Altered expression of a quality control protease in E. coli reshapes the in vivo mutational landscape of a model enzyme
Figures
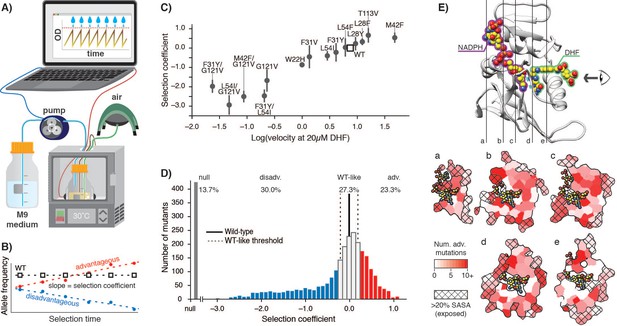
E. coli DHFR deep mutational scanning uncovers many advantageous mutations.
(A) Turbidostat schematic. Reoccurring dilutions with fresh medium keep the culture optical density (OD600) below 0.075. (B) The selection coefficient for each mutant is the slope of the linear regression of allele frequency over time. The wild-type (squares) value is normalized to zero. Advantageous (red) mutations increase and disadvantageous (blue) mutations decrease in frequency. (C) Selection coefficients from deep mutational scanning as a function of enzymatic velocity for purified DHFR point mutants measured in vitro. Velocities at 20 µM DHF were calculated from Michalis-Menten parameters. Error bars reflect the standard deviation from three biological replicates. (D) Histogram of selection coefficients. The wild-type value is indicated with a vertical black line. The median standard deviation over all mutations is the cut-off for WT-like behavior (Materials and methods, Figure 1—figure supplement 3, Figure 1—figure supplement 4) and is indicated with dashed lines. Mutation are colored as advantageous (red), disadvantageous (blue), WT-like (white), or null (grey). (E) Structural model of DHFR (PDB ID: 3QL3) with cross-section slices (a–e) indicated. The DHF substrate (green) and the NADPH cofactor (purple) are represented by spheres (yellow carbons and heteroatom coloring). An arrow indicates the perspective for each slice. (a–e) five cross-section slices. Color scale indicates numbers of advantageous mutations at each position. Crosshatching indicates residues with >20% solvent accessible surface area.
-
Figure 1—source data 1
Soluble DHFR expression levels in molecules per cell measured from lysate activity assays as described in Materials and methods.
The location of the DHFR gene is listed in parenthesis in the first column. Expression values corresponds to the cell strain in the column heading.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Selection coefficients for –Lon selection (Figure 1—source data 1) compared to monoculture growth rates measured in a plate reader in ER2566 ∆folA/∆thyA (–Lon) as described in Materials and methods.
For values listed as ND, no detectable change in OD was measured during a 30 hr growth period.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Michaelis-Menten kinetics for the set of DHFR mutants (Fierke and Benkovic, 1989; Huang et al., 1994; Reynolds et al., 2011) used to calibrate the selection are reported together with the reference from which the values were taken.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig1-data3-v2.xlsx
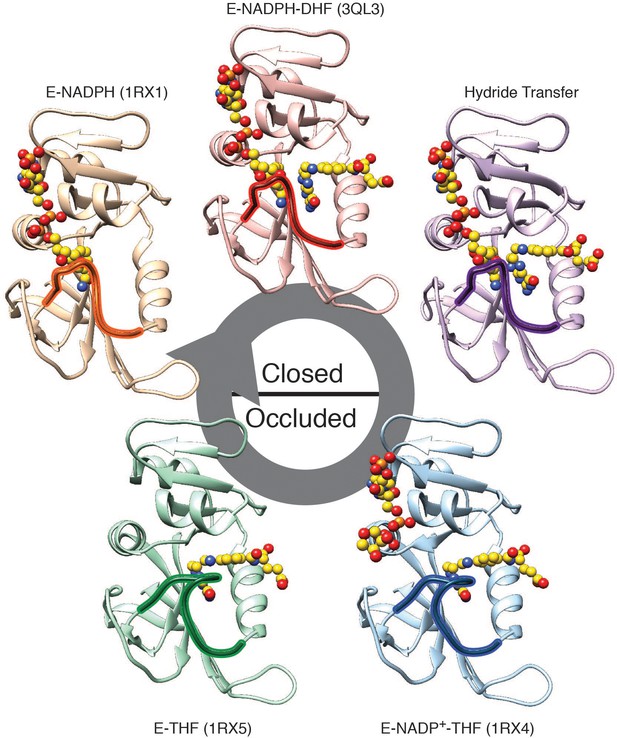
Conformations adopted during the DHFR catalytic cycle: 1RX1, 3QL3, 1RX4, and 1RX5) and a QMMM model of the hydride transfer step (Liu et al., 2013) represent the conformational states adopted by DHFR over the catalytic cycle.
The identity of each state is defined by the identity of the bound ligands (yellow spheres with heteroatom coloring) and the conformation of the M20 loop (outlined) that folds over the active site (closed or occluded). Upper models are in the closed state, and lower models are in the occluded state. All PDBs were downloaded from the PDB_REDO (Joosten et al., 2014).
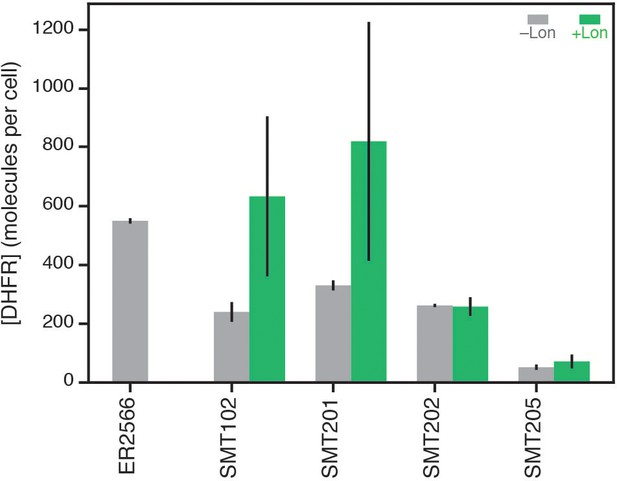
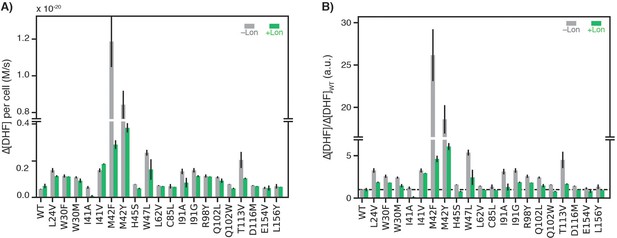
Soluble WT DHFR cellular abundance for endogenous (chromosomal) DHFR in the parental strain and DHFR expressed from plasmids in the selection system.
DHFR cellular abundance is calculated from lysate activity (see Materials and methods). ER2566 is the parental strain (–Lon). SMT102, SMT201, SMT202, SMT205 denote plasmid constructs with altered promoters and ribosome binding sites (see Figure 1—source data 3) in the ER2566 ∆folA/∆thyA strain. DHFR abundances in ER2566 and ER2566 ∆folA/∆thyA –Lon lysates are colored in grey. DHFR abundances in ER2566 ∆folA/∆thyA +Lon lysates are colored in green. Error bars represent the cumulative percent error (standard deviation) from three independent experiments for velocity and three biological replicates for lysate activity.
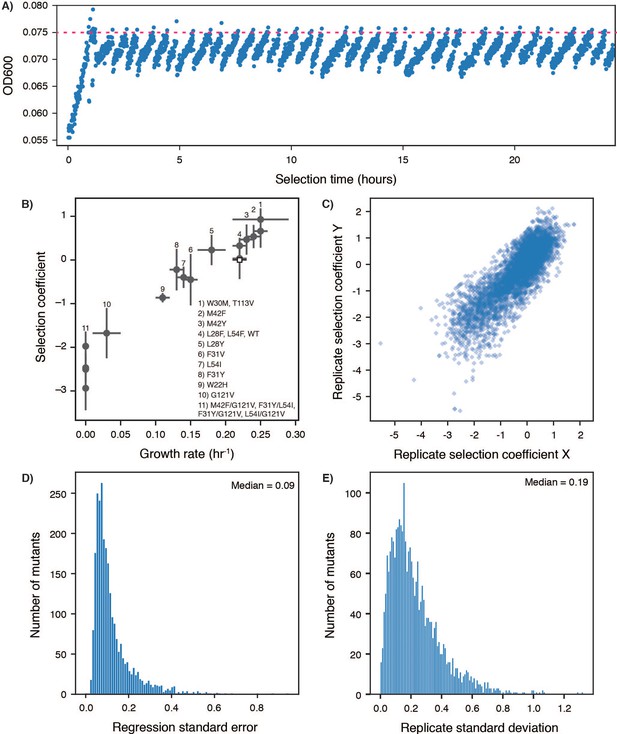
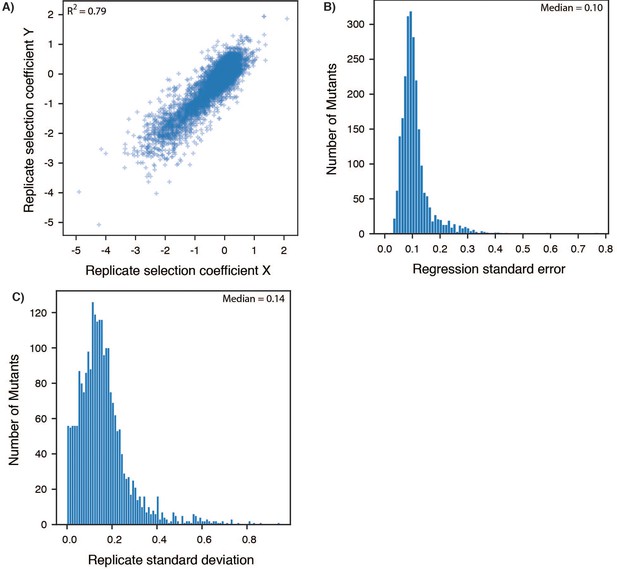
Determination of selection coefficients for DHFR.
(A) Example turbidostat trace from a selection experiment on a library of DHFR single point mutants. The OD600 value inferred from the voltage across an IR emitter-receiver pair is plotted as a function of time. The ‘clamp’ OD value (0.075) is shown as a dashed red line. Decreases in OD correspond to dilution from automatic addition of M9 medium. (B) Comparison of selection coefficients from Figure 1C with growth rates measured in a plate reader for monocultures of the selection strain transformed with the library plasmid (SMT205, Figure 1—source data 3) encoding a single DHFR variant (identified by number, in-Figure caption). Error bars reflect the standard deviation over at least three biological replicates. (C) Comparison of all pairwise replicates for selection coefficients from triplicate deep mutational scanning on DHFR. The Pearson correlation R2 value from linear regression was 0.70. (D) Distribution of standard errors for individual selection coefficients from a single replicate. Selection coefficients are the slope from a linear regression of allele frequency as a function of time in selection. The standard error here is the mean square of residuals. (E) Distribution of standard deviations of selection coefficients for individual point mutants over replicate experiments. Each mutant had a measured selection coefficient in at least 2 of the three replicates. The median of this distribution of standard deviations over all alleles was 0.2 and was used to determine the cut-offs for advantageous and disadvantageous mutations in Figure 1D.
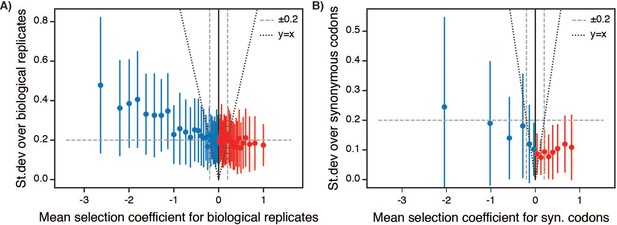
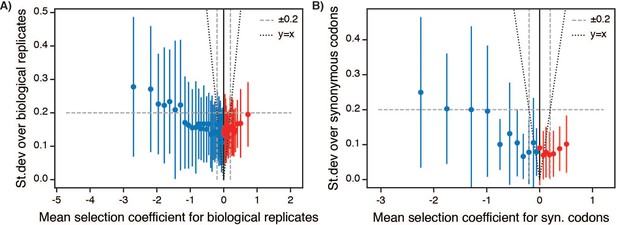
Variation in selection coefficients for –Lon selection.
(A) Standard deviation of selection coefficients over biological replicates. The data were plotted as a function of a sliding window over all single point mutants sorted by selection coefficient. Each point represents the mean error (biological replicate standard deviation) over 50 consecutive selection coefficients (after sorting by value) and the error bars represent ±1 standard deviation of the error. The dashed line represents median error over the entire dataset, which was used determine the for WT-like behavior in Figure 1D. The dotted line represents y = x for comparison between the magnitude of the error relative to the magnitude of the selection coefficient. (B) Standard deviation over synonymous codons coding for the same sequence, plotted as in A.

Residues previously known to have a functional role shown on the DHFR structure.
(A–C) Functionally important residues are colored green, labeled, and shown with slices of the –Lon heatmap (heatmap coloring by selection coefficient is as in Figure 2). The wild-type residue is outlined in black. Positions 22, 27, 35, 57, and 113 are Intolerant, and positions 20, 28, 31, 42, 54, 121, 122, and 148 are Deleterious. In A) the closed (upper, white, PDB ID: 3QL3) and occluded (lower, grey, PDB ID: 1RX4) conformation are shown to illustrate alternate stabilization of the two conformations by D122 (closed) (Miller and Benkovic, 1998) and S148 (occluded) (Miller et al., 2001). For all other panels, only the closed conformation is shown.
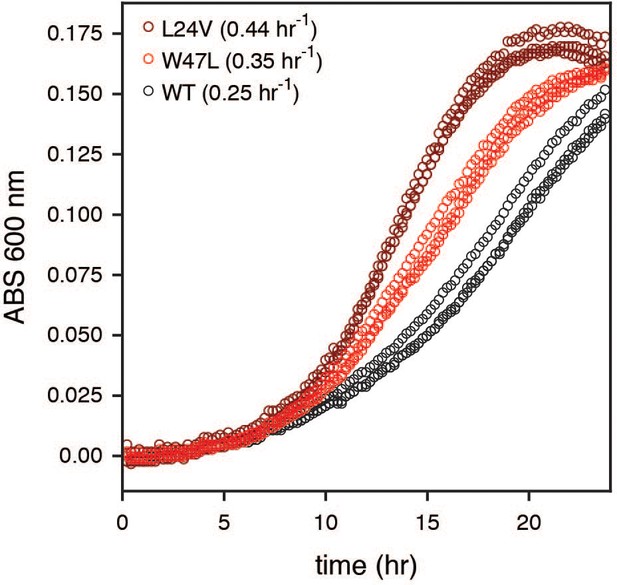
Growth curves for top advantageous mutations.
The absorbance (ABS) at 600 nm was monitored in 96-well plate format for monocultures of the selection strain transformed with strong advantageous mutants (L24V in dark red, W47L in bright red, wild-type in black). The doubling rates (top left in plot) were calculated from the early exponential phase of growth (see Methods). All growth curves are shown as sets of three biological replicates.
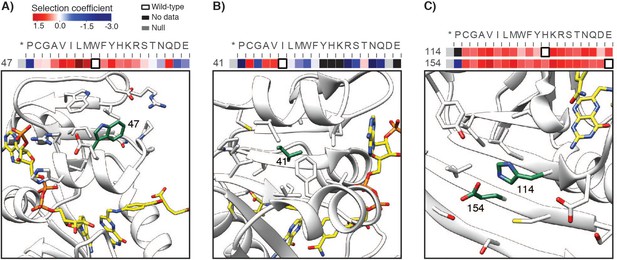
Example positions with multiple advantageous mutations hypothesized to be destabilizing, shown on the DHFR structure.
(A–C) Wild-type residues are colored in green on the DHFR structure (PDB ID: 3QL3) and depicted with slices of the –Lon heatmap (heatmap coloring is as in Figure 2). The wild-type residue is outlined in black on the heatmap. Positions 47, 114, and 154 are in the Beneficial category, and position 41 is in the Deleterious category. In the examples here, advantageous mutations appear to disrupt core packing and a surface salt bridge.
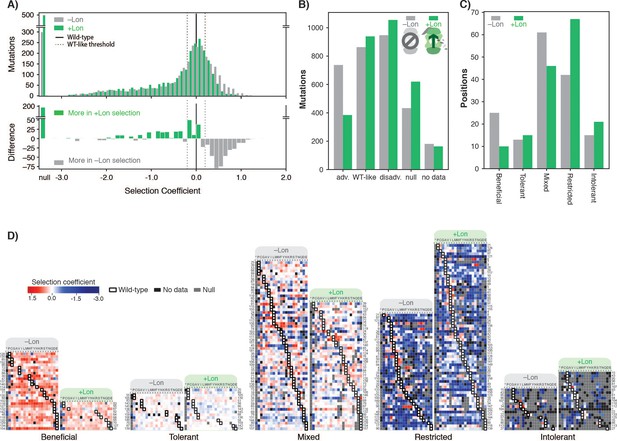
Lon protease expression reshapes the mutational landscape.
(A) Histogram of selection coefficients for mutations (top) in –Lon (grey) and +Lon selection (green). The difference of the histograms (bottom) is shown with grey indicating more mutants for –Lon selection and green indicating more mutants for +Lon selection. The threshold for classification for advantageous and disadvantageous mutations is as in Figure 1 and indicated with dashed lines. (B) Distribution of mutations classified by selection coefficients: 0.2 ≤ advantageous (adv.), 0.2 > WT like > –0.2, –0.2 ≥ disadvantageous (disadv.), null, and no data (a mutant was not detected in the library after transformation into the selection strain). Grey bars: –Lon selection; green bars: +Lon selection. (C) Distribution of sequence positions into the five mutational response categories: Beneficial, Tolerant, Mixed, Deleterious, Intolerant. Grey bars: –Lon selection; green bars: +Lon selection. (D) Heatmap of DHFR selection coefficients in the –Lon and +Lon strains, showing details of the distributions shown in C) (dotted border). Positions (rows) are grouped by their mutational response category for –Lon and +Lon as in C) and sorted by the wild-type amino acid. Amino acid residues (columns) are organized by physiochemical similarity and indicated by their one-letter amino acid code. An asterisk indicates a stop codon. Advantageous mutations are shown in shades of red, disadvantageous mutations in shades of blue, Null mutations in grey and ‘No data’ as defined in A) in black. Wild-type amino acid residues are outlined in black.
Quality of the selection under +Lon conditions.
(A) Comparison of all pairwise replicates for +Lon selection coefficients from triplicate deep mutational scanning on DHFR. The Pearson correlation R2 value from linear regression was 0.70. (B) Distribution of standard errors for individual +Lon selection coefficients from a single replicate. Selection coefficients are the slope from a linear regression of allele frequency as a function of time in selection. The standard error here is the mean square of residuals. (C) Distribution of standard deviations of selection coefficients for individual point mutants over replicate experiments. Each mutant had a measured selection coefficient in at least 2 of the three replicates.
Relationship between error and selection coefficient for +Lon selection.
(A) Standard deviation of selection coefficients over biological replicates. The data were plotted as a function of a sliding window over all single point mutants sorted by selection coefficient. Each point represents the mean error (biological replicate standard deviation) over 50 consecutive selection coefficients (after sorting by value) and the error bars represent ±1 standard deviation of the error. The dashed line represents median error over the entire dataset, which was used determine the for WT-like behavior in Figure 1D. The dotted line represents y = x for comparison between the magnitude of the error relative to the magnitude of the selection coefficient. (B) Standard deviation over synonymous codons coding for the same sequence, plotted as in A.
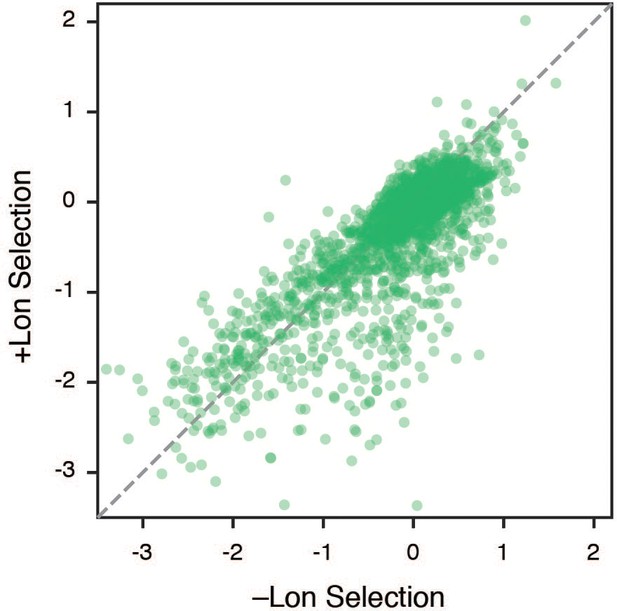
Comparison of selection coefficients ±Lon Scatterplot comparing selection coefficients in –Lon and +Lon selection, showing that mutations are generally repressed by Lon activity.
Despite this general trend, we note that some top advantageous mutations are not impacted by Lon activity.

Ranks of the wild-type amino acid residues in ±Lon selections.
(A) Boxplot showing the distribution of wild-type amino acid residue rankings for –Lon (grey) and +Lon (green) selection. The wild-type amino acid residue ranking at each position is also shown as a distribution of points. Box plots show the median (orange bar) and upper/lower quartiles. The median wild-type amino acid residue rank is eight for –Lon selection and five for +Lon selection. (B) Wild-type amino acid residue rankings from –Lon selection plotted against wild-type amino acid residue rankings from +Lon selection. Dashed lines show ±1 standard deviation for the change in rank between –Lon and +Lon selection.
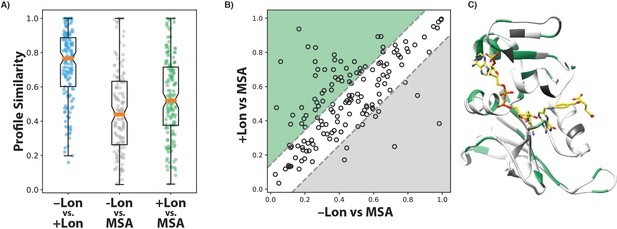
Comparison of DHFR per-position sequence preferences.
(A) Profile similarity (see Materials and methods) was calculated to compare the per-position distribution of amino acid frequencies between selection ±Lon (blue), between –Lon selection and an MSA of DHFR orthologues (grey), and between +Lon selection and the MSA. Each point represents a single position in DHFR. A profile similarity value of 1.0 indicates identical distributions at that position, and a value of 0 represents no overlap in the distributions. The box plot shows the median (orange line), the interval between the first and third quartiles (box), and the maximum and minimum (whiskers). (B) Scatterplot comparing the similarity of amino acid preferences in the MSA to selection ±Lon. Each dot represents a single position in the DHFR sequence. X-axis values represent the profile similarity score between the MSA and –Lon selection for amino acid preferences at each position. Y-axis values represent the profile similarity score between the MSA and +Lon selection for amino acid preferences at each position. The grey dashed lines represent y = x ±one standard deviation for |Similarity(–Lon vs. MSA)position – Similarity(+Lon vs. MSA)position|. Positions in the green region have amino acid preferences more similar to the MSA for +Lon selection, and positions in the grey region have amino acid preferences more similar to the MSA for –Lon selection. (C) Crystal structure model of DHFR (PDB ID: 3QL3) with positions colored by their location in the green, grey, and white regions from panel B).
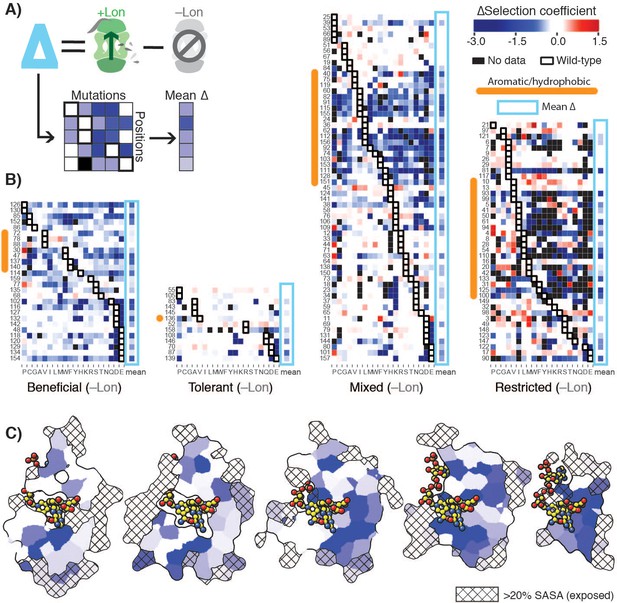
Delta selection coefficients show Lon impact.
(A) Conceptual diagram of ∆selection coefficients, calculated as the +Lon selection coefficient minus the –Lon selection coefficient (see Materials and methods). (B) Heatmap of ∆selection coefficient values for all positions not classified as Intolerant. ∆selection coefficients values between –0.2 and 0.2 are shown in white; ∆selection coefficients >0.2 are in shades of red and ∆selection coefficients <–0.2 in shades of blue. Amino acid residues (columns) are organized by physiochemical similarity and indicated by their one-letter amino acid code. The mean ∆selection coefficient (avg) at each position is shown as a separate column and outlined with a light blue box. Positions (rows) are sorted by the wild-type amino acid and grouped by their mutational response category from the –Lon selection in Figure 2C,D. Positions with a native VILMWF or Y amino acid are indicated with an orange bar to the left. (C) Per-position mean ∆selection coefficient displayed on the structural model of DHFR. The five cross-section slices of the DHFR structure are displayed as in Figure 1E, and the color scale is as in B).
-
Figure 3—source data 1
Burial classification for DHFR positions from the Getarea server (Fraczkiewicz and Braun, 1998) as described in Materials and methods.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig3-data1-v2.xlsx
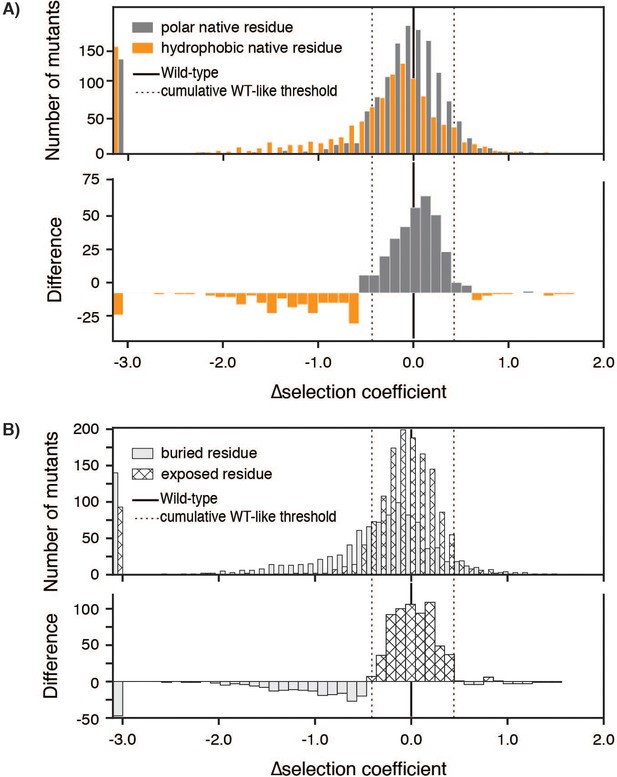
∆selection coefficients.
(A) Histogram of ∆selection coefficients (top) with mutants at positions with hydrophobic (AVILMWFY) wild-type amino acid residues in orange and at positions with polar (HKRSTNQDE) wild-type amino acid residues in grey. Selection coefficients for positions with a wild-type P, G, or C residue are not included. The difference of the histograms (bottom) is shown with grey indicating more mutants to positions with a wild-type polar residue and orange indicating more mutants to positions with a wild-type hydrophobic residue. Dotted lines indicate twice the median of standard deviations from Figure 1, Figure 1—figure supplement 3. (B) Histogram of ∆selection coefficients (top) with mutants at buried positions (solid) and at exposed positions (hatched) as listed in Figure 3—source data 1. Selection coefficients for positions that were Intolerant in –Lon selection are not included. The difference of the histograms (bottom) is shown with solid indicating more mutants to buried positions and hatched indicating more mutants to exposed positions.
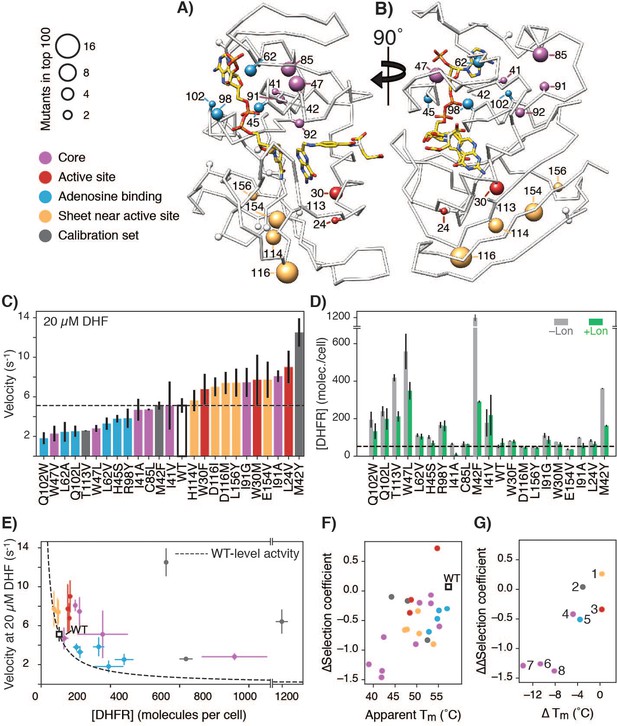
Advantageous mutations arise from an interplay of increased enzymatic velocity and increased abundance in the absence of Lon.
(A) DHFR structure with mutational hot-spots. For positions with two or more top 100 advantageous mutations in the absence of Lon, the beta carbon is depicted as a sphere scaled according to the number of top mutations. For mutants selected for in vitro characterization, the beta carbon is colored according to its location in the DHFR structure: core (purple), surface beta-sheet (gold), proximal to the adenine ring on NADPH (blue), or proximal to the active site and M20 loop (red). Positions for advantageous mutants from the calibration set are depicted in dark grey. (B) The structure from A) rotated 90° clockwise. (C) In vitro velocities of purified DHFR wild-type and point mutants measured at 20 µM DHF. Bars are colored in reference to the hot-spots in A). Error bars represent ±1 standard deviation from three independent experiments (Materials and methods). The dashed line represents the velocity of WT DHFR. (D) DHFR cellular abundance calculated from the lysate DHFR activity in Figure 4—figure supplement 2 and in vitro kinetics with purified enzyme (see Materials and methods). Error bars represent the cumulative percent error (standard deviation) from three independent experiments for velocity and three biological replicates for lysate activity. Data are shown in both the -Lon (light grey) and +Lon (green) conditions. The dashed line represents the WT expression level of DHFR in the –Lon background. Mutants are in the same order as in C) (see Figure 4—source data 2; four mutants were not measured). (E) Cellular abundance of DHFR vs. in vitro velocities of purified DHFR wild-type and point mutants measured at 20 µM DHF. Points are colored as in A). Error bars represent ±1 standard deviation from three independent experiments (Materials and methods). The dashed line represents WT-level DHFR activity, i.e. DHFR abundance/velocity pairs whose product is equivalent to [DHFR]WT • velocityWT. (F) Correlation between in vitro Tm values and in vivo ∆selection coefficients for DHFR wild-type and characterized mutants. Points are colored as in A). (G) ∆Tm values and ∆∆selection coefficient for mutations at the same position. Points representing comparison between mutants are numbered as follows: 1) D116I-M, 2) M42Y-F, 3) W30M-F, 4) I91G-A, 5) Q102W-L, 6) L62A-V, 7) I41A-V, 8) W47V-L.
-
Figure 4—source data 1
In vitro velocity for selected advantageous measured as described in Materials and methods at multiple concentrations of DHF are reported with the standard deviation over three independent experiments.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Soluble DHFR abundance levels in molecules per cell measured from lysate activity assays as described in Materials and methods.
All values are for the SMT205 plasmid transformed into the cell strain in the column heading. NM, not measured.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Apparent Tm values from thermal denaturation experiments monitored by CD signal at 225 nm are reported along with the ∆selection coefficient (Lon impact) value depicted in Figure 4D.
- https://cdn.elifesciences.org/articles/53476/elife-53476-fig4-data3-v2.xlsx
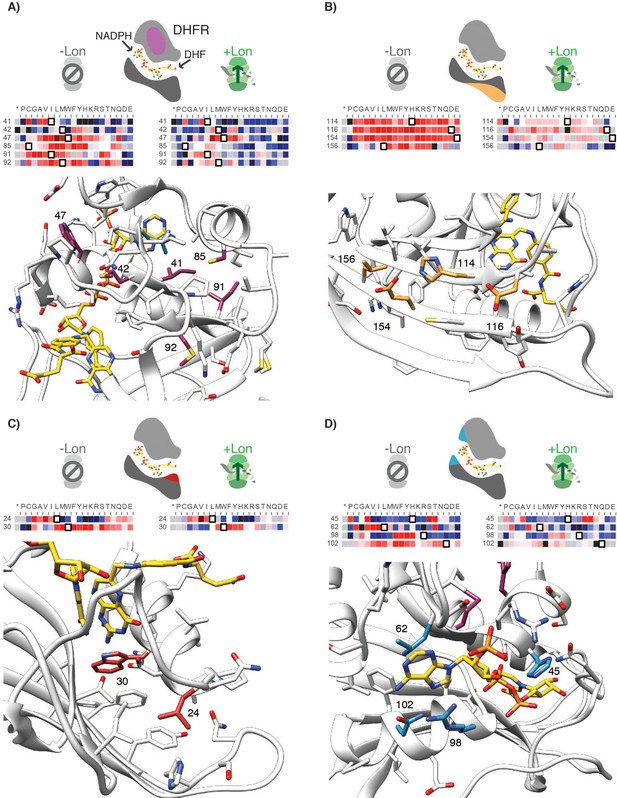
Structural context for hotspot residues from Figure 4.
(A–D) For each panel, the hot spot region is indicated on a cartoon of DHFR: globular core in purple (A), the beta-sheet surface below the active site in gold (B), the base of the M20 loop in red (C) and the adenosine binding site in blue (D). Slices of the –Lon and +Lon heatmaps are shown for each position within the hot spot region (heatmap coloring is as in Figure 2). The wild-type residue is outlined in black. Positions 30, 47, 85, 102, 114, 116, 154 are in the Beneficial category. Position 24, 25, 62, 91, 92, 156 are in the Mixed category. Positions 41, 42, and 98 are in the Deleterious category. For A-C) the structure shown is PDBID: 3QL3, and for D) the structure shown is PDB ID: 1RX1. In IRX1 (as in 1RX4), R98 is in proximity to the adenine ring. In 3QL3, R98 extends into bulk solvent. Residues within the hot spot cluster are labeled with their residue number.Figure 4—figure supplement 2.
Lysate activity for DHFR wild-type and point mutants on the selection plasmid.
(A) Lysate activity for DHFR variants under selection growth conditions (see Materials and methods) plotted as the rate of change in DHF concentration as a function of time monitored over the window of DHF concentration from 30 µM to 20 µM. DHFR activities in ER2566 ∆folA/∆thyA –Lon lysates are colored in grey. DHFR activities in ER2566 ∆folA/∆thyA +Lon lysates are colored in green. Error bars represent ±1 standard deviation from three biological replicates. (B) Relative lysate activities for DHFR variants. Lysate activities from A) normalized by WT-level of activity in the corresponding ±Lon cell lysate.
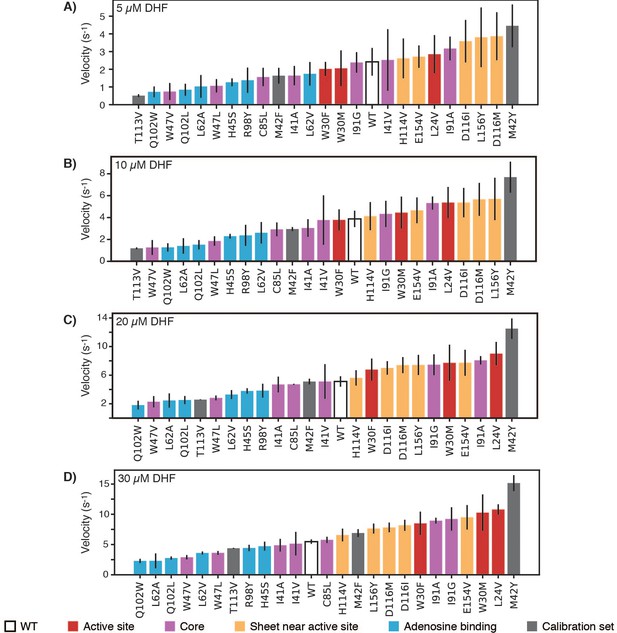
In vitro velocities of purified DHFR wild-type and point mutants.
Velocities were measured at (A) 5, (B) 10, (C) 20, and (D) 30 µM DHF (Figure 4—source data 1). For each mutant, the bar is colored by the mutation’s location within the hot spots from Figure 4 and Figure 4—figure supplement 1. Error bars represent ±1 standard deviation from three independent experiments.
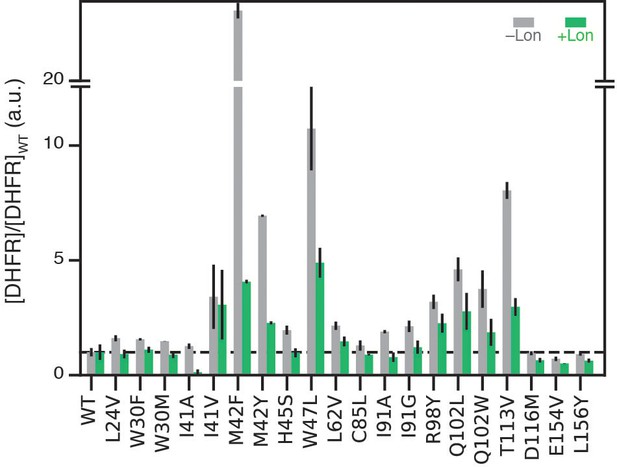
Soluble cellular abundance for DHFR wild-type and point mutants on the selection plasmid.
Relative expression of DHFR variants. DHFR abundances from Figure 4D normalized by WT-level of abundance in the corresponding ±Lon cell lysate. Relative DHFR abundances in ER2566 ∆folA/∆thyA –Lon lysates are colored in grey. Relative DHFR abundances in ER2566 ∆folA/∆thyA +Lon lysates are colored in green. Error bars represent the cumulative percent error (standard deviation) from three independent experiments for velocity and three biological replicates for lysate activity.
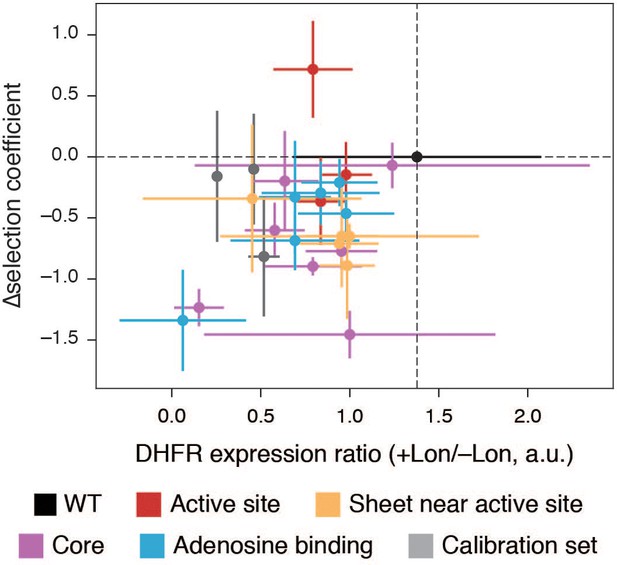
Lon impact as ∆selection coefficient versus change in DHFR abundance ±Lon.
Correlation between the ratio of cellular DHFR abundance (Figure 4D, Figure 4—source data 2, [DHFR]+Lon/[DHFR]–Lon) and in vivo ∆selection coefficients ±Lon for DHFR wild-type and point mutants. Points are colored by the mutation’s location within the hot spots from Figure 4 and Figure 4—figure supplement 1. X-axis error bars represent the cumulative percent error (standard deviation) from three measurements of DHFR concentration with and without Lon (Materials and methods). Y-axis error bars the cumulative error (standard deviation) from three biological replicates for selection with and without Lon (Materials and methods). The ratio of expression for WT is not 1.0 because there is an increase in WT DHFR expression in ER2566 ∆folA/∆thy +Lon relative to WT expression in ER2566 ∆folA/∆thy –Lon (see Figure 1—figure supplement 2, Figure 1—source data 1). The reason for the unusual behavior of L24V (positive ∆selection coefficient), a mutation in the active site, is unknown.
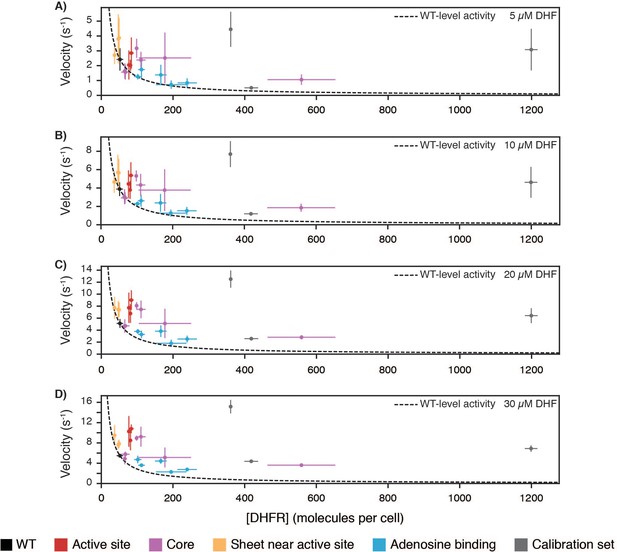
Cellular abundance versus in vitro velocity for DHFR wild-type and point mutants.
Cellular abundance of DHFR vs. in vitro velocities of purified DHFR measured at (A) 5, (B) 10, (C) 20, and (D) 30 µM DHF (see Figure 4—figure supplement 3, Figure 4—source data 1, Figure 4D, Figure 4—source data 2). Points are colored by the mutation’s location within the hot spots from Figure 4 and Figure 4—figure supplement 1. Error bars represent ±1 standard deviation from three independent experiments (Materials and methods). The dashed line represents WT equivalent DHFR activity, where [DHFR]WT • velocityWT = [DHFR]mut • velocitymut.
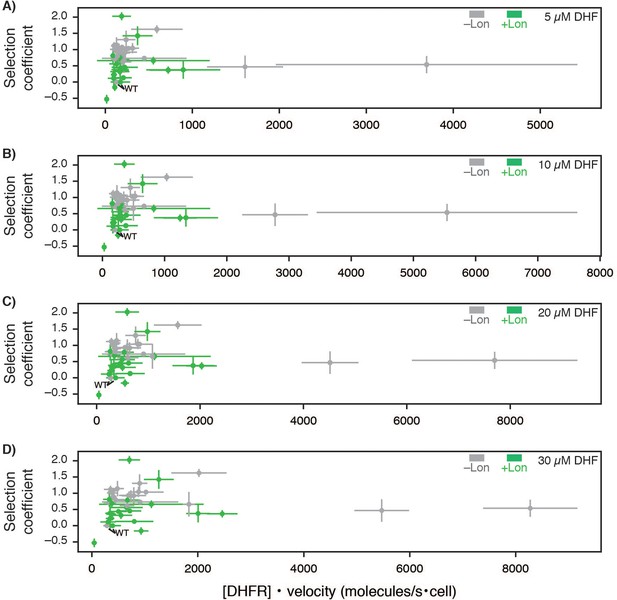
Selection coefficient compared to predictions of DHFR wild-type and point mutant activity from cellular abundance and in vitro velocity measurements.
Selection coefficients for –Lon selection (grey) and +Lon selection (green) plotted against DHFR activity calculated as cellular abundance of DHFR times in vitro velocities of purified DHFR variants ([DHFR] • velocity[DHF]) measured at (A) 5, (B) 10, (C) 20, and (D) 30 µM DHF (see Figure 4D, Figure 4—figure supplement 3, Figure 4—source data 1, Figure 4—source data 2). X-axis error bars represent the cumulative percent error (standard deviation) from three measurements of DHFR concentration with and without Lon and three independent experiments for velocity (Materials and methods). Y-axis error bars represent ±1 standard deviation from (Materials and methods).
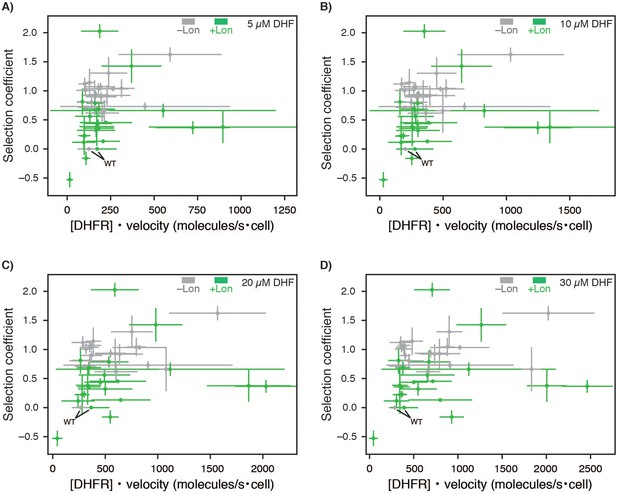
Zoom in for Selection coefficient compared to predictions of DHFR wild-type and point mutant activity from cellular abundance and in vitro velocity measurements.
Selection coefficients for –Lon selection (grey) and +Lon selection (green) plotted against DHFR activity calculated as cellular abundance of DHFR times in vitro velocities of purified DHFR variants ([DHFR] • velocity[DHF]) measured at (A) 5, (B) 10, (C) 20, and (D) 30 µM DHF (see Figure 4D, Figure 4—figure supplement 3, Figure 4—source data 1, Figure 4—source data 2). X-axis error bars represent the cumulative percent error (standard deviation) from three measurements of DHFR concentration with and without Lon and three independent experiments for velocity (Materials and methods).Y-axis error bars represent ±1 standard deviation from (Materials and methods).
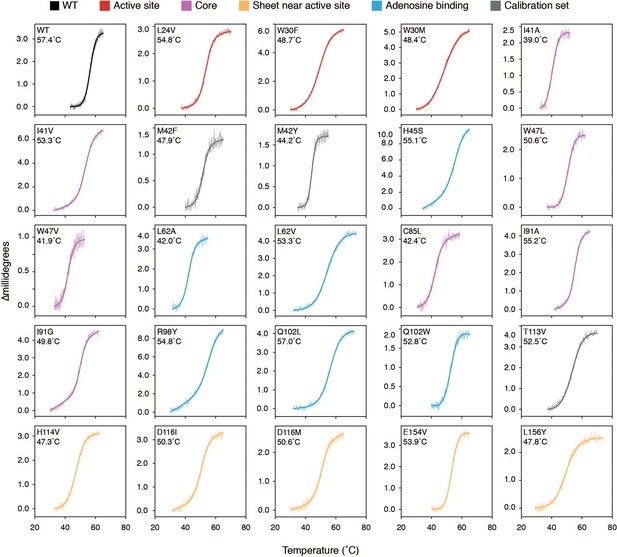
Thermal denaturation curves monitored by CD signal at 225 m for selected hotspot mutants.
The curves are colored by the mutation’s location within the hot spots from Figure 4 and Figure 4—figure supplement 1. The raw data are shown with thin lines and the fitted curves are shown as thick lines. For each plot, the mutant identity and apparent Tm value are listed in the top left corner.
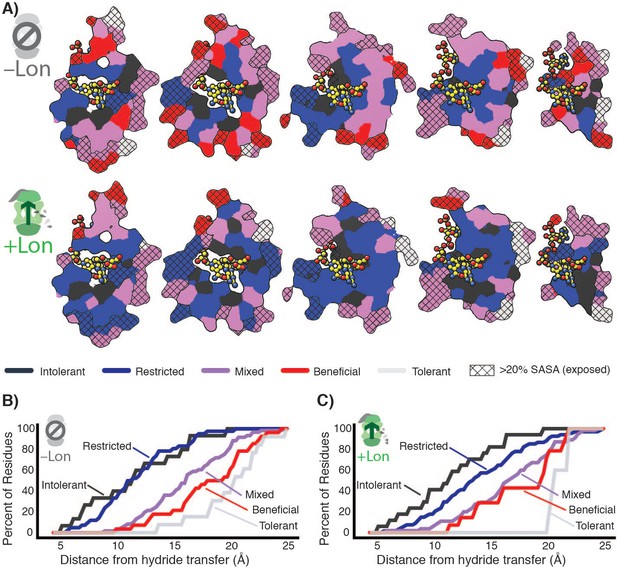
Structural characterization of multiple constraints on the DHFR mutational landscape.
(A) Mutational response categories from –Lon selection (top, categories in Figure 2C,D) and +Lon selection (bottom, categories as in Figure 2C,D) colored onto residues and displayed on slices as in Figure 1E. (B) Relationship between mutational response and distance from hydride transfer for –Lon selection. The percent of positions from each mutational response category are plotted as a function of distance from the site of hydride transfer. Each category colored as in A), top). (C) Relationship between mutational response and distance from hydride transfer for +Lon selection. Each category colored as in A), bottom).
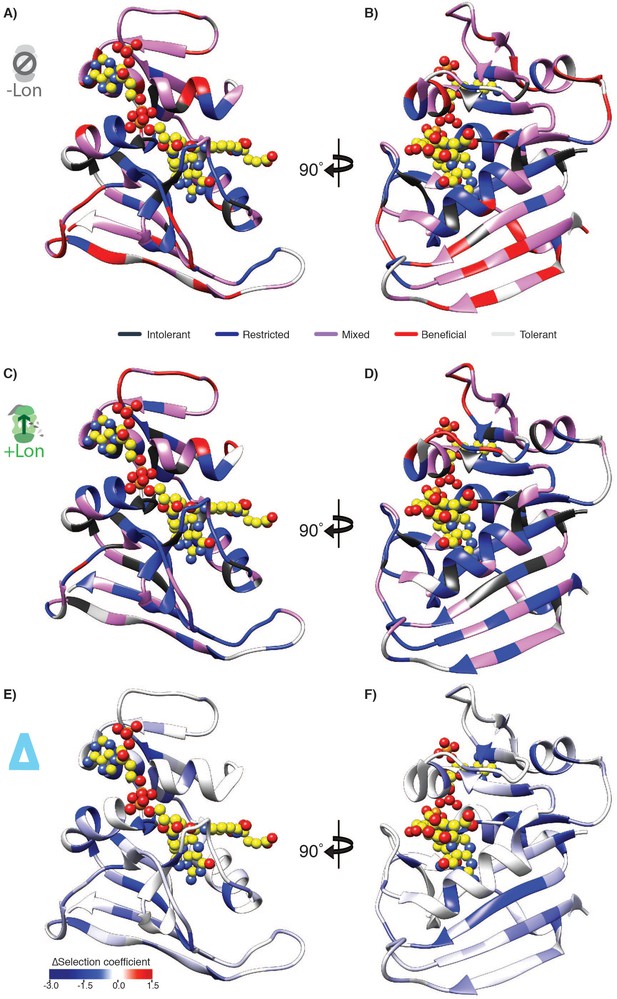
Selection coefficients under the two Lon expression regimes mapped on the DHFR structure.
Structural model of DHFR (PDB ID: 3QL3) in ribbon representation with the DHF substrate and the NADPH cofactor represented by spheres (yellow carbon and heteroatom coloring). The residues are colored in (A,B) by mutational response category from Figure 2C,D for –Lon selection, in (C,D) by mutational response category from +Lon selection, or in (E,F) by the per-position mean ∆selection coefficient from Figure 3.

Burial of residues within each mutation response category reported as the mean number of atomic neighbors.
Each point represents one amino acid side chain, and the y-axis reports the average number of heavy atom neighbors within an 8 Å shell for all heavy atoms in that side chain. Box plots are overlaid on the distribution to show the median (orange bar) and upper/lower quartiles. Mutational response categories are shown for both –Lon and +Lon selection. The green arrow highlights the absence of buried Beneficial positions in +Lon selection.

Residues in mutational response categories in the –Lon selection as a function of distance from several sites in the DHFR structure.
(A) Location of hydride transfer site, the M20 residue on the M20 loop (orange), and hot spot sites from Figure 4 (the core of the globular domain represented by I41, the beta-sheet surface below the active site represented by L112, and the adenine ring on NADPH) indicated on the DHFR structure (PDB ID: 3QL3). (B–F) The distance relationships between each site and the residues in each mutational response category in the –Lon selection are shown (left) as boxplots with points representing the individual mutants and (right) as curves showing the percent of sequence positions in each mutational response category as a function of distance from the site. Boxplots and curves are colored by mutational response categories from –Lon selection as in Figure 5.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | ER2566 | New England Biolabs | Cat# C2566I | Chemically competent cells |
| Strain, strain background (Escherichia coli) | ER2566 ∆folA/∆thyA (–Lon) | Reynolds et al. Cell 2011 | Chemically competent and electrocompetent cells | |
| Strain, strain background (Escherichia coli) | ER2566 ∆folA/∆thyA (+Lon) | This work | Chemically competent and electrocompetent cells | |
| Recombinant DNA reagent | SMT101 (plasmid) | This work | Dual expression of DHFR and TYMS, in vivo assays, chloramphenicol (35 µg/mL final concentration) | |
| Recombinant DNA reagent | SMT201 (plasmid) | This work | SMT101 with TET promter for TYMS, in vivo assays, Chloramphenicol (35 µg/mL final concentration) | |
| Recombinant DNA reagent | SMT205 (plasmid) | This work | SMT201 with mutated RBS for DHFR, in vivo assays, Chloramphenicol (35 µg/mL final concentration) | |
| Recombinant DNA reagent | SMT215 (plasmid) | This work | SMT205 with DHFR-FLAG-tag, western blot, Chloramphenicol (35 µg/mL final concentration) | |
| Recombinant DNA reagent | KR101/SMT301 (plasmid) | Reynolds et al. Cell 2011 | His8-tag, Heterologous expression, NiNTA purfication, kanamycin (50 µg/mL final concentration) | |
| Recombinant DNA reagent | pSIM6 (plasmid) | Blomfield et al., 1991 | Lambda Red recombinase expression, temperature-sensitive promoter, ampicillin/carbenicilin (100 µg/mL final concentration) | |
| Recombinant DNA reagent | pIB279 (plasmid) | Blomfield et al., 1991 | KAN-SacB cassette for positive/negative selection, ampicillin/carbenicilin (100 µg/mL final concentration) | |
| Sequence-based reagent | TetDuet1_sense | This work | Mutagenic PCR primer | ccgCTTAAGtcgaacagaaagtaatcgtattgtacatccctatc |
| Sequence-based reagent | TetDuet2_anti | This work | Mutagenic PCR primer | gatagggatgtcaatctctatcactgatagggatgtacaatacg |
| Sequence-based reagent | TetDuet3_sense | This work | Mutagenic PCR primer | agagattgacatccctatcagtgatagagatactgagcacatcag |
| Sequence-based reagent | TetDuet4_anti | This work | Mutagenic PCR primer | ctttaatgaattcggtcagtgcgtcctgctgatgtgctcagtatctc |
| Sequence-based reagent | TetDuet5_sense | This work | Mutagenic PCR primer | cactgaccgaattcattaaagaggagaaaggtaccatatggc |
| Sequence-based reagent | TetDuet_5flanking | This work | Mutagenic PCR primer | ccgcttaagtcgaacagaaag |
| Sequence-based reagent | TetDuet_3flanking | This work | Mutagenic PCR primer | cggagatctgccatatggtacc |
| Sequence-based reagent | WT_DHFR_pos2_fwd | This work | Mutagenic PCR primer | NNSAGTCTGATTGCGGCGTTAG |
| Sequence-based reagent | WT_DHFR_pos2_fwd2 | This work | Mutagenic PCR primer | NNSAGTCTGATTGCGGCGTTAG |
| Sequence-based reagent | WT_DHFR_pos3_fwd | This work | Mutagenic PCR primer | NNSCTGATTGCGGCGTTAGCG |
| Sequence-based reagent | WT_DHFR_pos4_fwd | This work | Mutagenic PCR primer | NNSATTGCGGCGTTAGCGGTA |
| Sequence-based reagent | WT_DHFR_pos5_fwd | This work | Mutagenic PCR primer | NNSGCGGCGTTAGCGGTAGAT |
| Sequence-based reagent | WT_DHFR_pos6_fwd | This work | Mutagenic PCR primer | NNSGCGTTAGCGGTAGATCGC |
| Sequence-based reagent | WT_DHFR_pos7_fwd | This work | Mutagenic PCR primer | NNSTTAGCGGTAGATCGCGTTATC |
| Sequence-based reagent | WT_DHFR_pos8_fwd | This work | Mutagenic PCR primer | NNSGCGGTAGATCGCGTTATCG |
| Sequence-based reagent | WT_DHFR_pos8_fwd2 | This work | Mutagenic PCR primer | NNSGCGGTAGATCGCGTTATCG |
| Sequence-based reagent | WT_DHFR_pos9_fwd | This work | Mutagenic PCR primer | NNSGTAGATCGCGTTATCGGCATG |
| Sequence-based reagent | WT_DHFR_pos10_fwd | This work | Mutagenic PCR primer | NNSGATCGCGTTATCGGCATGG |
| Sequence-based reagent | WT_DHFR_pos11_fwd | This work | Mutagenic PCR primer | NNSCGCGTTATCGGCATGGAAAA |
| Sequence-based reagent | WT_DHFR_pos12_fwd | This work | Mutagenic PCR primer | NNSGTTATCGGCATGGAAAACGC |
| Sequence-based reagent | WT_DHFR_pos13_fwd | This work | Mutagenic PCR primer | NNSATCGGCATGGAAAACGCC |
| Sequence-based reagent | WT_DHFR_pos14_fwd | This work | Mutagenic PCR primer | NNSGGCATGGAAAACGCCATG |
| Sequence-based reagent | WT_DHFR_pos15_fwd | This work | Mutagenic PCR primer | NNSATGGAAAACGCCATGCCG |
| Sequence-based reagent | WT_DHFR_pos16_fwd | This work | Mutagenic PCR primer | NNSGAAAACGCCATGCCGTGG |
| Sequence-based reagent | WT_DHFR_pos17_fwd | This work | Mutagenic PCR primer | NNSAACGCCATGCCGTGGAAC |
| Sequence-based reagent | WT_DHFR_pos18_fwd | This work | Mutagenic PCR primer | NNSGCCATGCCGTGGAACCTG |
| Sequence-based reagent | WT_DHFR_pos19_fwd | This work | Mutagenic PCR primer | NNSATGCCGTGGAACCTGCCT |
| Sequence-based reagent | WT_DHFR_pos20_fwd | This work | Mutagenic PCR primer | NNSCCGTGGAACCTGCCTGCC |
| Sequence-based reagent | WT_DHFR_pos21_fwd | This work | Mutagenic PCR primer | NNSTGGAACCTGCCTGCCGAT |
| Sequence-based reagent | WT_DHFR_pos22_fwd | This work | Mutagenic PCR primer | NNSAACCTGCCTGCCGATCTC |
| Sequence-based reagent | WT_DHFR_pos22_fwd2 | This work | Mutagenic PCR primer | NNSAACCTGCCTGCCGATCTC |
| Sequence-based reagent | WT_DHFR_pos23_fwd | This work | Mutagenic PCR primer | NNSCTGCCTGCCGATCTCGCC |
| Sequence-based reagent | WT_DHFR_pos24_fwd | This work | Mutagenic PCR primer | NNSCCTGCCGATCTCGCCTGG |
| Sequence-based reagent | WT_DHFR_pos25_fwd | This work | Mutagenic PCR primer | NNSGCCGATCTCGCCTGGTTT |
| Sequence-based reagent | WT_DHFR_pos26_fwd | This work | Mutagenic PCR primer | NNSGATCTCGCCTGGTTTAAACGC |
| Sequence-based reagent | WT_DHFR_pos27_fwd | This work | Mutagenic PCR primer | NNSCTCGCCTGGTTTAAACGCAACA |
| Sequence-based reagent | WT_DHFR_pos28_fwd | This work | Mutagenic PCR primer | NNSGCCTGGTTTAAACGCAACAC |
| Sequence-based reagent | WT_DHFR_pos29_fwd | This work | Mutagenic PCR primer | NNSTGGTTTAAACGCAACACCTTAAATAAAC |
| Sequence-based reagent | WT_DHFR_pos30_fwd | This work | Mutagenic PCR primer | NNSTTTAAACGCAACACCTTAAATAAACCCG |
| Sequence-based reagent | WT_DHFR_pos31_fwd | This work | Mutagenic PCR primer | NNSAAACGCAACACCTTAAATAAACCCGTG |
| Sequence-based reagent | WT_DHFR_pos32_fwd | This work | Mutagenic PCR primer | NNSCGCAACACCTTAAATAAACCCGT |
| Sequence-based reagent | WT_DHFR_pos33_fwd | This work | Mutagenic PCR primer | NNSAACACCTTAAATAAACCCGTGATTATGG |
| Sequence-based reagent | WT_DHFR_pos34_fwd | This work | Mutagenic PCR primer | NNSACCTTAAATAAACCCGTGATTATGGG |
| Sequence-based reagent | WT_DHFR_pos35_fwd | This work | Mutagenic PCR primer | NNSTTAAATAAACCCGTGATTATGGGCC |
| Sequence-based reagent | WT_DHFR_pos36_fwd | This work | Mutagenic PCR primer | NNSAATAAACCCGTGATTATGGGCC |
| Sequence-based reagent | WT_DHFR_pos37_fwd | This work | Mutagenic PCR primer | NNSAAACCCGTGATTATGGGCC |
| Sequence-based reagent | WT_DHFR_pos38_fwd | This work | Mutagenic PCR primer | NNSCCCGTGATTATGGGCCGC |
| Sequence-based reagent | WT_DHFR_pos39_fwd | This work | Mutagenic PCR primer | NNSGTGATTATGGGCCGCCATAC |
| Sequence-based reagent | WT_DHFR_pos40_fwd | This work | Mutagenic PCR primer | NNSATTATGGGCCGCCATACCT |
| Sequence-based reagent | WT_DHFR_pos41_fwd | This work | Mutagenic PCR primer | NNSATGGGCCGCCATACCTGG |
| Sequence-based reagent | WT_DHFR_pos42_fwd | This work | Mutagenic PCR primer | NNSGGCCGCCATACCTGGGAA |
| Sequence-based reagent | WT_DHFR_pos42_fwd2 | This work | Mutagenic PCR primer | NNSGGCCGCCATACCTGGGAATC |
| Sequence-based reagent | WT_DHFR_pos43_fwd | This work | Mutagenic PCR primer | NNSCGCCATACCTGGGAATCAATC |
| Sequence-based reagent | WT_DHFR_pos43_fwd2 | This work | Mutagenic PCR primer | NNSCGCCATACCTGGGAATCAATC |
| Sequence-based reagent | WT_DHFR_pos44_fwd | This work | Mutagenic PCR primer | NNSCATACCTGGGAATCAATCGGTC |
| Sequence-based reagent | WT_DHFR_pos45_fwd | This work | Mutagenic PCR primer | NNSACCTGGGAATCAATCGGTC |
| Sequence-based reagent | WT_DHFR_pos46_fwd | This work | Mutagenic PCR primer | NNSTGGGAATCAATCGGTCGTC |
| Sequence-based reagent | WT_DHFR_pos47_fwd | This work | Mutagenic PCR primer | NNSGAATCAATCGGTCGTCCGTTG |
| Sequence-based reagent | WT_DHFR_pos48_fwd | This work | Mutagenic PCR primer | NNSTCAATCGGTCGTCCGTTGC |
| Sequence-based reagent | WT_DHFR_pos49_fwd | This work | Mutagenic PCR primer | NNSATCGGTCGTCCGTTGCCA |
| Sequence-based reagent | WT_DHFR_pos50_fwd | This work | Mutagenic PCR primer | NNSGGTCGTCCGTTGCCAGGAC |
| Sequence-based reagent | WT_DHFR_pos51_fwd | This work | Mutagenic PCR primer | NNSCGTCCGTTGCCAGGACGC |
| Sequence-based reagent | WT_DHFR_pos52_fwd | This work | Mutagenic PCR primer | NNSCCGTTGCCAGGACGCAAA |
| Sequence-based reagent | WT_DHFR_pos53_fwd | This work | Mutagenic PCR primer | NNSTTGCCAGGACGCAAAAATATTATCC |
| Sequence-based reagent | WT_DHFR_pos54_fwd | This work | Mutagenic PCR primer | NNSCCAGGACGCAAAAATATTATCCTCAG |
| Sequence-based reagent | WT_DHFR_pos55_fwd | This work | Mutagenic PCR primer | NNSGGACGCAAAAATATTATCCTCAGCAG |
| Sequence-based reagent | WT_DHFR_pos56_fwd | This work | Mutagenic PCR primer | NNSCGCAAAAATATTATCCTCAGCAGTCAA |
| Sequence-based reagent | WT_DHFR_pos57_fwd | This work | Mutagenic PCR primer | NNSAAAAATATTATCCTCAGCAGTCAACCGG |
| Sequence-based reagent | WT_DHFR_pos58_fwd | This work | Mutagenic PCR primer | NNSAATATTATCCTCAGCAGTCAACCGGGTA |
| Sequence-based reagent | WT_DHFR_pos59_fwd | This work | Mutagenic PCR primer | NNSATTATCCTCAGCAGTCAACCG |
| Sequence-based reagent | WT_DHFR_pos60_fwd | This work | Mutagenic PCR primer | NNSATCCTCAGCAGTCAACCG |
| Sequence-based reagent | WT_DHFR_pos61_fwd | This work | Mutagenic PCR primer | NNSCTCAGCAGTCAACCGGGT |
| Sequence-based reagent | WT_DHFR_pos62_fwd | This work | Mutagenic PCR primer | NNSAGCAGTCAACCGGGTACG |
| Sequence-based reagent | WT_DHFR_pos63_fwd | This work | Mutagenic PCR primer | NNSAGTCAACCGGGTACGGAC |
| Sequence-based reagent | WT_DHFR_pos64_fwd | This work | Mutagenic PCR primer | NNSCAACCGGGTACGGACGAT |
| Sequence-based reagent | WT_DHFR_pos65_fwd | This work | Mutagenic PCR primer | NNSCCGGGTACGGACGATCGC |
| Sequence-based reagent | WT_DHFR_pos66_fwd | This work | Mutagenic PCR primer | NNSGGTACGGACGATCGCGTA |
| Sequence-based reagent | WT_DHFR_pos66_fwd2 | This work | Mutagenic PCR primer | NNSGGTACGGACGATCGCGTAAC |
| Sequence-based reagent | WT_DHFR_pos67_fwd | This work | Mutagenic PCR primer | NNSACGGACGATCGCGTAACG |
| Sequence-based reagent | WT_DHFR_pos67_fwd2 | This work | Mutagenic PCR primer | NNSACGGACGATCGCGTAACG |
| Sequence-based reagent | WT_DHFR_pos68_fwd | This work | Mutagenic PCR primer | NNSGACGATCGCGTAACGTGG |
| Sequence-based reagent | WT_DHFR_pos69_fwd | This work | Mutagenic PCR primer | NNSGATCGCGTAACGTGGGTG |
| Sequence-based reagent | WT_DHFR_pos70_fwd | This work | Mutagenic PCR primer | NNSCGCGTAACGTGGGTGAAG |
| Sequence-based reagent | WT_DHFR_pos71_fwd | This work | Mutagenic PCR primer | NNSGTAACGTGGGTGAAGTCGG |
| Sequence-based reagent | WT_DHFR_pos72_fwd | This work | Mutagenic PCR primer | NNSACGTGGGTGAAGTCGGTG |
| Sequence-based reagent | WT_DHFR_pos73_fwd | This work | Mutagenic PCR primer | NNSTGGGTGAAGTCGGTGGAT |
| Sequence-based reagent | WT_DHFR_pos73_fwd2 | This work | Mutagenic PCR primer | NNSTGGGTGAAGTCGGTGGATG |
| Sequence-based reagent | WT_DHFR_pos74_fwd | This work | Mutagenic PCR primer | NNSGTGAAGTCGGTGGATGAAGC |
| Sequence-based reagent | WT_DHFR_pos74_fwd2 | This work | Mutagenic PCR primer | NNSGTGAAGTCGGTGGATGAAGC |
| Sequence-based reagent | WT_DHFR_pos75_fwd | This work | Mutagenic PCR primer | NNSAAGTCGGTGGATGAAGCCAT |
| Sequence-based reagent | WT_DHFR_pos76_fwd | This work | Mutagenic PCR primer | NNSTCGGTGGATGAAGCCATC |
| Sequence-based reagent | WT_DHFR_pos77_fwd | This work | Mutagenic PCR primer | NNSGTGGATGAAGCCATCGCG |
| Sequence-based reagent | WT_DHFR_pos78_fwd | This work | Mutagenic PCR primer | NNSGATGAAGCCATCGCGGCG |
| Sequence-based reagent | WT_DHFR_pos79_fwd | This work | Mutagenic PCR primer | NNSGAAGCCATCGCGGCGTGT |
| Sequence-based reagent | WT_DHFR_pos80_fwd | This work | Mutagenic PCR primer | NNSGCCATCGCGGCGTGTGGT |
| Sequence-based reagent | WT_DHFR_pos80_fwd2 | This work | Mutagenic PCR primer | NNSGCCATCGCGGCGTGTGG |
| Sequence-based reagent | WT_DHFR_pos81_fwd | This work | Mutagenic PCR primer | NNSATCGCGGCGTGTGGTGAC |
| Sequence-based reagent | WT_DHFR_pos82_fwd | This work | Mutagenic PCR primer | NNSGCGGCGTGTGGTGACGTA |
| Sequence-based reagent | WT_DHFR_pos82_fwd2 | This work | Mutagenic PCR primer | NNSGCGGCGTGTGGTGACGTACCAGAAATC |
| Sequence-based reagent | WT_DHFR_pos83_fwd | This work | Mutagenic PCR primer | NNSGCGTGTGGTGACGTACCA |
| Sequence-based reagent | WT_DHFR_pos84_fwd | This work | Mutagenic PCR primer | NNSTGTGGTGACGTACCAGAAATCAT |
| Sequence-based reagent | WT_DHFR_pos84_fwd2 | This work | Mutagenic PCR primer | NNSTGTGGTGACGTACCAGAAATCATG |
| Sequence-based reagent | WT_DHFR_pos85_fwd | This work | Mutagenic PCR primer | NNSGGTGACGTACCAGAAATCATGG |
| Sequence-based reagent | WT_DHFR_pos86_fwd | This work | Mutagenic PCR primer | NNSGACGTACCAGAAATCATGGTGATTGG |
| Sequence-based reagent | WT_DHFR_pos87_fwd | This work | Mutagenic PCR primer | NNSGTACCAGAAATCATGGTGATTGGCGG |
| Sequence-based reagent | WT_DHFR_pos88_fwd | This work | Mutagenic PCR primer | NNSCCAGAAATCATGGTGATTGGCGG |
| Sequence-based reagent | WT_DHFR_pos89_fwd | This work | Mutagenic PCR primer | NNSGAAATCATGGTGATTGGCGGCG |
| Sequence-based reagent | WT_DHFR_pos89_fwd2 | This work | Mutagenic PCR primer | NNSGAAATCATGGTGATTGGCGGC |
| Sequence-based reagent | WT_DHFR_pos90_fwd | This work | Mutagenic PCR primer | NNSATCATGGTGATTGGCGGC |
| Sequence-based reagent | WT_DHFR_pos91_fwd | This work | Mutagenic PCR primer | NNSATGGTGATTGGCGGCGGTC |
| Sequence-based reagent | WT_DHFR_pos92_fwd | This work | Mutagenic PCR primer | NNSGTGATTGGCGGCGGTCGC |
| Sequence-based reagent | WT_DHFR_pos93_fwd | This work | Mutagenic PCR primer | NNSATTGGCGGCGGTCGCGTTTA |
| Sequence-based reagent | WT_DHFR_pos94_fwd | This work | Mutagenic PCR primer | NNSGGCGGCGGTCGCGTTTAT |
| Sequence-based reagent | WT_DHFR_pos95_fwd | This work | Mutagenic PCR primer | NNSGGCGGTCGCGTTTATGAA |
| Sequence-based reagent | WT_DHFR_pos95_fwd2 | This work | Mutagenic PCR primer | NNSGGCGGTCGCGTTTATGAAC |
| Sequence-based reagent | WT_DHFR_pos96_fwd | This work | Mutagenic PCR primer | NNSGGTCGCGTTTATGAACAGTTCTT |
| Sequence-based reagent | WT_DHFR_pos97_fwd | This work | Mutagenic PCR primer | NNSCGCGTTTATGAACAGTTCTTGC |
| Sequence-based reagent | WT_DHFR_pos98_fwd | This work | Mutagenic PCR primer | NNSGTTTATGAACAGTTCTTGCCAAAAGCGC |
| Sequence-based reagent | WT_DHFR_pos99_fwd | This work | Mutagenic PCR primer | NNSTATGAACAGTTCTTGCCAAAAGCGC |
| Sequence-based reagent | WT_DHFR_pos100_fwd | This work | Mutagenic PCR primer | NNSGAACAGTTCTTGCCAAAAGCGCAAAAAC |
| Sequence-based reagent | WT_DHFR_pos101_fwd | This work | Mutagenic PCR primer | NNSCAGTTCTTGCCAAAAGCGCAAAAAC |
| Sequence-based reagent | WT_DHFR_pos102_fwd | This work | Mutagenic PCR primer | NNSTTCTTGCCAAAAGCGCAAAAAC |
| Sequence-based reagent | WT_DHFR_pos103_fwd | This work | Mutagenic PCR primer | NNSTTGCCAAAAGCGCAAAAACTGTAT |
| Sequence-based reagent | WT_DHFR_pos104_fwd | This work | Mutagenic PCR primer | NNSCCAAAAGCGCAAAAACTGTATCTGA |
| Sequence-based reagent | WT_DHFR_pos104_fwd2 | This work | Mutagenic PCR primer | NNSCCAAAAGCGCAAAAACTGTATCTG |
| Sequence-based reagent | WT_DHFR_pos105_fwd | This work | Mutagenic PCR primer | NNSAAAGCGCAAAAACTGTATCTGACG |
| Sequence-based reagent | WT_DHFR_pos106_fwd | This work | Mutagenic PCR primer | NNSGCGCAAAAACTGTATCTGACG |
| Sequence-based reagent | WT_DHFR_pos107_fwd | This work | Mutagenic PCR primer | NNSCAAAAACTGTATCTGACGCATATCGAC |
| Sequence-based reagent | WT_DHFR_pos107_fwd2 | This work | Mutagenic PCR primer | NNSCAAAAACTGTATCTGACGCATATCG |
| Sequence-based reagent | WT_DHFR_pos108_fwd | This work | Mutagenic PCR primer | NNSAAACTGTATCTGACGCATATCGAC |
| Sequence-based reagent | WT_DHFR_pos109_fwd | This work | Mutagenic PCR primer | NNSCTGTATCTGACGCATATCGACG |
| Sequence-based reagent | WT_DHFR_pos110_fwd | This work | Mutagenic PCR primer | NNSTATCTGACGCATATCGACGCA |
| Sequence-based reagent | WT_DHFR_pos111_fwd | This work | Mutagenic PCR primer | NNSCTGACGCATATCGACGCAG |
| Sequence-based reagent | WT_DHFR_pos112_fwd | This work | Mutagenic PCR primer | NNSACGCATATCGACGCAGAAGT |
| Sequence-based reagent | WT_DHFR_pos113_fwd | This work | Mutagenic PCR primer | NNSCATATCGACGCAGAAGTGGAAG |
| Sequence-based reagent | WT_DHFR_pos114_fwd | This work | Mutagenic PCR primer | NNSATCGACGCAGAAGTGGAAG |
| Sequence-based reagent | WT_DHFR_pos115_fwd | This work | Mutagenic PCR primer | NNSGACGCAGAAGTGGAAGGC |
| Sequence-based reagent | WT_DHFR_pos116_fwd | This work | Mutagenic PCR primer | NNSGCAGAAGTGGAAGGCGAC |
| Sequence-based reagent | WT_DHFR_pos117_fwd | This work | Mutagenic PCR primer | NNSGAAGTGGAAGGCGACACC |
| Sequence-based reagent | WT_DHFR_pos118_fwd | This work | Mutagenic PCR primer | NNSGTGGAAGGCGACACCCAT |
| Sequence-based reagent | WT_DHFR_pos118_fwd2 | This work | Mutagenic PCR primer | NNSGTGGAAGGCGACACCCATTTC |
| Sequence-based reagent | WT_DHFR_pos119_fwd | This work | Mutagenic PCR primer | NNSGAAGGCGACACCCATTTCC |
| Sequence-based reagent | WT_DHFR_pos120_fwd | This work | Mutagenic PCR primer | NNSGGCGACACCCATTTCCCG |
| Sequence-based reagent | WT_DHFR_pos121_fwd | This work | Mutagenic PCR primer | NNSGACACCCATTTCCCGGATTAC |
| Sequence-based reagent | WT_DHFR_pos122_fwd | This work | Mutagenic PCR primer | NNSACCCATTTCCCGGATTACGA |
| Sequence-based reagent | WT_DHFR_pos123_fwd | This work | Mutagenic PCR primer | NNSCATTTCCCGGATTACGAGCC |
| Sequence-based reagent | WT_DHFR_pos124_fwd | This work | Mutagenic PCR primer | NNSTTCCCGGATTACGAGCCG |
| Sequence-based reagent | WT_DHFR_pos125_fwd | This work | Mutagenic PCR primer | NNSCCGGATTACGAGCCGGAT |
| Sequence-based reagent | WT_DHFR_pos126_fwd | This work | Mutagenic PCR primer | NNSGATTACGAGCCGGATGACTG |
| Sequence-based reagent | WT_DHFR_pos127_fwd | This work | Mutagenic PCR primer | NNSTACGAGCCGGATGACTGG |
| Sequence-based reagent | WT_DHFR_pos128_fwd | This work | Mutagenic PCR primer | NNSGAGCCGGATGACTGGGAA |
| Sequence-based reagent | WT_DHFR_pos129_fwd | This work | Mutagenic PCR primer | NNSCCGGATGACTGGGAATCG |
| Sequence-based reagent | WT_DHFR_pos130_fwd | This work | Mutagenic PCR primer | NNSGATGACTGGGAATCGGTATTCAG |
| Sequence-based reagent | WT_DHFR_pos131_fwd | This work | Mutagenic PCR primer | NNSGACTGGGAATCGGTATTCAGC |
| Sequence-based reagent | WT_DHFR_pos131_fwd2 | This work | Mutagenic PCR primer | NNSGACTGGGAATCGGTATTCAGC |
| Sequence-based reagent | WT_DHFR_pos132_fwd | This work | Mutagenic PCR primer | NNSTGGGAATCGGTATTCAGCGAATT |
| Sequence-based reagent | WT_DHFR_pos133_fwd | This work | Mutagenic PCR primer | NNSGAATCGGTATTCAGCGAATTCCAC |
| Sequence-based reagent | WT_DHFR_pos134_fwd | This work | Mutagenic PCR primer | NNSTCGGTATTCAGCGAATTCCAC |
| Sequence-based reagent | WT_DHFR_pos135_fwd | This work | Mutagenic PCR primer | NNSGTATTCAGCGAATTCCACGATG |
| Sequence-based reagent | WT_DHFR_pos135_fwd2 | This work | Mutagenic PCR primer | NNSGTATTCAGCGAATTCCACGATGC |
| Sequence-based reagent | WT_DHFR_pos136_fwd | This work | Mutagenic PCR primer | NNSTTCAGCGAATTCCACGATGC |
| Sequence-based reagent | WT_DHFR_pos136_fwd2 | This work | Mutagenic PCR primer | NNSTTCAGCGAATTCCACGATGC |
| Sequence-based reagent | WT_DHFR_pos137_fwd | This work | Mutagenic PCR primer | NNSAGCGAATTCCACGATGCTG |
| Sequence-based reagent | WT_DHFR_pos138_fwd | This work | Mutagenic PCR primer | NNSGAATTCCACGATGCTGATGC |
| Sequence-based reagent | WT_DHFR_pos139_fwd | This work | Mutagenic PCR primer | NNSTTCCACGATGCTGATGCG |
| Sequence-based reagent | WT_DHFR_pos140_fwd | This work | Mutagenic PCR primer | NNSCACGATGCTGATGCGCAG |
| Sequence-based reagent | WT_DHFR_pos140_fwd2 | This work | Mutagenic PCR primer | NNSCACGATGCTGATGCGCAG |
| Sequence-based reagent | WT_DHFR_pos141_fwd | This work | Mutagenic PCR primer | NNSGATGCTGATGCGCAGAACT |
| Sequence-based reagent | WT_DHFR_pos142_fwd | This work | Mutagenic PCR primer | NNSGCTGATGCGCAGAACTCTC |
| Sequence-based reagent | WT_DHFR_pos143_fwd | This work | Mutagenic PCR primer | NNSGATGCGCAGAACTCTCACAG |
| Sequence-based reagent | WT_DHFR_pos144_fwd | This work | Mutagenic PCR primer | NNSGCGCAGAACTCTCACAGC |
| Sequence-based reagent | WT_DHFR_pos145_fwd | This work | Mutagenic PCR primer | NNSCAGAACTCTCACAGCTATTGCTTTG |
| Sequence-based reagent | WT_DHFR_pos146_fwd | This work | Mutagenic PCR primer | NNSAACTCTCACAGCTATTGCTTTGAGATT |
| Sequence-based reagent | WT_DHFR_pos147_fwd | This work | Mutagenic PCR primer | NNSTCTCACAGCTATTGCTTTGAGATTCT |
| Sequence-based reagent | WT_DHFR_pos148_fwd | This work | Mutagenic PCR primer | NNSCACAGCTATTGCTTTGAGATTCTGG |
| Sequence-based reagent | WT_DHFR_pos149_fwd | This work | Mutagenic PCR primer | NNSAGCTATTGCTTTGAGATTCTGGAG |
| Sequence-based reagent | WT_DHFR_pos150_fwd | This work | Mutagenic PCR primer | NNSTATTGCTTTGAGATTCTGGAGCG |
| Sequence-based reagent | WT_DHFR_pos151_fwd | This work | Mutagenic PCR primer | NNSTGCTTTGAGATTCTGGAGCG |
| Sequence-based reagent | WT_DHFR_pos152_fwd | This work | Mutagenic PCR primer | NNSTTTGAGATTCTGGAGCGGC |
| Sequence-based reagent | WT_DHFR_pos153_fwd | This work | Mutagenic PCR primer | NNSGAGATTCTGGAGCGGCGG |
| Sequence-based reagent | WT_DHFR_pos154_fwd | This work | Mutagenic PCR primer | NNSATTCTGGAGCGGCGGTAA |
| Sequence-based reagent | WT_DHFR_pos155_fwd | This work | Mutagenic PCR primer | NNSCTGGAGCGGCGGTAACAG |
| Sequence-based reagent | WT_DHFR_pos156_fwd | This work | Mutagenic PCR primer | NNSGAGCGGCGGTAACAGGCG |
| Sequence-based reagent | WT_DHFR_pos157_fwd | This work | Mutagenic PCR primer | NNSCGGCGGTAACAGGCGTCG |
| Sequence-based reagent | WT_DHFR_pos158_fwd | This work | Mutagenic PCR primer | NNSCGGTAACAGGCGTCGACA |
| Sequence-based reagent | WT_DHFR_pos159_fwd | This work | Mutagenic PCR primer | NNSTAACAGGCGTCGACAAGCT |
| Sequence-based reagent | WT_DHFR_pos2_rev | This work | Mutagenic PCR primer | CATGGTATATCTCCTTATTAAAGTTAAA |
| Sequence-based reagent | WT_DHFR_pos2_rev2 | This work | Mutagenic PCR primer | CATGGTATATCTCATTATTAAAGTTAAAC |
| Sequence-based reagent | WT_DHFR_pos3_rev | This work | Mutagenic PCR primer | GATCATGGTATATCTCCTTATTAAAGTT |
| Sequence-based reagent | WT_DHFR_pos4_rev | This work | Mutagenic PCR primer | ACTGATCATGGTATATCTCCTTATTAAA |
| Sequence-based reagent | WT_DHFR_pos5_rev | This work | Mutagenic PCR primer | CAGACTGATCATGGTATATCTCCTTATT |
| Sequence-based reagent | WT_DHFR_pos6_rev | This work | Mutagenic PCR primer | AATCAGACTGATCATGGTATATCTCCTT |
| Sequence-based reagent | WT_DHFR_pos7_rev | This work | Mutagenic PCR primer | CGCAATCAGACTGATCATGGTATATCT |
| Sequence-based reagent | WT_DHFR_pos8_rev | This work | Mutagenic PCR primer | CGCCGCAATCAGACTGATC |
| Sequence-based reagent | WT_DHFR_pos8_rev2 | This work | Mutagenic PCR primer | CGCCGCAATCAGACTGATC |
| Sequence-based reagent | WT_DHFR_pos9_rev | This work | Mutagenic PCR primer | TAACGCCGCAATCAGACTGA |
| Sequence-based reagent | WT_DHFR_pos10_rev | This work | Mutagenic PCR primer | CGCTAACGCCGCAATCAG |
| Sequence-based reagent | WT_DHFR_pos11_rev | This work | Mutagenic PCR primer | TACCGCTAACGCCGCAAT |
| Sequence-based reagent | WT_DHFR_pos12_rev | This work | Mutagenic PCR primer | ATCTACCGCTAACGCCGC |
| Sequence-based reagent | WT_DHFR_pos13_rev | This work | Mutagenic PCR primer | GCGATCTACCGCTAACGC |
| Sequence-based reagent | WT_DHFR_pos14_rev | This work | Mutagenic PCR primer | AACGCGATCTACCGCTAAC |
| Sequence-based reagent | WT_DHFR_pos15_rev | This work | Mutagenic PCR primer | GATAACGCGATCTACCGCTAAC |
| Sequence-based reagent | WT_DHFR_pos16_rev | This work | Mutagenic PCR primer | GCCGATAACGCGATCTACC |
| Sequence-based reagent | WT_DHFR_pos17_rev | This work | Mutagenic PCR primer | CATGCCGATAACGCGATCTAC |
| Sequence-based reagent | WT_DHFR_pos18_rev | This work | Mutagenic PCR primer | TTCCATGCCGATAACGCG |
| Sequence-based reagent | WT_DHFR_pos19_rev | This work | Mutagenic PCR primer | GTTTTCCATGCCGATAACGC |
| Sequence-based reagent | WT_DHFR_pos20_rev | This work | Mutagenic PCR primer | GGCGTTTTCCATGCCGATAACG |
| Sequence-based reagent | WT_DHFR_pos21_rev | This work | Mutagenic PCR primer | CATGGCGTTTTCCATGCC |
| Sequence-based reagent | WT_DHFR_pos22_rev | This work | Mutagenic PCR primer | CGGCATGGCGTTTTCCAT |
| Sequence-based reagent | WT_DHFR_pos22_rev2 | This work | Mutagenic PCR primer | CGGCATGGCGTTTTCCATG |
| Sequence-based reagent | WT_DHFR_pos23_rev | This work | Mutagenic PCR primer | CCACGGCATGGCGTTTTC |
| Sequence-based reagent | WT_DHFR_pos24_rev | This work | Mutagenic PCR primer | GTTCCACGGCATGGCGTT |
| Sequence-based reagent | WT_DHFR_pos25_rev | This work | Mutagenic PCR primer | CAGGTTCCACGGCATGGC |
| Sequence-based reagent | WT_DHFR_pos26_rev | This work | Mutagenic PCR primer | AGGCAGGTTCCACGGCAT |
| Sequence-based reagent | WT_DHFR_pos27_rev | This work | Mutagenic PCR primer | GGCAGGCAGGTTCCACGG |
| Sequence-based reagent | WT_DHFR_pos28_rev | This work | Mutagenic PCR primer | ATCGGCAGGCAGGTTCCA |
| Sequence-based reagent | WT_DHFR_pos29_rev | This work | Mutagenic PCR primer | GAGATCGGCAGGCAGGTT |
| Sequence-based reagent | WT_DHFR_pos30_rev | This work | Mutagenic PCR primer | GGCGAGATCGGCAGGCAG |
| Sequence-based reagent | WT_DHFR_pos31_rev | This work | Mutagenic PCR primer | CCAGGCGAGATCGGCAGG |
| Sequence-based reagent | WT_DHFR_pos32_rev | This work | Mutagenic PCR primer | AAACCAGGCGAGATCGGC |
| Sequence-based reagent | WT_DHFR_pos33_rev | This work | Mutagenic PCR primer | TTTAAACCAGGCGAGATCGG |
| Sequence-based reagent | WT_DHFR_pos34_rev | This work | Mutagenic PCR primer | GCGTTTAAACCAGGCGAGAT |
| Sequence-based reagent | WT_DHFR_pos35_rev | This work | Mutagenic PCR primer | GTTGCGTTTAAACCAGGCGA |
| Sequence-based reagent | WT_DHFR_pos36_rev | This work | Mutagenic PCR primer | GGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | WT_DHFR_pos37_rev | This work | Mutagenic PCR primer | TAAGGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | WT_DHFR_pos38_rev | This work | Mutagenic PCR primer | ATTTAAGGTGTTGCGTTTAAACCAGG |
| Sequence-based reagent | WT_DHFR_pos39_rev | This work | Mutagenic PCR primer | TTTATTTAAGGTGTTGCGTTTAAACCAG |
| Sequence-based reagent | WT_DHFR_pos40_rev | This work | Mutagenic PCR primer | GGGTTTATTTAAGGTGTTGCGTTTAAAC |
| Sequence-based reagent | WT_DHFR_pos41_rev | This work | Mutagenic PCR primer | CACGGGTTTATTTAAGGTGTTGCGT |
| Sequence-based reagent | WT_DHFR_pos42_rev | This work | Mutagenic PCR primer | AATCACGGGTTTATTTAAGGTGTTGC |
| Sequence-based reagent | WT_DHFR_pos42_rev2 | This work | Mutagenic PCR primer | AATCACGGGTTTATTTAAGGTGTTGC |
| Sequence-based reagent | WT_DHFR_pos43_rev | This work | Mutagenic PCR primer | CATAATCACGGGTTTATTTAAGGTGTTG |
| Sequence-based reagent | WT_DHFR_pos43_rev2 | This work | Mutagenic PCR primer | CATAATCACGGGTTTATTTAAGGTGTTG |
| Sequence-based reagent | WT_DHFR_pos44_rev | This work | Mutagenic PCR primer | GCCCATAATCACGGGTTTATTTAAGG |
| Sequence-based reagent | WT_DHFR_pos45_rev | This work | Mutagenic PCR primer | GCGGCCCATAATCACGGG |
| Sequence-based reagent | WT_DHFR_pos46_rev | This work | Mutagenic PCR primer | ATGGCGGCCCATAATCAC |
| Sequence-based reagent | WT_DHFR_pos47_rev | This work | Mutagenic PCR primer | GGTATGGCGGCCCATAATC |
| Sequence-based reagent | WT_DHFR_pos48_rev | This work | Mutagenic PCR primer | CCAGGTATGGCGGCCCATA |
| Sequence-based reagent | WT_DHFR_pos49_rev | This work | Mutagenic PCR primer | TTCCCAGGTATGGCGGCC |
| Sequence-based reagent | WT_DHFR_pos50_rev | This work | Mutagenic PCR primer | TGATTCCCAGGTATGGCGGC |
| Sequence-based reagent | WT_DHFR_pos51_rev | This work | Mutagenic PCR primer | GATTGATTCCCAGGTATGGCGG |
| Sequence-based reagent | WT_DHFR_pos52_rev | This work | Mutagenic PCR primer | ACCGATTGATTCCCAGGTATG |
| Sequence-based reagent | WT_DHFR_pos53_rev | This work | Mutagenic PCR primer | ACGACCGATTGATTCCCAG |
| Sequence-based reagent | WT_DHFR_pos54_rev | This work | Mutagenic PCR primer | CGGACGACCGATTGATTCC |
| Sequence-based reagent | WT_DHFR_pos55_rev | This work | Mutagenic PCR primer | CAACGGACGACCGATTGATTC |
| Sequence-based reagent | WT_DHFR_pos56_rev | This work | Mutagenic PCR primer | TGGCAACGGACGACCGAT |
| Sequence-based reagent | WT_DHFR_pos57_rev | This work | Mutagenic PCR primer | TCCTGGCAACGGACGACC |
| Sequence-based reagent | WT_DHFR_pos58_rev | This work | Mutagenic PCR primer | GCGTCCTGGCAACGGACG |
| Sequence-based reagent | WT_DHFR_pos59_rev | This work | Mutagenic PCR primer | TTTGCGTCCTGGCAACGG |
| Sequence-based reagent | WT_DHFR_pos60_rev | This work | Mutagenic PCR primer | ATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | WT_DHFR_pos61_rev | This work | Mutagenic PCR primer | AATATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | WT_DHFR_pos62_rev | This work | Mutagenic PCR primer | GATAATATTTTTGCGTCCTGGCAAC |
| Sequence-based reagent | WT_DHFR_pos63_rev | This work | Mutagenic PCR primer | GAGGATAATATTTTTGCGTCCTGGC |
| Sequence-based reagent | WT_DHFR_pos64_rev | This work | Mutagenic PCR primer | GCTGAGGATAATATTTTTGCGTCCTG |
| Sequence-based reagent | WT_DHFR_pos65_rev | This work | Mutagenic PCR primer | ACTGCTGAGGATAATATTTTTGCGTCCT |
| Sequence-based reagent | WT_DHFR_pos66_rev | This work | Mutagenic PCR primer | TTGACTGCTGAGGATAATATTTTTGCG |
| Sequence-based reagent | WT_DHFR_pos66_rev2 | This work | Mutagenic PCR primer | TTGACTGCTGAGGATAATATTTTTGC |
| Sequence-based reagent | WT_DHFR_pos67_rev | This work | Mutagenic PCR primer | CGGTTGACTGCTGAGGATAATATTTTTG |
| Sequence-based reagent | WT_DHFR_pos67_rev2 | This work | Mutagenic PCR primer | CGGTTGACTGCTGAGGATAATATTTTTG |
| Sequence-based reagent | WT_DHFR_pos68_rev | This work | Mutagenic PCR primer | ACCCGGTTGACTGCTGAG |
| Sequence-based reagent | WT_DHFR_pos69_rev | This work | Mutagenic PCR primer | CGTACCCGGTTGACTGCT |
| Sequence-based reagent | WT_DHFR_pos70_rev | This work | Mutagenic PCR primer | GTCCGTACCCGGTTGACT |
| Sequence-based reagent | WT_DHFR_pos71_rev | This work | Mutagenic PCR primer | ATCGTCCGTACCCGGTTG |
| Sequence-based reagent | WT_DHFR_pos72_rev | This work | Mutagenic PCR primer | GCGATCGTCCGTACCCGG |
| Sequence-based reagent | WT_DHFR_pos73_rev | This work | Mutagenic PCR primer | TACGCGATCGTCCGTACC |
| Sequence-based reagent | WT_DHFR_pos73_rev2 | This work | Mutagenic PCR primer | TACGCGATCGTCCGTACC |
| Sequence-based reagent | WT_DHFR_pos74_rev | This work | Mutagenic PCR primer | CGTTACGCGATCGTCCGT |
| Sequence-based reagent | WT_DHFR_pos74_rev2 | This work | Mutagenic PCR primer | CGTTACGCGATCGTCCGTAC |
| Sequence-based reagent | WT_DHFR_pos75_rev | This work | Mutagenic PCR primer | CCACGTTACGCGATCGTC |
| Sequence-based reagent | WT_DHFR_pos76_rev | This work | Mutagenic PCR primer | CACCCACGTTACGCGATC |
| Sequence-based reagent | WT_DHFR_pos77_rev | This work | Mutagenic PCR primer | CTTCACCCACGTTACGCG |
| Sequence-based reagent | WT_DHFR_pos78_rev | This work | Mutagenic PCR primer | CGACTTCACCCACGTTACG |
| Sequence-based reagent | WT_DHFR_pos79_rev | This work | Mutagenic PCR primer | CACCGACTTCACCCACGTTAC |
| Sequence-based reagent | WT_DHFR_pos80_rev | This work | Mutagenic PCR primer | ATCCACCGACTTCACCCACGTTAC |
| Sequence-based reagent | WT_DHFR_pos80_rev2 | This work | Mutagenic PCR primer | ATCCACCGACTTCACCCAC |
| Sequence-based reagent | WT_DHFR_pos81_rev | This work | Mutagenic PCR primer | TTCATCCACCGACTTCACCCA |
| Sequence-based reagent | WT_DHFR_pos82_rev | This work | Mutagenic PCR primer | GGCTTCATCCACCGACTTCAC |
| Sequence-based reagent | WT_DHFR_pos82_rev2 | This work | Mutagenic PCR primer | GGCTTCATCCACCGACTTCAC |
| Sequence-based reagent | WT_DHFR_pos83_rev | This work | Mutagenic PCR primer | GATGGCTTCATCCACCGAC |
| Sequence-based reagent | WT_DHFR_pos84_rev | This work | Mutagenic PCR primer | CGCGATGGCTTCATCCAC |
| Sequence-based reagent | WT_DHFR_pos84_rev2 | This work | Mutagenic PCR primer | CGCGATGGCTTCATCCAC |
| Sequence-based reagent | WT_DHFR_pos85_rev | This work | Mutagenic PCR primer | CGCCGCGATGGCTTCATC |
| Sequence-based reagent | WT_DHFR_pos86_rev | This work | Mutagenic PCR primer | ACACGCCGCGATGGCTTC |
| Sequence-based reagent | WT_DHFR_pos87_rev | This work | Mutagenic PCR primer | ACCACACGCCGCGATGGC |
| Sequence-based reagent | WT_DHFR_pos88_rev | This work | Mutagenic PCR primer | GTCACCACACGCCGCGAT |
| Sequence-based reagent | WT_DHFR_pos89_rev | This work | Mutagenic PCR primer | TACGTCACCACACGCCGC |
| Sequence-based reagent | WT_DHFR_pos89_rev2 | This work | Mutagenic PCR primer | TACGTCACCACACGCCG |
| Sequence-based reagent | WT_DHFR_pos90_rev | This work | Mutagenic PCR primer | TGGTACGTCACCACACGC |
| Sequence-based reagent | WT_DHFR_pos91_rev | This work | Mutagenic PCR primer | TTCTGGTACGTCACCACACGC |
| Sequence-based reagent | WT_DHFR_pos92_rev | This work | Mutagenic PCR primer | GATTTCTGGTACGTCACCACACG |
| Sequence-based reagent | WT_DHFR_pos93_rev | This work | Mutagenic PCR primer | CATGATTTCTGGTACGTCACCACAC |
| Sequence-based reagent | WT_DHFR_pos94_rev | This work | Mutagenic PCR primer | CACCATGATTTCTGGTACGTCACC |
| Sequence-based reagent | WT_DHFR_pos95_rev | This work | Mutagenic PCR primer | AATCACCATGATTTCTGGTACGTCA |
| Sequence-based reagent | WT_DHFR_pos95_rev2 | This work | Mutagenic PCR primer | AATCACCATGATTTCTGGTACGTC |
| Sequence-based reagent | WT_DHFR_pos96_rev | This work | Mutagenic PCR primer | GCCAATCACCATGATTTCTGGTAC |
| Sequence-based reagent | WT_DHFR_pos97_rev | This work | Mutagenic PCR primer | GCCGCCAATCACCATGATTT |
| Sequence-based reagent | WT_DHFR_pos98_rev | This work | Mutagenic PCR primer | ACCGCCGCCAATCACCATGATTTC |
| Sequence-based reagent | WT_DHFR_pos99_rev | This work | Mutagenic PCR primer | GCGACCGCCGCCAATCAC |
| Sequence-based reagent | WT_DHFR_pos100_rev | This work | Mutagenic PCR primer | AACGCGACCGCCGCCAAT |
| Sequence-based reagent | WT_DHFR_pos101_rev | This work | Mutagenic PCR primer | ATAAACGCGACCGCCGCC |
| Sequence-based reagent | WT_DHFR_pos102_rev | This work | Mutagenic PCR primer | TTCATAAACGCGACCGCC |
| Sequence-based reagent | WT_DHFR_pos103_rev | This work | Mutagenic PCR primer | CTGTTCATAAACGCGACCG |
| Sequence-based reagent | WT_DHFR_pos104_rev | This work | Mutagenic PCR primer | GAACTGTTCATAAACGCGACC |
| Sequence-based reagent | WT_DHFR_pos104_rev2 | This work | Mutagenic PCR primer | GAACTGTTCATAAACGCGACCG |
| Sequence-based reagent | WT_DHFR_pos105_rev | This work | Mutagenic PCR primer | CAAGAACTGTTCATAAACGCGAC |
| Sequence-based reagent | WT_DHFR_pos106_rev | This work | Mutagenic PCR primer | TGGCAAGAACTGTTCATAAACGC |
| Sequence-based reagent | WT_DHFR_pos107_rev | This work | Mutagenic PCR primer | TTTTGGCAAGAACTGTTCATAAACG |
| Sequence-based reagent | WT_DHFR_pos107_rev2 | This work | Mutagenic PCR primer | TTTTGGCAAGAACTGTTCATAAACG |
| Sequence-based reagent | WT_DHFR_pos108_rev | This work | Mutagenic PCR primer | CGCTTTTGGCAAGAACTGTTCATAAA |
| Sequence-based reagent | WT_DHFR_pos109_rev | This work | Mutagenic PCR primer | TTGCGCTTTTGGCAAGAACT |
| Sequence-based reagent | WT_DHFR_pos110_rev | This work | Mutagenic PCR primer | TTTTTGCGCTTTTGGCAAGAAC |
| Sequence-based reagent | WT_DHFR_pos111_rev | This work | Mutagenic PCR primer | CAGTTTTTGCGCTTTTGGCAAG |
| Sequence-based reagent | WT_DHFR_pos112_rev | This work | Mutagenic PCR primer | ATACAGTTTTTGCGCTTTTGGCAA |
| Sequence-based reagent | WT_DHFR_pos113_rev | This work | Mutagenic PCR primer | CAGATACAGTTTTTGCGCTTTTGG |
| Sequence-based reagent | WT_DHFR_pos114_rev | This work | Mutagenic PCR primer | CGTCAGATACAGTTTTTGCGCTTTT |
| Sequence-based reagent | WT_DHFR_pos115_rev | This work | Mutagenic PCR primer | ATGCGTCAGATACAGTTTTTGCG |
| Sequence-based reagent | WT_DHFR_pos116_rev | This work | Mutagenic PCR primer | GATATGCGTCAGATACAGTTTTTGCG |
| Sequence-based reagent | WT_DHFR_pos117_rev | This work | Mutagenic PCR primer | GTCGATATGCGTCAGATACAGTTTTTG |
| Sequence-based reagent | WT_DHFR_pos118_rev | This work | Mutagenic PCR primer | TGCGTCGATATGCGTCAGATA |
| Sequence-based reagent | WT_DHFR_pos118_rev2 | This work | Mutagenic PCR primer | TGCGTCGATATGCGTCAGATAC |
| Sequence-based reagent | WT_DHFR_pos119_rev | This work | Mutagenic PCR primer | TTCTGCGTCGATATGCGTCA |
| Sequence-based reagent | WT_DHFR_pos120_rev | This work | Mutagenic PCR primer | CACTTCTGCGTCGATATGCG |
| Sequence-based reagent | WT_DHFR_pos121_rev | This work | Mutagenic PCR primer | TTCCACTTCTGCGTCGATATG |
| Sequence-based reagent | WT_DHFR_pos122_rev | This work | Mutagenic PCR primer | GCCTTCCACTTCTGCGTC |
| Sequence-based reagent | WT_DHFR_pos123_rev | This work | Mutagenic PCR primer | GTCGCCTTCCACTTCTGC |
| Sequence-based reagent | WT_DHFR_pos124_rev | This work | Mutagenic PCR primer | GGTGTCGCCTTCCACTTC |
| Sequence-based reagent | WT_DHFR_pos125_rev | This work | Mutagenic PCR primer | ATGGGTGTCGCCTTCCAC |
| Sequence-based reagent | WT_DHFR_pos126_rev | This work | Mutagenic PCR primer | GAAATGGGTGTCGCCTTCC |
| Sequence-based reagent | WT_DHFR_pos127_rev | This work | Mutagenic PCR primer | CGGGAAATGGGTGTCGCC |
| Sequence-based reagent | WT_DHFR_pos128_rev | This work | Mutagenic PCR primer | ATCCGGGAAATGGGTGTC |
| Sequence-based reagent | WT_DHFR_pos129_rev | This work | Mutagenic PCR primer | GTAATCCGGGAAATGGGTGTC |
| Sequence-based reagent | WT_DHFR_pos130_rev | This work | Mutagenic PCR primer | CTCGTAATCCGGGAAATGGG |
| Sequence-based reagent | WT_DHFR_pos131_rev | This work | Mutagenic PCR primer | CGGCTCGTAATCCGGGAA |
| Sequence-based reagent | WT_DHFR_pos131_rev2 | This work | Mutagenic PCR primer | CGGCTCGTAATCCGGGAAATG |
| Sequence-based reagent | WT_DHFR_pos132_rev | This work | Mutagenic PCR primer | ATCCGGCTCGTAATCCGG |
| Sequence-based reagent | WT_DHFR_pos133_rev | This work | Mutagenic PCR primer | GTCATCCGGCTCGTAATCC |
| Sequence-based reagent | WT_DHFR_pos134_rev | This work | Mutagenic PCR primer | CCAGTCATCCGGCTCGTA |
| Sequence-based reagent | WT_DHFR_pos135_rev | This work | Mutagenic PCR primer | TTCCCAGTCATCCGGCTC |
| Sequence-based reagent | WT_DHFR_pos135_rev2 | This work | Mutagenic PCR primer | TTCCCAGTCATCCGGCTC |
| Sequence-based reagent | WT_DHFR_pos136_rev | This work | Mutagenic PCR primer | CGATTCCCAGTCATCCGG |
| Sequence-based reagent | WT_DHFR_pos136_rev2 | This work | Mutagenic PCR primer | CGATTCCCAGTCATCCGGC |
| Sequence-based reagent | WT_DHFR_pos137_rev | This work | Mutagenic PCR primer | TACCGATTCCCAGTCATCCG |
| Sequence-based reagent | WT_DHFR_pos138_rev | This work | Mutagenic PCR primer | GAATACCGATTCCCAGTCATCC |
| Sequence-based reagent | WT_DHFR_pos139_rev | This work | Mutagenic PCR primer | GCTGAATACCGATTCCCAGTC |
| Sequence-based reagent | WT_DHFR_pos140_rev | This work | Mutagenic PCR primer | TTCGCTGAATACCGATTCCCA |
| Sequence-based reagent | WT_DHFR_pos140_rev2 | This work | Mutagenic PCR primer | TTCGCTGAATACCGATTCCCAG |
| Sequence-based reagent | WT_DHFR_pos141_rev | This work | Mutagenic PCR primer | GAATTCGCTGAATACCGATTCCC |
| Sequence-based reagent | WT_DHFR_pos142_rev | This work | Mutagenic PCR primer | GTGGAATTCGCTGAATACCGATTC |
| Sequence-based reagent | WT_DHFR_pos143_rev | This work | Mutagenic PCR primer | ATCGTGGAATTCGCTGAATACC |
| Sequence-based reagent | WT_DHFR_pos144_rev | This work | Mutagenic PCR primer | AGCATCGTGGAATTCGCTG |
| Sequence-based reagent | WT_DHFR_pos145_rev | This work | Mutagenic PCR primer | ATCAGCATCGTGGAATTCGC |
| Sequence-based reagent | WT_DHFR_pos146_rev | This work | Mutagenic PCR primer | CGCATCAGCATCGTGGAATT |
| Sequence-based reagent | WT_DHFR_pos147_rev | This work | Mutagenic PCR primer | CTGCGCATCAGCATCGTG |
| Sequence-based reagent | WT_DHFR_pos148_rev | This work | Mutagenic PCR primer | GTTCTGCGCATCAGCATC |
| Sequence-based reagent | WT_DHFR_pos149_rev | This work | Mutagenic PCR primer | AGAGTTCTGCGCATCAGC |
| Sequence-based reagent | WT_DHFR_pos150_rev | This work | Mutagenic PCR primer | GTGAGAGTTCTGCGCATCAG |
| Sequence-based reagent | WT_DHFR_pos151_rev | This work | Mutagenic PCR primer | GCTGTGAGAGTTCTGCGC |
| Sequence-based reagent | WT_DHFR_pos152_rev | This work | Mutagenic PCR primer | ATAGCTGTGAGAGTTCTGCG |
| Sequence-based reagent | WT_DHFR_pos153_rev | This work | Mutagenic PCR primer | GCAATAGCTGTGAGAGTTCTGC |
| Sequence-based reagent | WT_DHFR_pos154_rev | This work | Mutagenic PCR primer | AAAGCAATAGCTGTGAGAGTTCTG |
| Sequence-based reagent | WT_DHFR_pos155_rev | This work | Mutagenic PCR primer | CTCAAAGCAATAGCTGTGAGAGTTC |
| Sequence-based reagent | WT_DHFR_pos156_rev | This work | Mutagenic PCR primer | AATCTCAAAGCAATAGCTGTGAGAGTTC |
| Sequence-based reagent | WT_DHFR_pos157_rev | This work | Mutagenic PCR primer | CAGAATCTCAAAGCAATAGCTGTGAGAG |
| Sequence-based reagent | WT_DHFR_pos158_rev | This work | Mutagenic PCR primer | CTCCAGAATCTCAAAGCAATAGCTG |
| Sequence-based reagent | WT_DHFR_pos159_rev | This work | Mutagenic PCR primer | CCGCTCCAGAATCTCAAAGC |
| Sequence-based reagent | SL1_FWD | This work | Round one amplicon PCR primer | CACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNACTTTAATAACGAGATATACCATG |
| Sequence-based reagent | SL1_REV | This work | Round one amplicon PCR primer | TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNGTATGGCGGCCCATAAT |
| Sequence-based reagent | SL2_FWD | This work | Round one amplicon PCR primer | CACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNACACCTTAAATAAACCCGTG |
| Sequence-based reagent | SL2_REV | This work | Round one amplicon PCR primer | TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNCACGCCGCGATGGC |
| Sequence-based reagent | SL3_FWD | This work | Round one amplicon PCR primer | CACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNTGAAGTCGGTGGATGAA |
| Sequence-based reagent | SL3_REV | This work | Round one amplicon PCR primer | TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNGAAATGGGTGTCGCC |
| Sequence-based reagent | SL4_FWD | This work | Round one amplicon PCR primer | CACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNCGACGCAGAAGTGGAA |
| Sequence-based reagent | SL4_REV | This work | Round one amplicon PCR primer | TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNGCTTGTCGACGCCTG |
| Sequence-based reagent | D501 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACTATAGCCTACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D502 | Illumina/Reynolds et al. Cell 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACATAGAGGCACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D503 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACCCTATCCTACACTCTTT CCCTACACGAC |
| Sequence-based reagent | D504 | Illumina/Reynolds et al. Cell 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACGGCTCTGAACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D505 | Illumina/Reynolds et al. Cell 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACAGGCGAAGACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D506 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACTAATCTTAACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D507 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACCAGGACGTACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D508 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | AATGATACGGCGACCACCGAGATCTACACGTACTGACACACTCTTTCCCTACACGAC |
| Sequence-based reagent | D701 | Illumina/Reynolds et al. Cell 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D702 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D703 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D704 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D705 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D706 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D707 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D708 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAG ATGCGCATTAGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D709 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D710 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D711 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | D712 | Illumina/Reynolds et al., 2011 | Round two amplicon PCR primer | CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTG |
| Sequence-based reagent | KanSacB_round1_fwd | This work | PCR primer | caggcatctggtgaataaTCCTTTTATGATTTTCTATCAAACAAAAGAGG |
| Sequence-based reagent | KanSacB_round1_rev | This work | PCR primer | tcaatgcgttcagaacgctcaggattcatGCTTGGTCGGTCATTTCGAAC |
| Sequence-based reagent | KanSacB_round2_fwd/Anderson_promoter_outer_fwd | This work | PCR primer | gtcaaagcaaaccgttgctgatttatggcaagccggaagcgcaacaggcatctggtgaataa |
| Sequence-based reagent | KanSacB_round2_rev/Anderson_promoter_outer_rev | This work | PCR primer | ccaccacatcgcgcagcggcaatacggggatttcaatgcgttcagaacgctcaggattcat |
| Sequence-based reagent | Anderson_promoter_outer_fwd/KanSacB_round2_fwd | This work | PCR primer | same as KanSacB_round2_fwd/Anderson_promoter_outer_fwd |
| Sequence-based reagent | Anderson_promoter_inner_fwd | This work | PCR primer | CCTAGGACTGAGCTAGCTGTCAAcgtcagtatatggggatgtttcccc |
| Sequence-based reagent | Anderson_promoter_inner_rev | This work | PCR primer | GCTAGCTCAGTCCTAGGTATAATGCTAGCAGGAtacctggcggaaattaaactaagagag |
| Sequence-based reagent | Anderson_promoter_outer_rev/KanSacB_round2_rev | This work | PCR primer | same as KanSacB_round2_rev/Anderson_promoter_outer_rev |
Additional files
-
Supplementary file 1
Selection coefficients for –Lon selection measured as described in Materials and methods are reported with the standard deviation between biological replicates and the standard error from linear regression (as calculated by Enrich2; Rubin et al., 2017).
Values are reported as calculated, but based on the selection calibration, differences between selection coefficients with values below ~–2.5 are not interpretable.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp1-v2.csv
-
Supplementary file 2
Selection coefficients for +Lon selection measured as described in Materials and methods are reported with the standard deviation between biological replicates and the standard error from linear regression.
Values are reported as calculated, but based on the selection calibration, differences between selection coefficients with values below ~–2.5 are not interpretable.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp2-v2.csv
-
Supplementary file 3
Raw deep sequencing counts for the calibration set of mutants –Lon selection.
Counts are recorded for all turbidostat timepoints over three repeats.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp3-v2.csv
-
Supplementary file 4
Raw deep sequencing counts for single point mutants in –Lon selection.
Counts are recorded for a sample collected from the transformation rescue medium, a sample from overnight outgrowth in supplemented M9, and for all turbidostat timepoints over six experiments. In each experiment, 2 of 4 sublibraries were screened as described in Materials and methods, for a total of 3 repeats over the full library.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp4-v2.csv
-
Supplementary file 5
Raw deep sequencing counts for single point mutants in +Lon selection.
Counts are recorded as in Supplementary file 4.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp5-v2.csv
-
Supplementary file 6
DHFR reaction velocities as a function of DHF concentration used for the measurement of soluble DHFR abundance from lysate activities as described in Materials and methods.
For each mutant (column 1), three technical repeats are included (column 2). There are two lines for each repeat reaction, one for DHF concentration and one for the reaction velocity at that concentration (column 3). All data in columns 4+ are the experimental values.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp6-v2.csv
-
Supplementary file 7
Multiple sequence alignment of bacterial DHFR sequences generated as described in Materials and methods and used for bioinformatics analyses.
- https://cdn.elifesciences.org/articles/53476/elife-53476-supp7-v2.csv
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53476/elife-53476-transrepform-v2.docx






