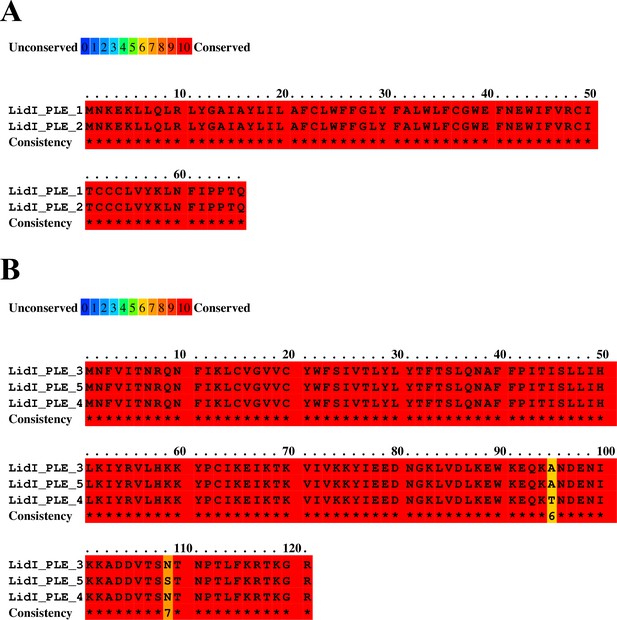
PRALINE alignment of LidI homologs.
LidI homologs cluster neatly in two groups: (A) LidIPLE 1 and LidIPLE 2 are short 66 amino acid predicted proteins with four synonymous SNPs between them, while (B) lidIPLE 3, lidIPLE 4, and lidIPLE 5 encode larger proteins at 121 amino acids in length with three SNPs that result in two amino acid substitutions.

Complete Western Blot.
Tagged LidIPLE 1 was visualized via western blot. EV and FLAG-LidIPLE 1 expressed in trans served as controls showing FLAG-LidIPLE 1 specificity as well as the presence of background bands. In PLE 1 V. cholerae, there is no detectable band similar in size to tagged LidIPLE 1 (as indicated by the arrow). When LidIPLE 1 is tagged in the native locus and strains are infected with ICP1, FLAG-LidIPLE 1 is visible late in infection (18 and 20 minutes post-infection).

Redundancy in the PLE.
(A) PLE one and PLE 1 ΔlidI were infected with ICP1 (MOI = 1) followed by superinfection (MOSI = 5; arrow) showing lysis inhibition is collapsed by PLE in the face of exogenously added phage. Every predicted PLE ORF was knocked out and each resulting strain was tested for accelerated lysis after ICP1 infection (MOI = 5) (B) and ability to inhibit plaque formation (C).
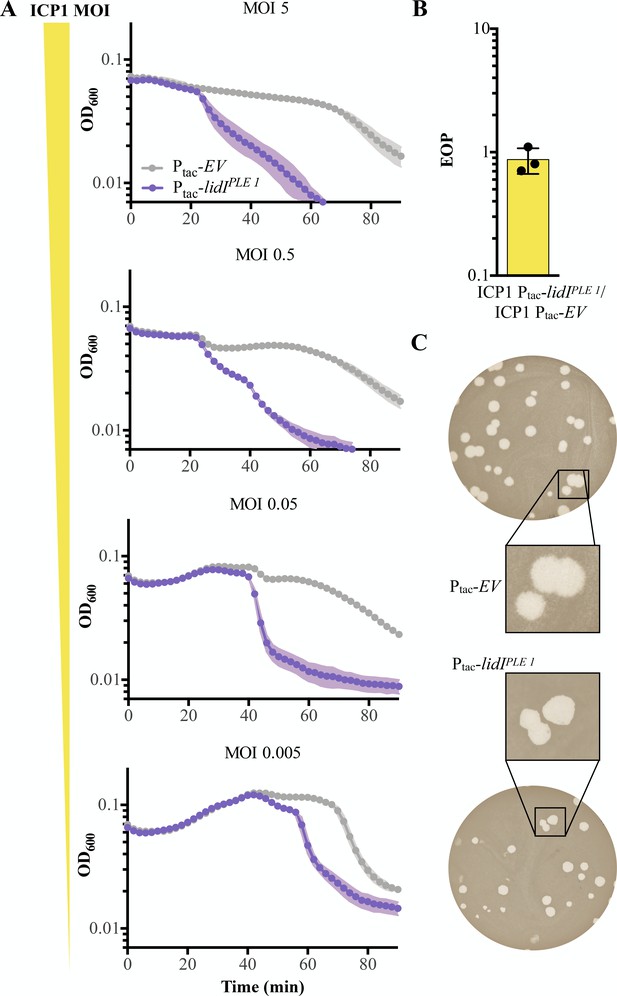
LidI functions through lysis inhibition disruption.
(A) Optical density (D600) of empty vector (EV) or LidIPLE 1 expressing cultures after infection with different initial multiplicities of infection (highest MOI top to lowest MOI bottom). Data points represent the average reading of n = 3 biological replicates and shaded regions display the standard deviation of experiments. (B) Efficiency of plaquing (EOP) for V. cholerae harboring induced lidIPLE 1 in comparison to an empty vector control (EV). Points represent individual replicates; bar height is the average; error bars display the standard deviation of samples. (C) Plaque edge phenotypes were determined for ICP1 plaques on EV (top) and lidIPLE 1 (bottom) PLE (-) V. cholerae.
-
Figure 5—source data 1
This spreadsheet contains the data used to create Figure 5.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig5-data1-v1.xlsx
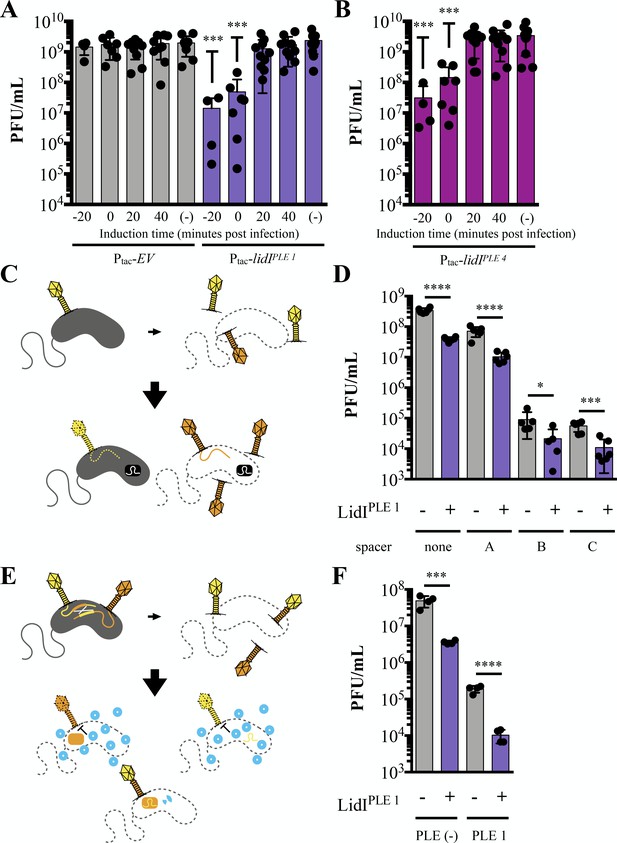
LidI puts a bottleneck on phage populations.
(A and B) Phage infection yields measured in plaque forming units per mL (PFU/mL) were determined from V. cholerae with empty vector (EV), lidIPLE 1, and lidIPLE 4 constructs induced at the specified time with respect to phage infection at multiplicity of infection of 5 or left uninduced (-). All samples were mechanically lysed 100 minutes after infection and titers were compared via one-way ANOVA followed by Dunnett’s multiple comparison test comparing all values to titers from the EV cultures. Only significant differences are noted. (C) Schematic of evolution as probed by overcoming V. cholerae-encoded CRISPR targeting. Top: cultures with induced EV and lidIPLE 1 constructs are infected with ICP1 (MOI = 0.1) and allowed to produce progeny phage before mechanical lysis. The majority of phages will be wild type (yellow) while some phages will harbor mutations (orange). Bottom: The phage populations were then plaqued on V. cholerae strains with a CRISPR-Cas system (black rectangle) and different spacers. Wild type phage infections do not result in plaques because the injected phage DNA is degraded (dotted line) while mutants that can overcome the targeting, produce progeny, lyse the cells and form plaques. Expanded schematic in Figure 6—figure supplement 2. (D) Quantification of the number of phages that could overcome targeting by CRISPR-Cas from populations of progeny phages with and without LidIPLE 1. Significance was determined by unpaired one-tailed t-tests between EV and lidI-expressing hosts. (E) Schematic of evolution by successful homologous recombination between coinfecting phages. Top: cultures with induced EV and lidIPLE 1constructs are coinfected with ICP1 harboring mutated, non-functional CRISPR-Cas systems: CRISPR*-Cas ICP1 (orange) and CRISPR-Cas* ICP1 (yellow) at MOI 0.01. Bottom: The progeny phage from these infections were then plated on PLE 1 V. cholerae. PLE (blue circles) inhibit phage production by all phages that did not successfully recombine to restore a functional CRISPR-Cas system (two-colored phage). Expanded schematic in Figure 6—figure supplement 3. (F) Progeny phage from hosts with and without LidIPLE 1 co-infected with CRISPR*-Cas ICP1 and CRISPR-Cas* ICP1 were harvested via mechanical lysis. These were then plaqued on permissive (PLE (-) V. cholerae) and restrictive (PLE 1 V. cholerae) hosts. Significance was determined via one-tailed t-tests between EV and lidI-expressing hosts. For all graphs, data points represent individual values, bar height represents the average value, and error bars represent the standard deviation. Additional efficiency of plaquing analysis of 6D and 6F can be found in Figure 6—figure supplement 4. *p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001.
-
Figure 6—source data 1
This spreadsheet contains the data used to create Figure 6.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig6-data1-v1.xlsx
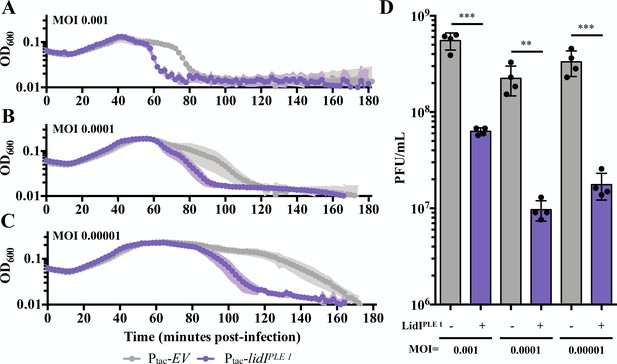
Accelerated lysis and phage yield after low MOI infections.
(A, B and C) Optical density (OD600) of empty vector (EV) or LidIPLE 1-expressing cultures after infection at MOI = 0.001 (A), MOI = 0.0001 (B), and MOI 0.00001 (C). Data points represent the average reading of n = 4 biological replicates and shaded regions display the standard deviation of experiments. (D) Quantification of progeny phage produced from the low MOI infections in panels A-C. Points represent individual replicates; bar height is the average; error bars display the standard deviation of samples. Significance was determined via two-tail t-test. **p≤0.01, ***p≤0.001.
-
Figure 6—figure supplement 1—source data 1
This spreadsheet contains the data used to create Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig6-figsupp1-data1-v1.xlsx
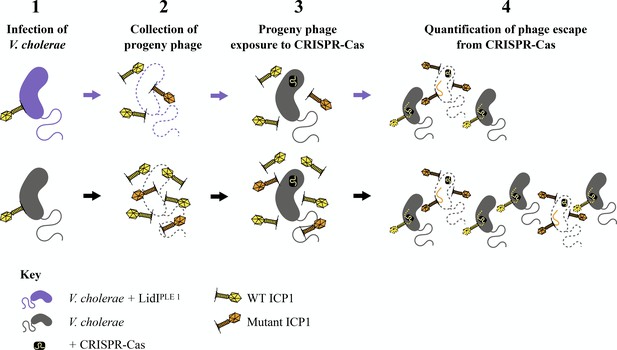
Diversity of progeny phage from infections as measured by phage escape from CRISPR-Cas – expanded schematic.
1. V. cholerae strains with and without LidIPLE 1 (top/purple and bottom/grey, respectively) were exposed to ICP1 (yellow). 2. 90 minutes post-infection, infected cultures were mechanically lysed to release progeny phage (yellow and orange phage). 3. These progeny phage were plated on V. cholerae encoding a Type I-E CRISPR-Cas system (black rectangle) with a spacer against ICP1 (white and yellow line). 4. Only progeny phage that had mutated (orange) to escape targeting by the spacer could successfully infect, avoid degradation, replicate, and lyse cells to form plaques. Quantifying these plaques gives insight into how many mutant phage resulted from the initial infection.

Successful recombination of progeny phage from infections as measured by phage-encoded CRISPR-Cas overcoming PLE – expanded schematic.
A We engineered strains of ICP1 encoding a Type I-F CRISPR-Cas system to lack nuclease activity by deleting Cas2-3 in the CRISPR-Cas* phage (yellow) and disrupt targeting by deleting the spacers and mutating Cas1 so new spacers could not be incorporated in the CRISPR*-Cas phage (red). When these phage infect PLE (-) V. cholerae (A1), the resulting progeny phage (A2) consist of phages with the parental genotypes of the phages (yellow and red) or homologous recombination upon coinfection can produce recombinant progeny phage (orange). Some recombinant progeny phage will have reconstituted a functional CRISPR-Cas system. We can test this by exposing the progeny phage to PLE (+) V. cholerae (A3) where only phages that can defend against PLE with CRISPR-Cas will be able to form plaques (middle). B To test if LidIPLE 1 expression can impact evolution through homologous recombination, we coinfected V. cholerae with (purple) and without (grey) LidIPLE 1 with the CRISPR-Cas deficient phages (B1). After lysing the infected cultures (B2) we quantified how many progeny phage recombined to reconstitute a functional CRISPR-Cas system by exposing them to PLE (B3).

EOP of evolution experiments in the presence of LidIPLE 1.
(A) Efficiency of plaquing (EOP) was determined between plaque forming units (PFUs) from LidIPLE 1V. cholerae infections and PFUs from EV V. cholerae infections plaqued on CRISPR-Cas (+) V. cholerae harboring each of the specified spacers. No significant differences were detected via one-way ANOVA. (B) EOP was determined between PFUs of a population plaqued on PLE 1 V. cholerae in comparison to the PFUs of the same population on PLE (-) V. cholerae. Points represent individual replicates; bar height is the average; error bars display the standard deviation of samples. No significant difference was detected by two-tailed t-test.
-
Figure 6—figure supplement 4—source data 1
This spreadsheet contains the data used to create Figure 6—figure supplement 4.
- https://cdn.elifesciences.org/articles/53200/elife-53200-fig6-figsupp4-data1-v1.xlsx
Tables
Acronyms.
All the acronyms used in this work are listed in alphabetical order.
| Acronym | Meaning |
|---|---|
| DiOC2(3) | 3,3’-diethloxacarbocyanine iodide |
| DNP | 2,4-dinitrophenol |
| EOP | efficiency of plaquing |
| EV | empty vector |
| gp | gene product |
| IM | inner membrane |
| LIN | lysis inhibition |
| MGE | mobile genetic element |
| MOI | multiplicity of infection |
| MOSI | multiplicity of superinfection |
| OD | optical density |
| OM | outer membrane |
| ORF | open reading frame |
| PG | peptidoglycan |
| PLE | phage-inducible chromosomal island-like element |
| SaPI | Staphylococcus aureus pathogenicity island |
| WT | wild type |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Vibrio cholerae) | lidIPLE 1 (PLE 1 ORF20.1) | * | ||
| Gene (Vibrio cholerae) | lidIPLE 2 (PLE 2 ORF24.1) | * | ||
| Gene (Vibrio cholerae) | lidIPLE 3 (PLE 3 ORF24.1) | * | ||
| Gene (Vibrio cholerae) | lidIPLE 4 (PLE 4 ORF26) | (O'Hara et al., 2017) | ||
| Gene (Vibrio cholerae) | lidIPLE 5 (PLE 5 ORF26) | (O'Hara et al., 2017) | ||
| Gene (bacteriophage ICP1) | teaAICP1 (gp137) | (Angermeyer et al., 2018),* | ||
| Gene (bacteriophage ICP1) | arrAICP1 (gp138) | (Angermeyer et al., 2018),* | ||
| Strain (Vibrio cholerae) | PLE (-) V. cholerae (E7946) | (Levine et al., 1982) | KDS 6 | |
| Strain (Vibrio cholerae) | PLE 1 V. cholerae (PLE 1 E7946) | (O'Hara et al., 2017) | KDS 36 | |
| Strain (Vibrio cholerae) | ΔlacZ::Ptac-EV (E7946) | (McKitterick and Seed, 2018) | KDS 116 | |
| Strain (Vibrio cholerae) | ΔlacZ::Ptac-lidIPLE 1 (E7946) | * | KDS 139 | |
| Strain (Vibrio cholerae) | ΔlacZ::Ptac-lidIPLE 4 (E7946) | * | KDS 267 | |
| Strain (Vibrio cholerae) | PLE 1 FLAG-LidIPLE 1 (E7946) | * | KDS 268 | |
| Strain (Vibrio cholerae) | PLE 1 ΔlidI (E7946) | * | KDS 170 | |
| Strain (Vibrio cholerae) | PLE 4 ΔlidI (E7946) | * | KDS 269 | |
| Strain (Vibrio cholerae) | CRISPR-Cas (+) (E7946) | (Box et al., 2016) | KDS 112 | Inducible Cas Proteins |
| Recombinant DNA reagent (plasmid) | Ptac-Empty Vector (pKL06 in E7946) | (McKitterick and Seed, 2018) | KDS 196 | Empty Vector Control |
| Recombinant DNA reagent (plasmid) | Ptac-lidIPLE 1 (plasmid in E7946) | * | KDS 219 | Inducible lidIPLE 1 |
| Recombinant DNA reagent (plasmid) | Ptac-lidIPLE 4 (plasmid in E7946) | * | KDS 270 | Inducible lidIPLE 4 |
| Recombinant DNA reagent (plasmid) | Ptac-teaAICP1 (plasmid in E7946) | * | KDS 271 | Inducible teaAICP1 |
| Recombinant DNA reagent (plasmid) | Ptac-arrAICP1 (plasmid in E7946) | * | KDS 272 | Inducible arrAICP1 |
| Recombinant DNA reagent (plasmid) | Ptac-tT4 (plasmid in E7946) | * | KDS 273 | Inducible tT4 |
| Recombinant DNA reagent (plasmid) | Ptac-anti-gp138 spacer (plasmid in E7946) | * | KDS 274 | CRISPR array containing anti-gp138 spacer |
| Recombinant DNA reagent (plasmid) | Ptac-anti-gp138 spacer and repair template | * | KDS 275 | CRISPR array containing anti-gp138 spacer and repair template |
| Recombinant DNA reagent (plasmid) | Ptac-FLAG-lidIPLE 1 | * | KDS 276 | Inducible FLAG-tagged blot control |
| Recombinant DNA reagent (plasmid) | Ptac-none (CRISPR array with no spacers against WT ICP1) | (McKitterick et al., 2019b) | KDS 277 | Spacer control |
| Recombinant DNA reagent (plasmid) | Ptac-spacer A | * | KDS 278 | Spacer A against ICP1 |
| Recombinant DNA reagent (plasmid) | Ptac-spacer B | * | KDS 279 | Spacer B against ICP1 |
| Recombinant DNA reagent (plasmid) | Ptac-spacer C | * | KDS 280 | Spacer C against ICP1 |
| Strain (bacteriophage ICP1) | ICP1 (ICP1 2006E ΔCRISPR ΔCas) | (McKitterick and Seed, 2018) | ||
| Strain (bacteriophage ICP1) | ΔarrA ICP1 (ICP1 2006E ΔarrA/gp137) | * | SGH Φ 61 | |
| Strain (bacteriophage ICP1) | CRISPR*-Cas ICP1 (ICP1 2011A Δspacer2-9 Cas1D244A) | (McKitterick et al., 2019b) | ACM Φ 232 | |
| Strain (bacteriophage ICP1) | CRISPR-Cas* ICP1 (ICP1 2011A Δcas2-3) | * | SGH Φ 62 | |
| Chemical compound | Isopropyl-beta-D-thiogalactoside (IPTG) | GoldBio | 12481C5 | |
| Chemical compound | Theophylline | Sigma-Aldrich | T1633-100G | |
| Chemical compound | 2,4-Dinitrophenol (DNP) | Sigma-Aldrich | D198501-100G | |
| Chemical compound | 3,3-Diethyloxacarbocyanine iodide (DiOC2(3)) | Sigma-Aldrich | 320684–1G | |
| Antibody | Rabbit anti-FLAG polyclonal antibody | Sigma-Aldrich | RRID:SAB4301135 | |
| Antibody | Goat anti-Rabbit IgG antibody, peroxidase conjugated | Sigma-Aldrich | RRID:AP132P |
-
*Identified or created in this work.
Additional files
-
Source data 1
This spreadsheet contains the data used to create Supplementary files 2 and 3.
- https://cdn.elifesciences.org/articles/53200/elife-53200-data1-v1.xlsx
-
Supplementary file 1
ICP1_2006_E gene product (gp) GenBank References.
The gene products referred to in this work relate to open reading frames (ORFs) as noted in the ‘Locus Tag Note’.
- https://cdn.elifesciences.org/articles/53200/elife-53200-supp1-v1.docx
-
Supplementary file 2
TeaA homologs.
BLASTP was used to find homologs that share 30% identity with TeaA over 85% of the query. The GenBank ID, description, number of transmembrane domains (TMD) as predicted by TMHMM Server 2.0, and organism is listed for each homolog. Whether or not an ArrA homolog was found in the same organism is noted in the ‘ArrA’ column. Additionally, the adjacent upstream and downstream genes were analyzed for TMDs. GenBank descriptions are color coded. Due to the number of homologs analyzed, this table is only available as a spreadsheet as Source data 1.
- https://cdn.elifesciences.org/articles/53200/elife-53200-supp2-v1.docx
-
Supplementary file 3
ArrA homologs.
BLASTP was used to find proteins with 20% identity to ArrA over 75% of the query. The GenBank ID, description, number of transmembrane domains (TMD) as predicted by TMHMM Server 2.0, and organism is listed for each homolog. Whether or not a TeaA homolog was found in the same organism is noted in the ‘TeaA’ column. Additionally, the adjacent upstream and downstream genes of each homolog were analyzed for TMDs. GenBank descriptions are color coded. The source data for this table is available in Source data 1.
- https://cdn.elifesciences.org/articles/53200/elife-53200-supp3-v1.docx
-
Supplementary file 4
Primer Table.
Primers used in this work are provided with a description, identifier, and sequence.
- https://cdn.elifesciences.org/articles/53200/elife-53200-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53200/elife-53200-transrepform-v1.docx





