Gating and selectivity mechanisms for the lysosomal K+ channel TMEM175
Figures
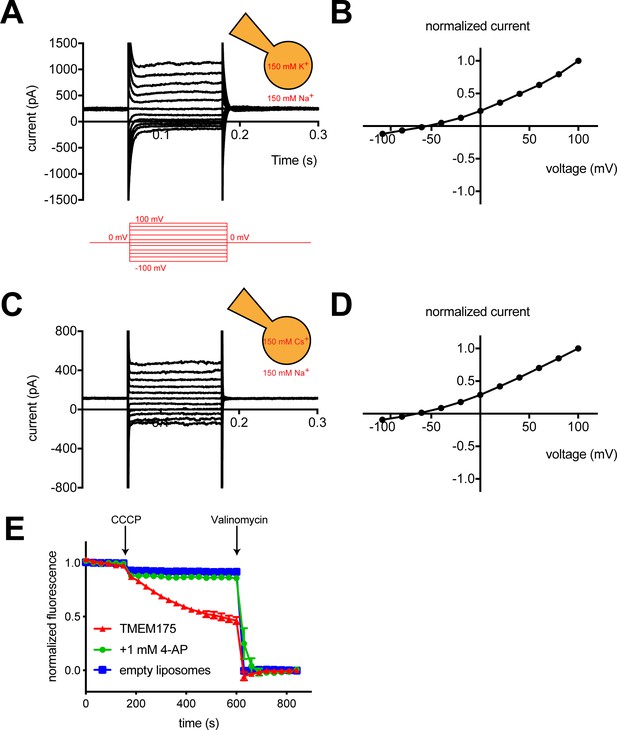
hTMEM175 is a K+ selective channel.
(A, C) Representative whole-cell electrical recordings of hTMEM175-transfected HEK293T cells. In bi-ionic conditions of 150 mM K+ (intracellular) and 150 mM Na+ (extracellular) (A) or 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular) (C), currents were measured using the following protocol (red): from a holding potential of 0 mV, the voltage was stepped to voltages between −100 and +100 mV, in 20 mV increments, then returned to 0 mV. (B, D) Normalized current-voltage relationships of three independent whole-cell patch clamp recordings of hTMEM175-transfected HEK293T cells in bi-ionic conditions of 150 mM K+ (intracellular) and 150 mM Na+ (extracellular) (B) or 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular) (D). (E) K+ efflux from purified hTMEM175 reconstituted into liposomes in the presence or absence of 1 mM 4-aminopyridine and from empty liposomes was monitored using a fluorescence-based flux assay. Arrows mark addition of the proton ionophore CCCP to initiate K+ flux and addition of the K+ ionophore valinomycin to measure total flux capacity of the liposomes. All experiments were performed in triplicate and error bars represent SEM.
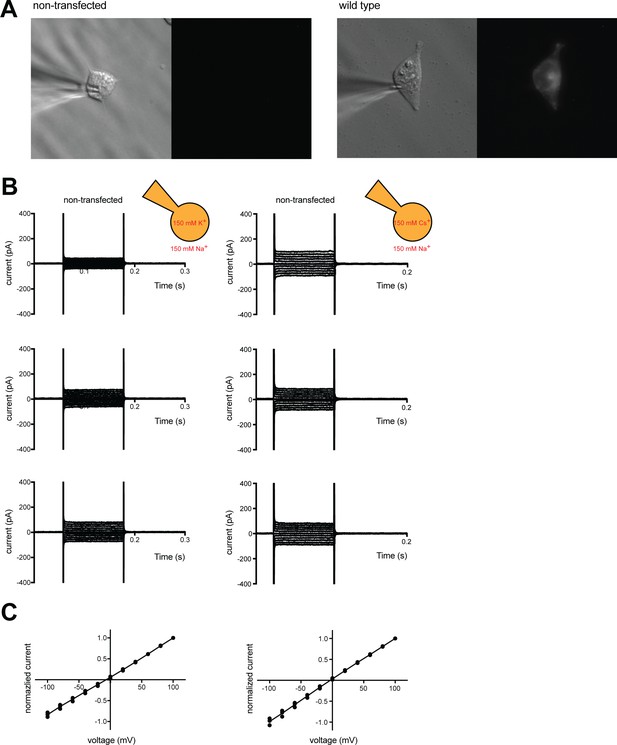
Electrophysiological analysis of hTMEM175 in HEK293T cells.
(A) Representative differential interference contrast microscopy (left) and GFP fluorescence microscopy (right) of non-transfected and hTMEM175 transfected HEK293T cells used for whole-cell patch clamp. (B) Three representative whole-cell electrical recordings of non-transfected HEK293T cells in bi-ionic conditions of 150 mM K+ (intracellular) and 150 mM Na+ (extracellular) (left) or 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular) (right). (C) Normalized current-voltage relationships of experiments shown in B. Currents were normalized to the maximum current of each experiment (at +100 mV). Error bars are shown in SEM. Recording protocol is same as in Figure 1A.
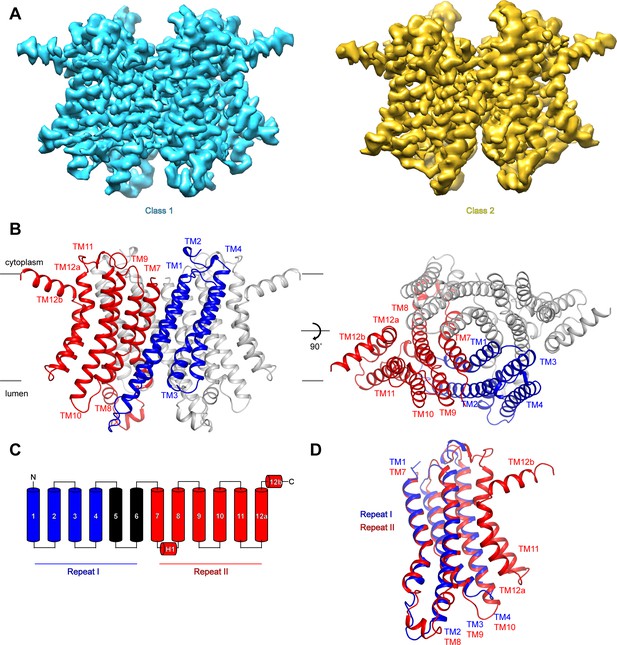
Structure of hTMEM175.
(A) Cryo-EM density maps of class 1 (cyan) and class 2 (gold) hTMEM175 in KCl depicted from within the membrane. (B) Structure of class 1 hTMEM175 depicted from within the membrane (left) and from the cytoplasm (right). TM1-TM4 (repeat I) and TM7-TM12 (repeat II) of protomer A are shown in blue and red, respectively. Protomer B is shown in grey. Approximate width of the lipid bilayer is shown as grey bars. (C) Topology of hTMEM175. TM1-TM4 (repeat I) and TM7-TM12 (repeat II) are shown in blue and red, respectively. Unmodelled helices TM5 and TM6 are shown in black. (D) Superposition of class 1 hTMEM175 repeat I (blue) with repeat II (red).
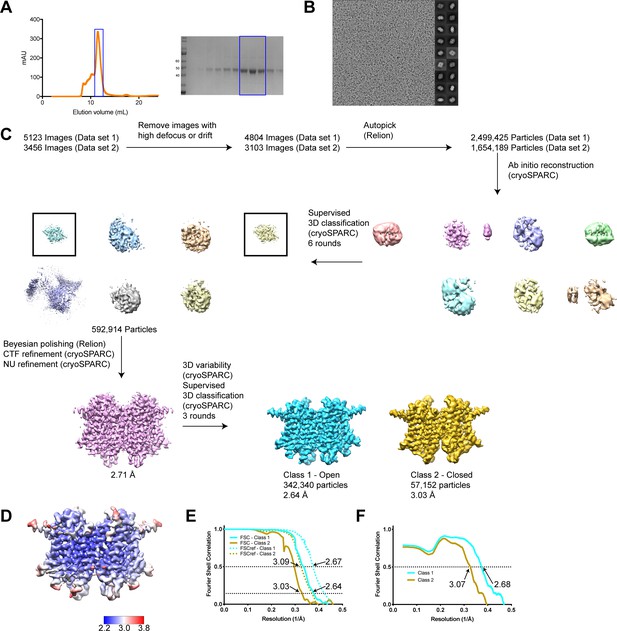
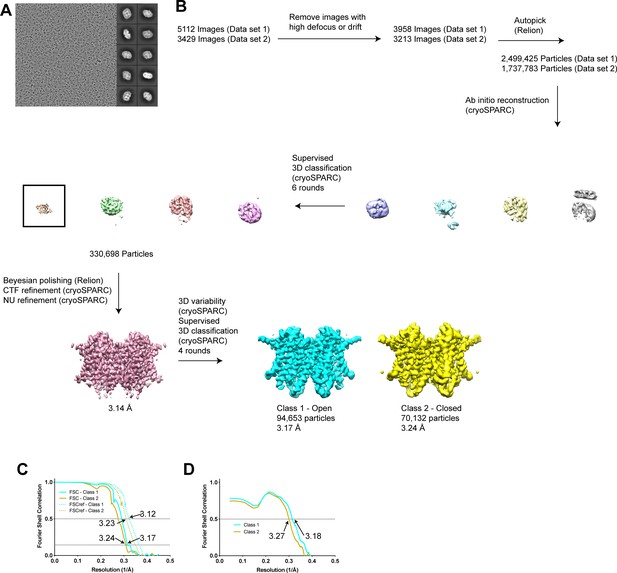
Cryo-EM analysis of hTMEM175 in KCl.
(A) Superdex 200 size exclusion profile of hTMEM175 (left) and Coomassie-stained PAGE analysis of purified hTMEM175 purified in LMNG and 150 mM KCl (right). Peak fractions from gel filtration are highlighted in the image of the Coomassie-stained gel by a blue box (B) Representative image and 2D class averages of hTMEM175 in 150 mM KCl. (C) Simplified image processing workflow. (D) Density map of class 1 colored by local resolution. (E) Fourier shell correlation (FSC) of two unfiltered half-maps for class 1 (solid cyan) and class 2 (solid gold) and cross correlation plot of two unfiltered half-maps following density modification for class 1 (dotted cyan) and class 2 (dotted gold). (F) Fourier shell correlation (FSC) of refined class 1 model compared with density modified map (cyan) and refined class 2 model compared with density modified map (gold).

Cryo-EM densities in hTMEM175.
(A) Section through 6 Å lowpass-filtered class 1 (left, cyan) and class 2 (right, gold) density maps revealing weak density for poorly ordered transmembrane helices TM5 and TM6. (B–C) Density-modified class 1 (B) and class 2 (C) cryo-EM density maps with atomic model in sticks for TM1, TM2, TM7 and TM8, thresholded at 10 σ and 3 σ, respectively.
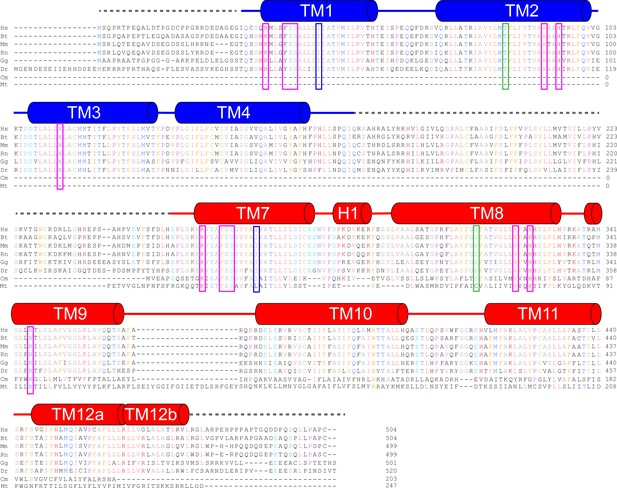
Sequence alignment of TMEM175 channels.
Hs – human TMEM175, Bt – cow TMEM175, Mm – mouse TMEM175, Rn – Rat TMEM175, Gg – chicken TMEM175, Dr – Zebrafish TMEM175, Cm - Chamaesiphon minutus TMEM175, Mt - Marivirga tractuosa TMEM175. Identical residues are colored by amino acid. RxxxFSD motif interaction network residues are highlighted by magenta boxes, isoleucine constriction residues are highlighted by blue boxes and residues whose side chains coordinate water molecules that stabilize the kinks in TM1 and TM7 are highlighted by green boxes.

TMEM175 channels share a common fold.
(A) Superposition of class 1 hTMEM175 (cyan) with TMEM175Cm (left, brown - main-chain RMSD = 3.2 Å) and TMEM175Mt (right, magenta - main-chain RMSD = 4.0 Å). (B) Superposition of TM7-TM12 of class 1 hTMEM175 (cyan) with monomeric structures of TMEM175Cm (left, brown - main-chain RMSD = 2.3 Å) and TMEM175Mt (right, magenta - main-chain RMSD = 3.9 Å).
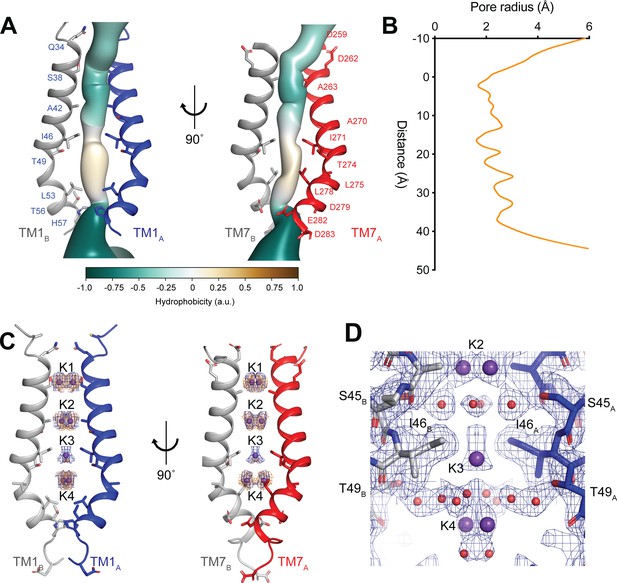
hTMEM175 ion conduction pathway.
(A) Ion permeation pathway of class 1 hTMEM175. Pore-lining helices TM1 from protomers A (blue) and B (grey) are shown at left and TM7 from protomers A (red) and B (grey) are shown at right with all other helices removed for clarity. Pore-lining residues are shown as sticks. Surface representation of the ion permeation pathway colored by hydrophobicity calculated using the class 1 structure without ions and water molecules calculated using CHAP (Klesse et al., 2019). (B) Dimensions of the ion conduction pathway in class 1 calculated using CHAP (Klesse et al., 2019). (C) Overlapping non-protein density peaks in the ion permeation pathway of class 1 in the presence of K+ (blue mesh, 12 σ threshold) and Cs+ (gold mesh, 8 σ threshold). hTMEM175 is shown as in A. K+ ions are shown as violet spheres. (D) Density map near the isoleucine constriction displayed as blue mesh and contoured at 12 σ threshold. K+ ions are shown as violet spheres and water molecules are shown as red spheres.
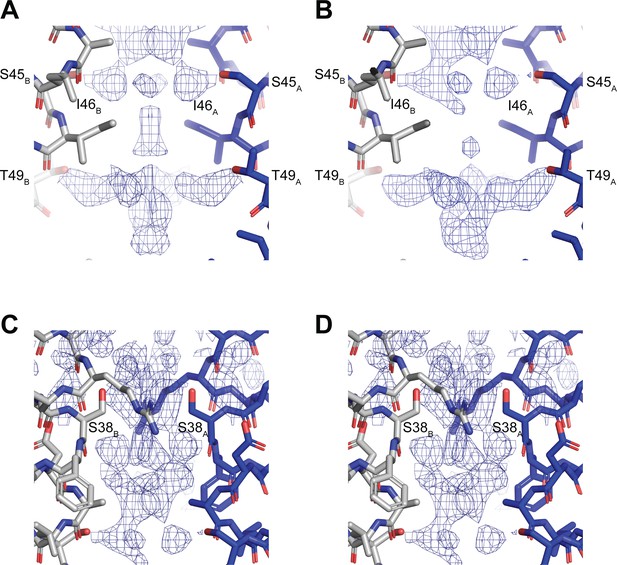
Non-protein densities in hTMEM175 density maps.
(A–B) Non-protein densities near the isoleucine constriction for class 1 with C2 symmetry imposed (A) and without imposing symmetry (B). (C–D) Non-protein densities in the cytoplasmic region of the pore for class 1 with C2 symmetry imposed (C) and without imposing symmetry (D). Density is shown as blue mesh contoured at 12 σ threshold for C2 map and at 8 σ threshold for C1 map.
Cryo-EM analysis of hTMEM175 in CsCl.
(A) Representative image and 2D class averages of hTMEM175 in 150 mM CsCl. (B) Simplified image processing workflow. (C) Fourier shell correlation (FSC) of two unfiltered half-maps for class 1 (solid cyan) and class 2 (solid gold) and cross correlation plot of two unfiltered half-maps following density modification for class 1 (dotted cyan) and class 2 (dotted gold). (D) Fourier shell correlation (FSC) of refined class 1 model compared with density modified map (cyan) and refined class 2 model compared with density modified map (gold).
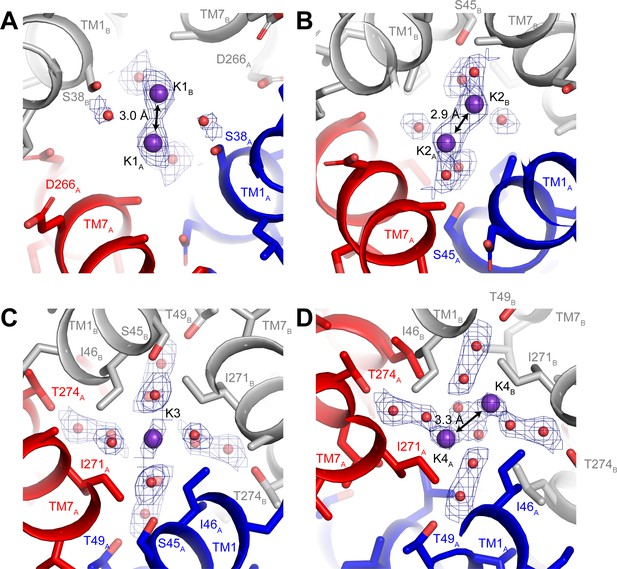
Ion-binding sites in hTMEM175.
Structure of the K1 (A), K2 (B), K3 (C) and K4 (D) binding sites in class 1. K+ ions are shown as violet spheres and water molecules are shown as red spheres. Density for K+ and water molecules shown as blue mesh and contoured at 12 σ threshold.
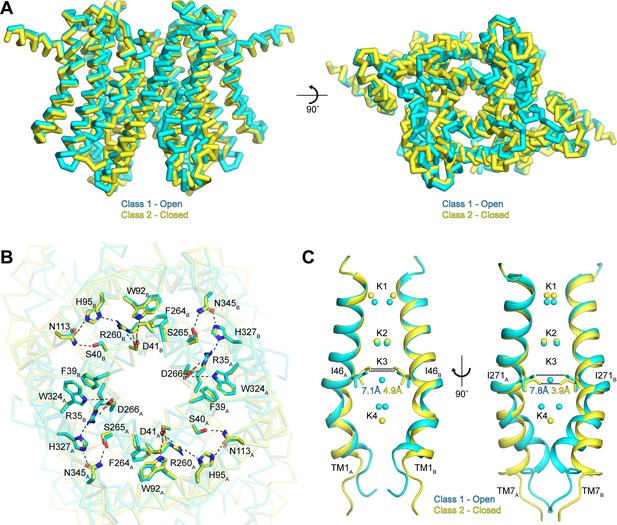
Gating in hTMEM175.
(A) Superposition of class 1 (cyan) and class 2 (gold) viewed from within the membrane (left) and from the lysosomal lumen (right). (B) Alignment of the RxxxFSD inter- and intra-subunit interaction networks in class 1 (cyan) and class 2 (gold) depicted as sticks and viewed from the cytosol. Ionic and polar interaction are shown as dashed lines. (C) Ion conduction pathways of class 1 (cyan) and class 2 (gold). TM1 is shown at left and TM7 is shown at right with all other helices removed for clarity. K+ ion binding sites are shown as spheres. Dotted lines correspond to minimum distance between opposing residues at the isoleucine constriction.
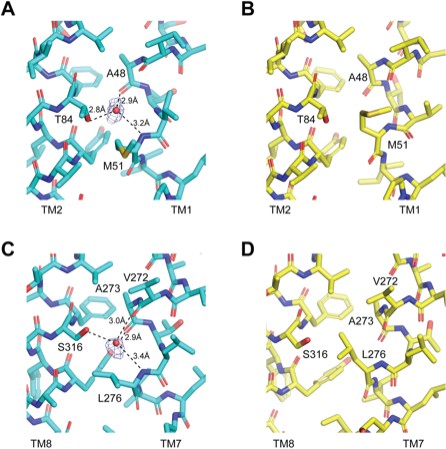
Water molecules stabilize kinked conformation of TM1 and TM7 in class 1.
(A) Order water molecule stabilizing the kinked TM1 in class 1 is coordinated by Ala48, Met51 and Thr84. (B) Side chain of Met51 occupies the position of the water molecule in class 2 and TM1 adopts a straight configuration. (C) Order water molecule stabilizing the kinked TM7 in class 1 is coordinated by Val272, Ala273, Leu276 and Ser316. (D) Side chain of Leu276 occupies the position of the water molecule in class 2 and TM7 adopts a straight configuration.
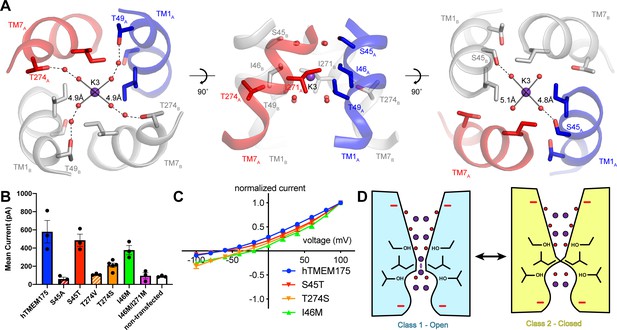
Permeation and selectivity through the isoleucine constriction.
(A) The isoleucine constriction is flanked by two layers of ordered water molecules. The cytosolic layer of waters is partially coordinated by Ser45, while the luminal layer is partially coordinated by Thr49 and Thr274. (B) Mean current recorded from HEK293T cells transfected with hTMEM175 (blue), S45A (red dashed), S45T (red), T274V (orange dashed), T274S (orange), I46M (green), I46M/I271M (magenta) and non-transfected (white) at +100 mV in a bi-ionic condition of 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular). (C) Normalized I-V relationship of whole-cell patch clamp of hTMEM175 transfected (blue), S45T transfected (red), T274S transfected (orange) and I46M transfected (green) HEK293T cells in a bi-ionic condition of 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular). All experiments were performed at least three times and error bars represent SEM. (D) Model for ion selectivity and gating in hTMEM175. In the open state, ions are transiently dehydrated through the isoleucine constriction, favoring permeation of K+ ions. In the closed state, the isoleucine constriction closes, preventing ion permeation.
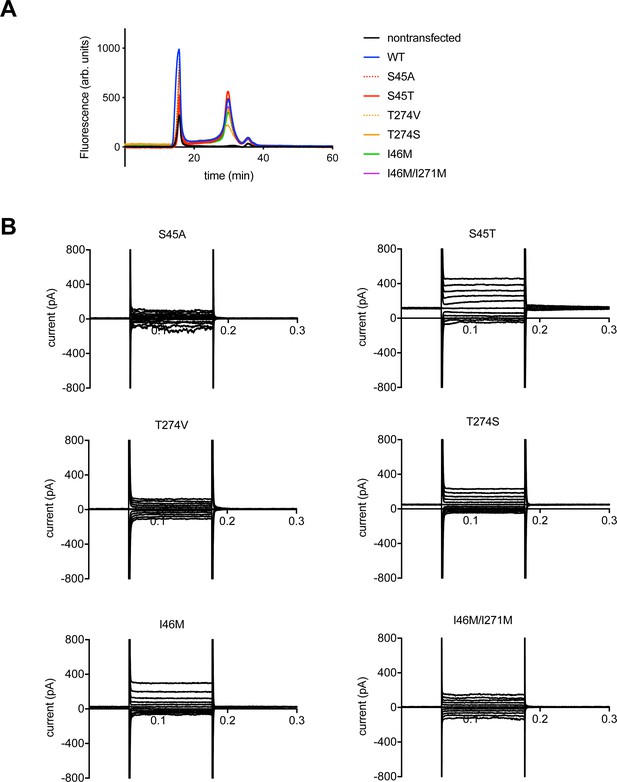
Functional analysis of hTMEM175 mutants.
(A) Superose 6 fluorescence size exclusion chromatography traces of hTMEM175 (blue), S45A (red dashed), S45T (red), T274V (orange dashed), T274S (orange), I46M (green), I46M/I271M (magenta) and non-transfected (black). (B) Representative whole-cell electrical recordings of HEK293T cells transfected with S45A, S45T, T274V, T274S, I46M or I46M/I271M mutants in bi-ionic condition of 150 mM Cs+ (intracellular) and 150 mM Na+ (extracellular). Recording protocol is same as in Figure 1C.
Videos
Morph between open class 1 and closed class 2.
Tables
Cryo-EM data acquisition, reconstruction and model refinement statistics.
| hTMEM175 | hTMEM175 | hTMEM175 | hTMEM175 | |
| Class 1 K+ | Class 2 K+ | Class 1 Cs+ | Class 2 Cs+ | |
| Cryo-EM acquisition and processing | ||||
| EMDB accession # | 21603 | 21604 | 21605 | 21606 |
| Magnification | 22,500x | 22,500x | 22,500x | 22,500x |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Total electron exposure (e- / Å2) | 61 | 61 | 61 | 61 |
| Exposure time (s) | 8 | 8 | 8 | 8 |
| Defocus range (uM) | -1.0 to -2.5 | -1.0 to -2.5 | -1.0 to -2.5 | -1.0 to -2.5 |
| Pixel size (Å) | 1.088 | 1.088 | 1.088 | 1.088 |
| Symmetry imposed | C2 | C2 | C2 | C2 |
| Initial particles | 4,153,614 | 4,153,614 | 4,275,219 | 4,275,219 |
| Final particles | 342,340 | 57,152 | 94,653 | 70,132 |
| Resolution (masked FSC = 0.143, Å) | 2.64 | 3.03 | 3.17 | 3.24 |
| Density modified CC (0.5, Å) | 2.67 | 3.09 | 3.12 | 3.23 |
| Model Refinement | ||||
| PDB ID | 6WC9 | 6WCA | 6WCB | 6WCC |
| Model resolution (FSC = 0.50/0.143Å) | 2.68 / 2.32 | 3.07 / 2.67 | 3.18 / 2.71 | 3.27 / 2.84 |
| Model refinement resolution | 300-2.6 | 300-3.0 | 300-3.2 | 300-3.2 |
| RMS deviations | ||||
| Bond length (Å) | 0.005 | 0.002 | 0.004 | 0.003 |
| Bond angle (°) | 0.532 | 0.507 | 0.406 | 0.506 |
| Ramachandran plot | ||||
| Favored (%) | 96.13 | 96.42 | 99.17 | 99.17 |
| Allowed (%) | 3.87 | 3.58 | 0.83 | 0.83 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
| Rotamer Outliers (%) | 2.27 | 1.61 | 1.29 | 2.26 |
| Validation | ||||
| MolProbity score | 1.71 | 1.74 | 1.12 | 1.49 |
| Clashscore | 3.88 | 6.28 | 2.50 | 4.39 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | hTMEM175 | Synbio technologies | ||
| Cell line (H. sapiens) | HEK-293T | ATCC | CRL-3216 RRID:CVCL_0063 | |
| Cell line (H. sapiens) | HEK-293S GnTi- | ATCC | CRL-3022 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | 850757 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) | Avanti Polar Lipids | 840457 | |
| Chemical compound, drug | carbonyl cyanide m-chlorophenylhydrazone (CCCP) | Thermo Fisher Scientific | 215911250 MG | |
| Chemical compound, drug | 9-amino-6-chloro-2-methoxyacridine (ACMA) | Thermo Fisher Scientific | A1324 | |
| Chemical compound, drug | valinomycin | Sigma | V0627 | |
| Chemical compound, drug | Polyethylenimine, Linear, MW 25000, Transfection Grade (PEI 25K) | Polysciences, Inc | 23966–1 | |
| Chemical compound, drug | Sodium Butyrate | Sigma | 8451440100 | |
| Chemical compound, drug | lauryl maltoside neopentyl glycol | Anatrace | NG310 | |
| Chemical compound, drug | n-Octyl-β-D-Maltopyranoside | Anatrace | O310S | |
| Software, algorithm | MotionCor2 | Zheng et al., 2017 | RRID:SCR_016499 | |
| Software, algorithm | CtfFind 4.1.10 | Rohou and Grigorieff, 2015 | RRID:SCR_016731 | |
| Software, algorithm | RELION 3.1 | Scheres, 2016 | http://www2.mrc-lmb.cam.ac.uk/relion RRID:SCR_016274 | |
| Software, algorithm | SerialEM | Mastronarde, 2005 | RRID:SCR_017293 | |
| Software, algorithm | cryoSPARC v2 | Structura Biotechnology | https://cryosparc.com/ RRID:SCR_016501 | |
| Software, algorithm | PHENIX | Liebschner et al., 2019 | https://www.phenix-online.org/ RRID:SCR_014224 | |
| Software, algorithm | COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ RRID:SCR_014222 | |
| Software, algorithm | PyMOL | Schrödinger, 2020 | https://pymol.org/2/ RRID:SCR_000305 | |
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera RRID:SCR_004097 | |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software | ||
| Software, algorithm | SoftMax Pro 6 | Molecular Devices | ||
| Software, algorithm | Axon Digidata 1550B digitizer | Molecular Devices | ||
| Software, algorithm | Clampex 10.6 | Molecular Devices | ||
| Software, algorithm | CHAP | Klesse et al., 2019 | https://www.channotation.org/ | |
| Software, algorithm | Clampfit 10.6 | Molecular Devices | ||
| Others | QUANTIFOIL R1.2/1.3 holey carbon grids | Quantifoil | ||
| Others | FEI Vitrobot Mark IV | FEI Thermo Fisher |





