Determining the scale at which variation in a single gene changes population yields
Figures
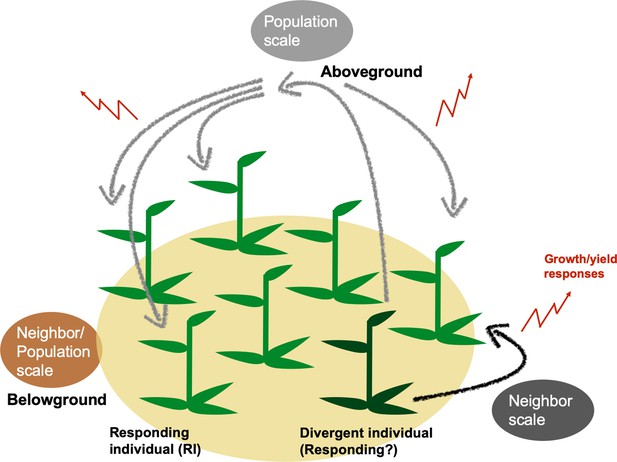
Genetically divergent plants can alter population yields through plant neighbor responses generated at neighbor or population scales.
Genetic variation within a population can change population growth and yield by altering individuals’ outputs either in localized areas within populations (neighbor scale) or across all plants of a population (population scale). At the neighbor scale (black), a divergent individual (dark green) may elicit responses only in immediate neighbors (responding individuals, RIs). RIs’ responses at either spatial scale may include changes in growth and yield, which can cumulatively change a population’s growth and yield (red). Responses to divergent individuals could be caused by above- (black, gray) and/or belowground interactions (brown) among plants in population.
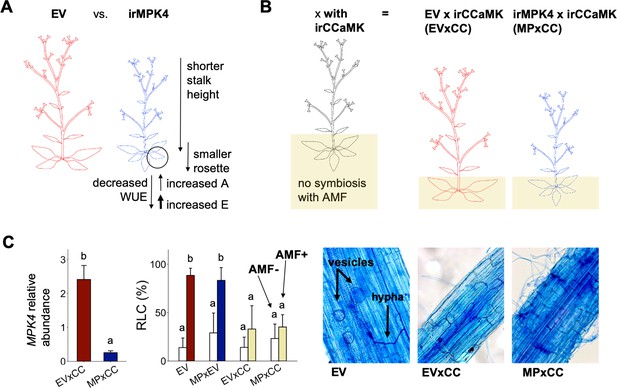
Characterization of EV, irMPK4, irMPK4xEV, EV x irCCaMK and irMPK4 x irCCaMK plants.
(A) A schematic summary of the findings of Hettenhausen et al. (2012): irMPK4 Nicotiana attenuata plants, silenced in mitogen-activated protein kinase 4, have disrupted stomatal control resulting in increased rates of leaf transpiration (E) which surpass the smaller increases in rates of leaf carbon assimilation (A) and therefore decrease water-use efficiency (WUE) in comparison to empty-vector (EV) plants. irMPK4 stalks and rosettes are smaller than those of EV. (B) A schematic demonstrating a method established by Groten et al. (2015) to control arbuscular mycorrhizal association in the field: irCCaMK N. attenuata plants, silenced in calcium and calmodulin-dependent protein kinase, are crossed with EV and irMPK4 to create EV x irCCaMK (EVxCC) and irMPK4 x irCCaMK (MPxCC) lines hemizygous for each of the transgenes and are not able to associate with arbuscular mycorrhizal fungi (AMF). (C) Mitogen-activated protein kinase 4 (MPK4) transcript abundances, calculated relative to a housekeeping gene, in hemizygous MPxCC and EVxCC plants (left panel, mean + CI, n = 9 for EV, 13 for irMPK4) compare with those of homozygous irMPK4 and EV plants (Figure 2—figure supplement 1). EVxCC and MPxCC roots inoculated with an arbuscular mycorrhizal fungus, Rhizophagus irregularis (AMF+), did not show significant increases in comparison to un-inoculated counterparts (AMF-) in root length colonization (RLC; center panel, mean + CI, n = 7 for EVxCC, n = 8 for MPxCC), in contrast to the strong colonization of EV plants (n = 8) and control hemizygous irMPK4 crosses: irMPK4xEV (MPxEV, n = 7–8). Vesicles and hyphae are visible in trypan blue-stained AMF+ EV roots, but not in AMF+ EVxCC and MPxCC roots.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig2-data1-v2.xlsx
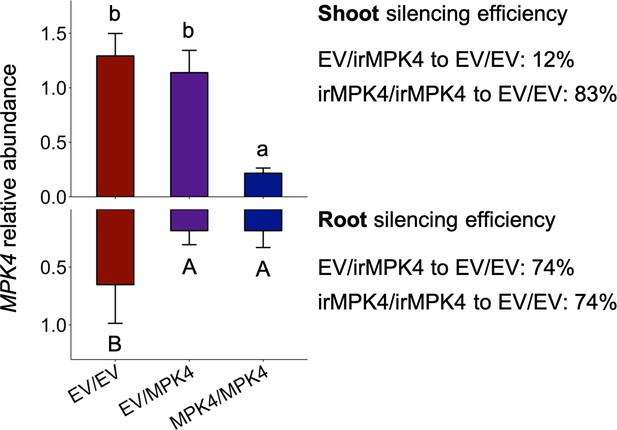
MPK4 transcript abundance relative to a housekeeping gene (+ 95% CI) and silencing efficiency in homozygous EV and irMPK4 shoots (top) and roots (bottom) which were grafted together for the glasshouse grafting pair experiment (n = 8 for EV/EV, 9 for EV/irMPK4, 6 for irMPK4/irMPK4).
Transcript accumulation data is also used to compare to the MPK4 silencing efficiency in hemizygous EV and irMPK4 crosses (Figure 2C).
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig2-figsupp1-data1-v2.xlsx
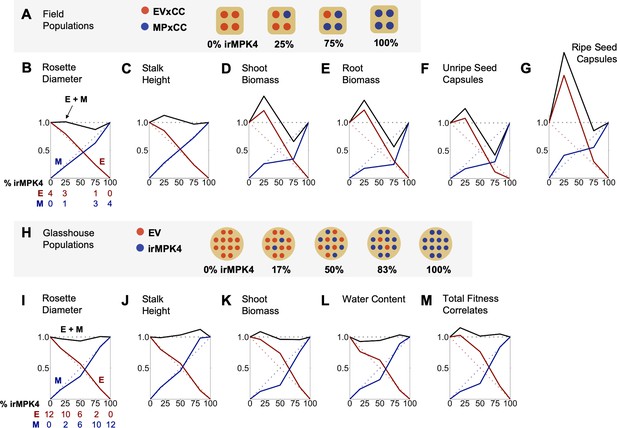
In the field and glasshouse, populations with low percentages of MPK4-deficient plants show overyielding.
(A) Field populations of four plants around a central water dripper were planted with varying percentages of EV and irMPK4 (MP) plants crossed with irCCaMK (CC) to abolish interaction with arbuscular mycorrhizal networks: four EVxCC (0%, n = 12), three EVxCC and one MPxCC (25%, n = 20), one EVxCC and three MPxCC (75%, n = 20), or four MPxCC (100%, n = 20) plants (for additional details of the experimental set-up see Figure 3—figure supplement 1). (B – G) Replacement diagrams show relative (B) rosette diameters (n = 11–35); (C) stalk heights (n = 6–29); (D) shoot biomasses (n = 10–31); (E) root biomasses (n = 8–31); (F) unripe seed capsules (n = 4–14); and (G) ripe seed capsules (n = 3–17) of EVxCC (E, red) and MPxCC (M, blue) plants in 0–100% irMPK4 field populations. Relative growth and yield for each genotype was calculated as: (trait mean in mixture * # of plants in mixture)/(trait mean in monoculture * 4). Means and error structures are shown in Figure 3—figure supplement 2. Relative yield totals of the populations (RYT, black) are calculated as E + M. Dotted lines indicate predicted yields from plants in monocultures. (H) Glasshouse populations of 12 plants were planted with varying percentages of irMPK4 plants: 12 EV (0%), 10 EV and 2 irMPK4 (17%), 6 EV and 6 irMPK4 (50%), 2 EV and 10 irMPK4 (83%), or 12 irMPK4 (100%). Each population was watered in proportion to its daily water consumption to ensure equal water availability across all populations (for additional details of the experimental set-up see Figure 3—figure supplement 5B and Water treatments in Materials and methods). (I – M) Replacement diagrams show relative (I) rosette diameters (n = 11–35); (J) stalk heights (n = 21–41); (K) shoot biomasses (n = 21–41); (L) water contents (n = 22–41); and M) total reproductive yield measured as counts of fitness correlates (n = 19–44) from EV (E, red) and irMPK4 (M, blue) plants in 0–100% irMPK4 glasshouse populations. Relative growth and yield for each genotype was calculated as: (trait mean in mixture * # of plants in mixture)/(trait mean in monoculture * 12). Means and error structures are shown in Figure 3—figure supplement 6. RYTs (black) are calculated as E + M. Dotted lines indicate predicted yields from monocultures.
-
Figure 3—source data 1
Source Data for Figure 3.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-data1-v2.xlsx
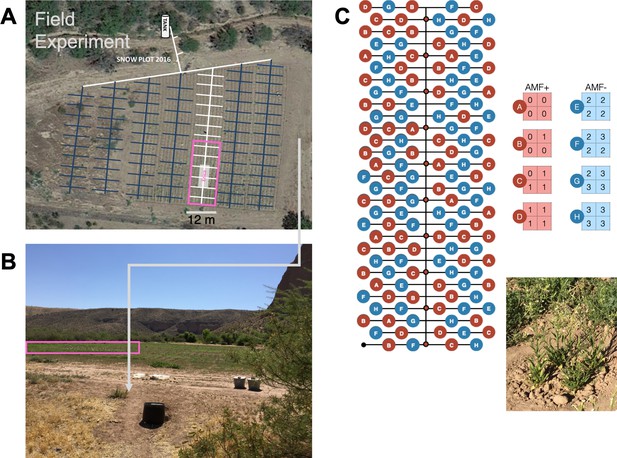
Layout of field population experiment at the Lytle Ranch Reserve field plot (‘Snow Plot’, Utah) from an (A) aerial view (pink, Google Images), from a (B) side view (pink, picture by E. M.), and as a (C) schematic of all planted populations, with an example population at harvest, 53 days post planting (dpp; inset, picture by EM).
© 2013 Google. All rights reserved. Panel A is taken from Google Earth. It is not covered by the CC-BY 4.0 licence and further reproduction of this panel may need permission from the copyright holder.
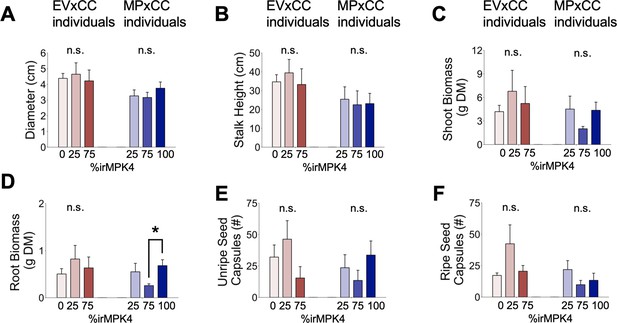
Growth and yield of EV and irMPK4 individuals crossed with irCCaMK (EVxCC and MPxCC, respectively) compared when planted in the varying field population types (see Figure 3A): A) rosette diameter (mean + CI, n = 11–35; 13 dpp), (B) stalk height (mean + CI, n = 6–29; 23 dpp), (C) shoot biomass (mean + SE, n = 10–31; 46–53 dpp), (D) root biomass (mean + SE, n = 8–31; 46–53 dpp), (E) unripe seed capsules (mean + SE, n = 4–14; 46–53 dpp) and F) ripe seed capsules (mean + SE, n = 3–17; 46–53 dpp).
Significant differences are presented within genotypes (*: p<0.05).
-
Figure 3—figure supplement 2—source data 1
Source Data for Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp2-data1-v2.xlsx
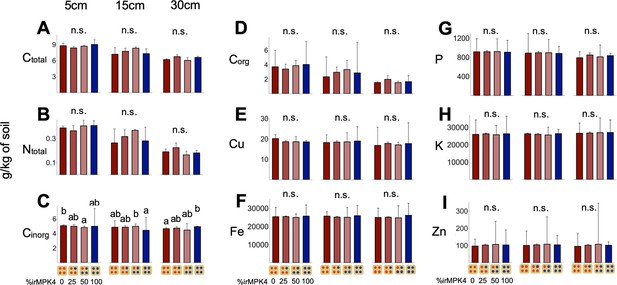
Soil element concentration (mean + CI, y-axis, n = 3) of (A) total carbon, (B) total nitrogen, (C) inorganic carbon, (D) organic carbon, (E) copper, (F) iron, (G) phosphorus, (H) potassium, and (I) zinc, taken from soil cores 5, 15 and 30 cm below the center dripper of each population type (x-axis).
-
Figure 3—figure supplement 3—source data 1
Source Data for Figure 3—figure supplement 3.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp3-data1-v2.xlsx
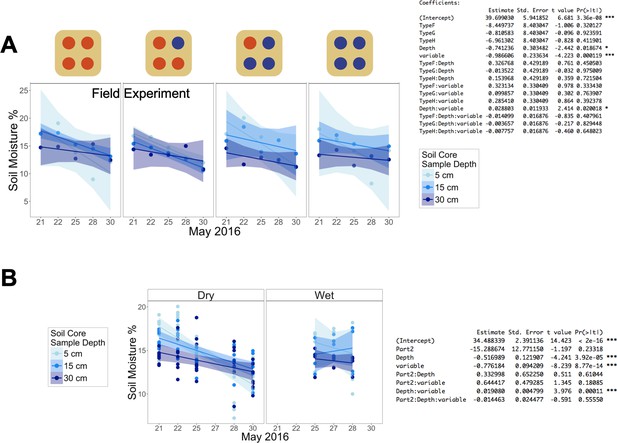
Soil moisture (%) of soil cores taken 5, 15 and 30 cm below the center dripper of each population type over 9 days, either (A) cumulatively across the plot or (B) divided into the ‘Dry’ and ‘Wet’ subsections (see Water treatments in Materials and methods), with corresponding regression analysis results.
-
Figure 3—figure supplement 4—source data 1
Source Data for Figure 3—figure supplement 4.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp4-data1-v2.xlsx
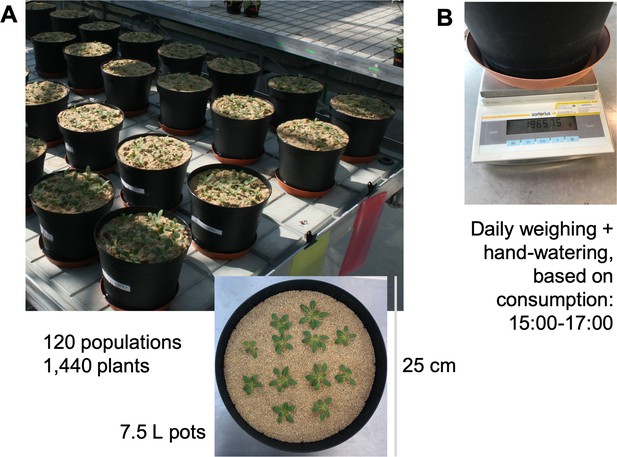
Layout of glasshouse population experiment as pictured from (A) above the glasshouse table (picture by DM), and above a single example pot (inset, picture by EM).
Each 7.5L pot was (B) weighed daily to determine how much water was lost during the day before as well as provide consumption-based watering based on daily water loss (picture by EM).
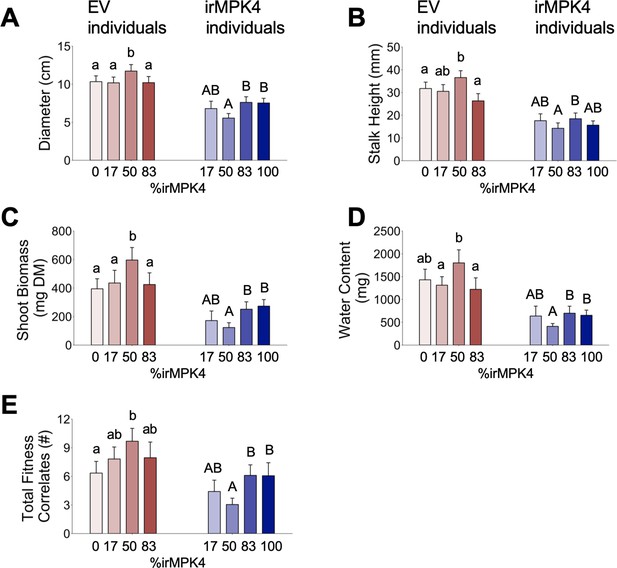
Growth and yield of EV and irMPK4 individuals compared when planted among the varying glasshouse population types (see Figure 3H): (A) rosette diameter (mean + CI, n = 11–35; 30 dpp), (B) stalk height (mean + CI, n = 21–41; 30 dpp), (C) shoot biomass (mean + CI, n = 21–41; 50 dpp), (D) water content (mean + CI, n = 22–41; 50 dpp), (E) total reproductive yield measured as counts of fitness correlates (buds, flowers, unripe and ripe seed capsules; mean + CI, n = 19–44; 50 dpp).
Significant differences are presented within genotypes.
-
Figure 3—figure supplement 6—source data 1
Source Data for Figure 3—figure supplement 6.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp6-data1-v2.xlsx
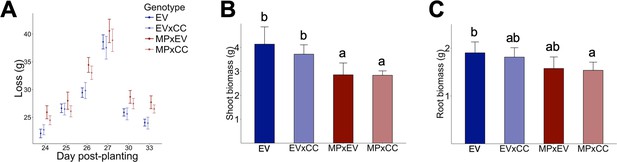
Characterization experiment comparing EV to EV x irCCaMK (EVxCC) and irMPK4xEV (MPxEV) to irMPK4 x irCCaMK (MPxCC) in (A) water loss per day (g ± CI, n = 15–16), (B) fresh shoot biomass (g + CI, n = 5–7), and (C) fresh root biomass (g + CI, n = 6–7).
-
Figure 3—figure supplement 7—source data 1
Source data for Figure 3—figure supplement 7.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp7-data1-v2.xlsx
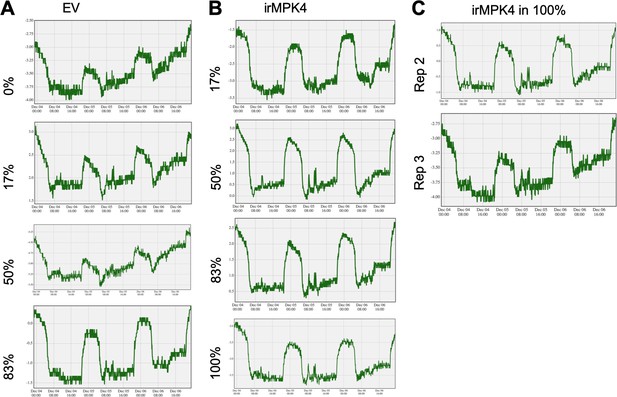
Yara ZIM-probe leaf turgor measurements (mean kPa, representative recordings of three replicate measurements are shown).
Leaf turgor was recorded over the 48 hr period from 00:00 December 4th to 00:00 December 7th on (A) EV and (B) irMPK4 individuals in all glasshouse population types. Note: relative turgor pressure values (y-axis) are not comparable among recordings due to the different clamping pressure at the initialization of recording (see Leaf turgor and potential effects of controlled watering on diurnal rhythms in Materials and methods). (C) Two additional replicates of irMPK4 plants in 100% populations demonstrate that within treatment group variance in peak-trough values exceeds among treatment group variance.
-
Figure 3—figure supplement 8—source data 1
Source Data for Figure 3—figure supplement 8.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig3-figsupp8-data1-v2.xlsx
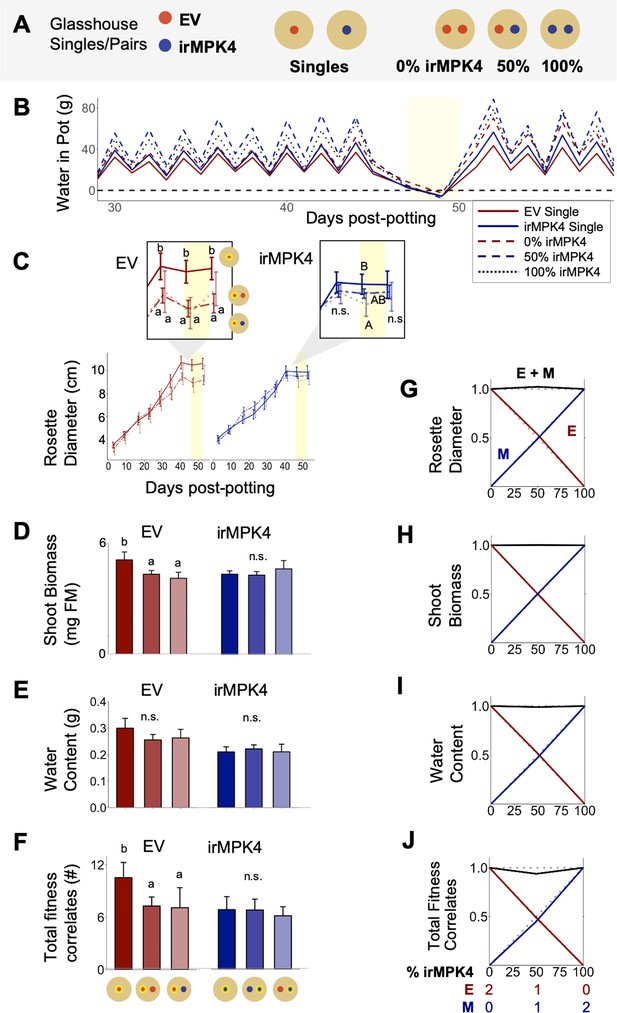
In the glasshouse under equal water availability, EV but not irMPK4 plants have reduced growth in the presence of a neighbor.
(A) EV and irMPK4 were planted either as singles or in mono- or mixed-culture pairs. EV monoculture pairs have 0%, mixed-cultures have 50%, and irMPK4 monocultures have 100% irMPK4 plants. (B) All pots were watered based on daily individual consumption to ensure equal water availability (see Water treatments in Materials and methods): mean g of water per pot for each potting type (red, solid: EV Single; red, dashed: 0% irMPK4; blue, solid: irMPK4 Single; blue, dashed: 100% irMPK4; black, dotted: 50% irMPK4) immediately following a watering event (graphical peaks) and immediately preceding the next watering event (graphical troughs) are displayed. Withholding water for 2 days caused all pots to reach a state of no available water in the pot (yellow shading). (C – F) EV (red) and irMPK4 (blue) individual means in each pot type (Single: solid line; 0%/100%: dashed line; 50%: dotted line) for (C) rosette diameter (mean cm ±95% CI, n = 11–24; 3–53 days post potting, dpp), (D) shoot biomass (mean + CI, n = 10–22; 71dpp), (E) water content (mean + CI, n = 8–22; 71dpp), and (F) total reproductive yield measured as counts of fitness correlates (buds, flowers, unripe and ripe seed capsules; mean + CI, n = 9–22; 71dpp). Significant differences are presented within genotypes. Inset of (C): Significant differences in EV and irMPK4 rosette diameters among planting types are indicated for the last three time points of the main panel, within each genotype. To evaluate growth effects of the equal water availability (yellow shading), growth values before and after water was withheld are highlighted in the inset. (G – I) Replacement diagrams show (G) rosette diameters (n = 11–24; 53 dpp); (H) shoot biomasses (n = 10–22); (I) water contents (n = 8–22); and (J) total reproductive yield measured as counts of fitness correlates (n = 9–22) from EV (E, red) and irMPK4 (M, blue) plants in 0–100% irMPK4 glasshouse pairs, calculated as (trait mean in mixture*# of plants)/(trait mean in monoculture*2). Relative yield totals (RYT, black) are calculated as E + M. Means and error structures can be found in panels (C – F). Dotted lines indicate no deviations from yields in monocultures.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig4-data1-v2.xlsx
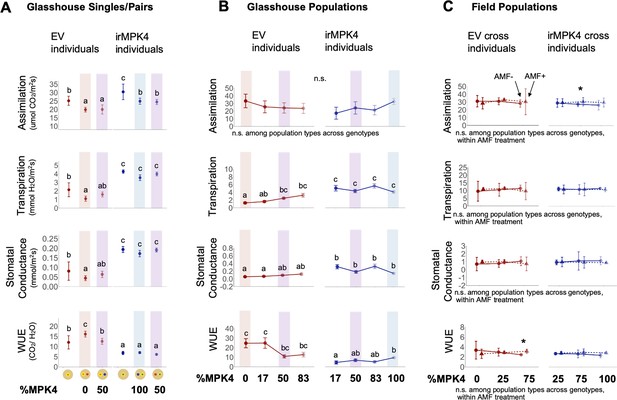
irMPK4 plants have low WUE in glasshouse, but not field experiments, regardless of AMF associations.
(A) Assimilation rate, transpiration rate, stomatal conductance and water-use efficiency (WUE; mean ±CI, n = 3–8) of EV (red) and irMPK4 (blue) individuals from each planting type in the paired glasshouse experiment (see Figure 4) at 48 days post potting (dpp). To facilitate comparison of data to the population glasshouse experiment, EV in 0% irMPK4 populations (red shading), irMPK4 plants in 100% irMPK4 populations (blue shading) and both genotypes in 50% irMPK4 populations (purple shading) are highlighted. Significant differences are presented across genotypes. (B) Assimilation rate, transpiration rate, stomatal conductance and WUE (mean ±CI, n = 11–32) of EV (red) and irMPK4 (blue) individuals from each planting type in the population glasshouse experiment (Figure 3H) at 32 dpp. Measurements were taken between 12:00-14:00; additional pre-dawn measurements (4:00-6:00) are included in Figure 5—figure supplement 1. For comparison to the paired glasshouse experiment, EV and irMPK4 in 0% (red), 50% (purple) and 100% (blue) irMPK4 populations are highlighted. Significant differences are presented across genotypes. (C) Assimilation rate, transpiration rate, stomatal conductance and WUE (mean ±CI, n = 3) of EV (red, circle) and irMPK4xEV (blue, circle) individuals with the ability to associate with arbuscular mycorrhizal fungi (AMF, solid line), or EVxCC (red, triangle) and irMPK4xCC (blue, triangle) individuals without the ability to associate with AMF from the field population experiment. Measurements were performed at 34 dpp on irrigated plants (‘Wet’, see Water treatments in Materials and methods). Significant differences are presented both across genotypes, within AMF treatments (text below panels), or within the genotype and planting type, between AMF treatments (*: p<0.05).
-
Figure 5—source data 1
Source Data for Figure 5.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig5-data1-v2.xlsx
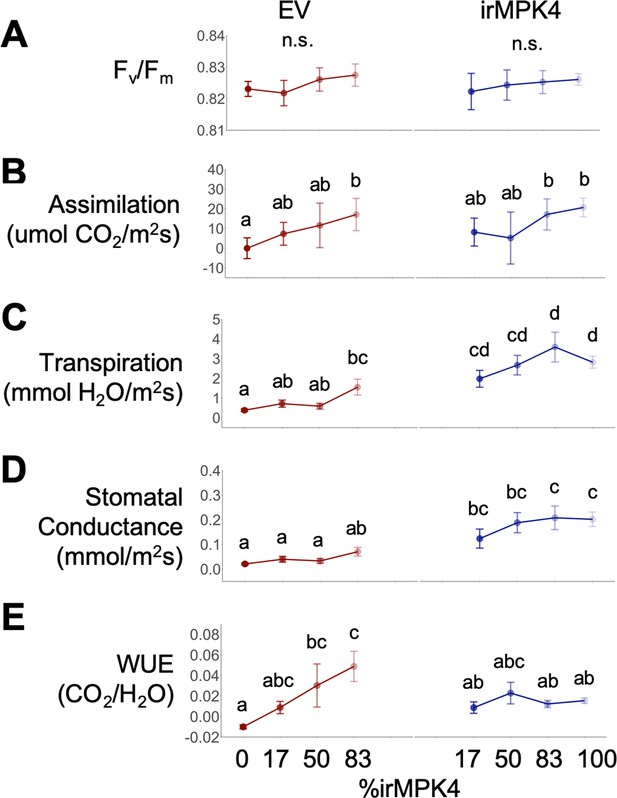
Pre-dawn photosynthetic measurements of EV (red) and irMPK4 (blue) plants in the various planting types of the population glasshouse experiment.
(A) Fv/Fm, the maximum yield of the photosynthetic systems, (B) assimilation rates, (C) transpiration rates, (D) stomatal conductances, and (E) water-use efficiencies (WUEs; mean ±CI, n = 11–32) were measured 32 days post-planting (dpp). Significant differences are presented across genotypes and planting types.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig5-figsupp1-data1-v2.xlsx
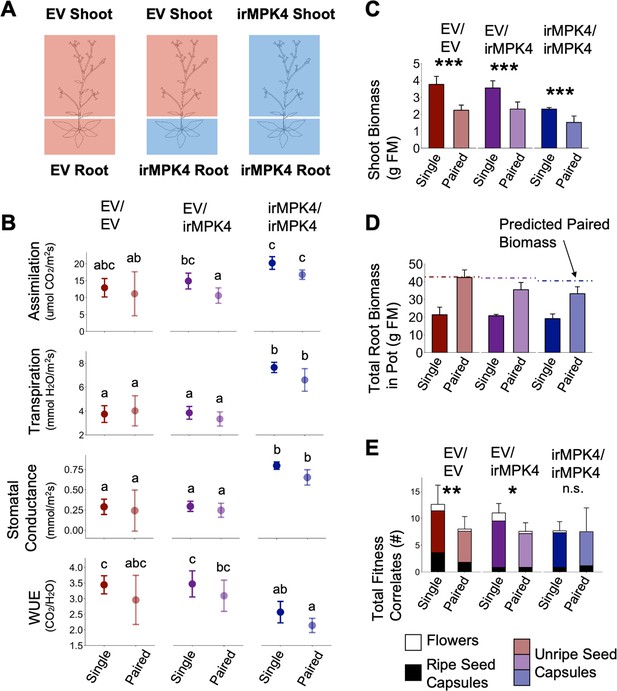
Expression of MPK4 in the shoots mediates changes in N. attenuata reproductive output in response to neighbors.
(A) EV shoots were micro-grafted onto irMPK4 roots, producing plants deficient in MPK4 in the root but not in the shoot (EV/irMPK4, Figure 2—figure supplement 1). These were compared to EV/EV and irMPK4/irMPK4 homografts as controls. All three graft types were grown both as singles and in pairs with an ungrafted EV neighbor, under conditions of equal water availability in a glass house experiment (Figure 6—figure supplement 1A). (B) Assimilation rates, transpiration rates, stomatal conductance and water-use efficiency (WUE; mean ±CI, n = 3–7) of single and paired plants of each grafting type (EV/EV: red; EV/irMPK4: purple; irMPK4/irMPK4: blue) were measured at 37 dpp. Significant differences are presented across all graft and potting types. (C) Shoot biomass (mean + CI, n = 4–6) of EV/EV (red), EV/irMPK4 (purple) and irMPK4/irMPK4 (blue) individuals in each potting type (Single and Paired) was recorded at 50 dpp. Significant differences within genotypes are indicated (***: p<0.001). (D) Total root biomass in each pot (mean + CI, n = 4–6) was recorded for each potting type at 50 dpp. Dashed lines indicate the total pot root biomass predicted from the summed root biomasses of the respective genotype + EV/EV when planted as single plants in pots. (E) Counts (#) of fitness correlates (mean + CI, n = 5–7) of EV/EV (red), EV/irMPK4 (purple) and irMPK4/irMPK4 (blue) individuals in each pot type were recorded at 50 dpp. Statistical analyses were only performed for the total fitness correlates, although each bar is dissected into its contributing parts: flowers (white), unripe seed capsules (color), and ripe seed capsules (black). Significant differences within genotypes are indicated (*: p<0.05; **: p<0.01).
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig6-data1-v2.xlsx
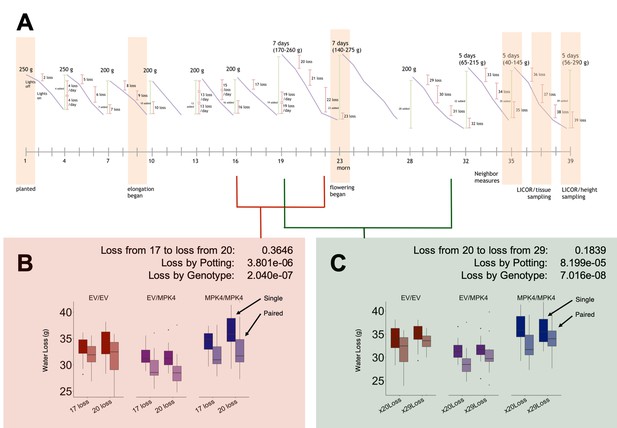
Watering regime of the glasshouse grafted pair experiment.
(A) The amount of water (g) given to each pot on each day of watering is indicated above the watering event (green). Orange shading highlights key experimental events (developmental changes and sampling times). Two analyses on whether (B) increasing the amount of water given to a pot during a watering event changes the water loss per day of the pot (y-axis) from experimental day 17 (given 200 g water) to 20 (given 170–260 g water, x-axis) or whether (C) decreasing water given to a pot during a watering event changes the water loss per day of the pot (y-axis) from experimental day 20 (given 170–260 g water) to 29 (given 200 g water, x-axis), are presented. Each analysis is accompanied with respective p-values from an ANOVA conducted on the variables: loss between days, loss between potting type (single: dark colors; paired: faded colors), or loss by genotype of the grafted plant in the pot (red: EV/EV; purple: EV/irMPK4; blue: irMPK4/irMPK4).
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig6-figsupp1-data1-v2.xlsx
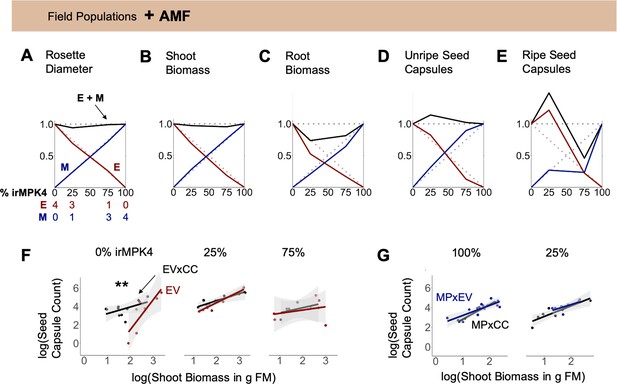
Interaction with arbuscular mycorrhizal fungi (AMF) abolishes overyielding in biomass, but not seed capsule production of populations with low percentages of MPK4-deficient plants.
(A – E) EV and irMPK4 individuals crossed with EV instead of irCCaMK (EVxEV: EV; irMPK4xEV: MPxEV) can associate with arbuscular mycorrhizal fungi. Field populations were varied in percentages of EV and MPxEV plants (Figure 3A, Figure 3—figure supplement 1). Replacement diagrams show (A) rosette diameters (n = 12–28); (B) shoot biomasses (n = 6–12 excl. M in 75% = 2); (C) root biomasses (n = 7–12 excl. M in 75% = 2); (D) unripe seed capsules (n = 6–14 excl. M in 75% = 2); (E) ripe seed capsules (n = 6–16) of EV (E, red) and MPxEV (M, blue) plants in 0–100% irMPK4 field populations. Relative growth and yield for each genotype is calculated as (trait mean in mixture*# of plants)/(trait mean in monoculture*4). Relative yield totals (RYT, black) are calculated as E + M. Dotted lines indicate no deviations from yields in monocultures. (F – G) Allometric trajectories of (F) EV x irCCaMK (EVxCC, black) plants compared to EV (red) plants in 0%, 25% and 75% irMPK4 populations, as well as (G) irMPK4 x irCCaMK (MPxCC, black) plants compared to irMPK4xEV (MPxEV, blue) plants in 100% and 25% irMPK4 populations. Asterisks indicate significant differences within population types (lstrends, pairwise comparisons of slopes of fitted lines: ***p<0.001, **p<0.01, *p<0.05).
-
Figure 7—source data 1
Source Data for Figure 7.
- https://cdn.elifesciences.org/articles/53517/elife-53517-fig7-data1-v2.xlsx
Tables
emmeans contrasts of EV individuals in varying potting types for Figure 4C–F.*
| Model | Contrast | Trait | t-value | p-value |
|---|---|---|---|---|
| LM | EV Single(n = 9) – EV Mono(n = 22) | Water Content | 2.378 | 0.0614 |
| EV Single(n = 9) –EV Mix(n = 11) | Water Content | 1.751 | 0.2047 | |
| LM | EV Single(n = 12) – EV Mono(n = 24) | Rosette Diameter | 5.131 | <0.0001† |
| EV Single(n = 12) – EV Mix(n = 12) | Rosette Diameter | 4.979 | <0.0001† | |
| GLS | EV Single(n = 11) –EV Mono(n = 22) | Shoot Biomass | 4.196 | 0.0004† |
| EV Single(n = 11) –EV Mix(n = 10) | Shoot Biomass | 4.531 | 0.0002† | |
| LM | EV Single(n = 11) –EV Mono(n = 21) | Total Fitness Correlates | 3.848 | 0.0017‡ |
| EV Single(n = 11) –EV Mix(n = 11) | Total Fitness Correlates | 3.323 | 0.0066‡ |
-
*extracted from linear (LM) or generalized least squares (GLS) models with significant ANOVA results.
†p value < 0.001; ‡p value < 0.01.
emmeans contrasts of EV to irMPK4 individuals planted as singles for Figure 5A*.
| Model | Contrast | Trait | t-value | p-value |
|---|---|---|---|---|
| LME | EV Single(n = 4) – irMPK4 Single(n = 3) | Assimilation | −3.947 | 0.0134‡ |
| EV Mono(n = 7) – EV Mix(n = 4) | Assimilation | −0.123 | 1.0000 | |
| rMPK4 Mono(n = 8) – irMPK4 Mix(n = 4) | Assimilation | 0.396 | 0.9985 | |
| LME | EV Single(n = 4) – irMPK4 Single(n = 4) | Transpiration | −8.089 | <0.0001† |
| EV Mono(n = 8) – EV Mix(n = 4) | Transpiration | −3.171 | 0.0527 | |
| irMPK4 Mono(n = 8) – irMPK4 Mix(n = 4) | Transpiration | −2.776 | 0.1104 | |
| LME | EV Single(n = 3) – irMPK4 Single(n = 4) | SC | −8.089 | <0.0001† |
| EV Mono(n = 8) – EV Mix(n = 4) | SC | −3.171 | 0.0527 | |
| irMPK4 Mono(n = 8) – irMPK4 Mix(n = 4) | SC | −2.776 | 0.1104 | |
| LME | EV Single(n = 4) – irMPK4 Single(n = 4) | WUE | 6.394 | 0.0001† |
| EV Mono(n = 8) – EV Mix(n = 4) | WUE | 3.723 | 0.0205‡ | |
| irMPK4 Mono(n = 8) – irMPK4 Mix(n = 4) | WUE | 3.203 | 0.0544 |
-
*extracted from linear-mixed effect (LME) models with significant ANCOVA results SC = Stomatal Conductance; WUE = Water Use Efficiency.
†p value < 0.001; ‡p value < 0.05.
Statistical emmeans contrasts within planting treatments for Figure 6B*.
| Parameter | Model | Contrast | T value | P value |
|---|---|---|---|---|
| Assimilation | LM | S: irMPK4/irMPK4(n = 6) to EV/EV(n = 5) | −5.718 | 0.0001† |
| S: irMPK4/irMPK4(n = 6) to EV/irMPK4(n = 7) | −4.537 | 0.0014‡ | ||
| P: irMPK4/irMPK4(n = 5) to EV/EV(n = 3) | −3.666 | 0.0127§ | ||
| P: irMPK4/irMPK4(n = 5) to EV/irMPK4(n = 6) | −4.837 | 0.0007† | ||
| Transpiration | LM | S: irMPK4/irMPK4(n = 5) to EV/EV(n = 5) | −10.979 | <0.0001† |
| S: irMPK4/irMPK4(n = 5) to EV/irMPK4(n = 6) | −11.163 | <0.0001† | ||
| P: irMPK4/irMPK4(n = 4) to EV/EV(n = 4) | −6.506 | <0.0001† | ||
| P: irMPK4/irMPK4(n = 4) to EV/irMPK4(n = 6) | −9.008 | <0.0001† | ||
| Stomatal | LM | S: irMPK4/irMPK4(n = 4) to EV/EV(n = 5) | −9.429 | <0.0001† |
| conductance | S: irMPK4/irMPK4(n = 4) to EV/irMPK4(n = 7) | −9.971 | <0.0001† | |
| P: irMPK4/irMPK4(n = 6) to EV/EV(n = 3) | −7.209 | <0.0001† | ||
| P: irMPK4/irMPK4(n = 6) to EV/irMPK4(n = 7)–9.079 < 0.0001*** | ||||
| WUE | LM | S: irMPK4/irMPK4(n = 6) to EV/EV(n = 4) | 3.696 | 0.0109‡ |
| S: irMPK4/irMPK4(n = 6) to EV/irMPK4(n = 7) | 4.000 | 0.0051‡ | ||
| P: irMPK4/irMPK4(n = 6) to EV/EV(n = 5) | 3.240 | 0.0329§ | ||
| P: irMPK4/irMPK4(n = 6) to EV/irMPK4(n = 6) | 4.376 | 0.0019‡ |
-
*extracted from linear (LM) or generalized least squares (GLS) models with significant ANOVA results S: Singles; P: Paired.
†p value < 0.001; ‡p value < 0.01; §p value < 0.05.
Statistical emmeans contrasts within planting treatments for Figure 6C,E*.
| Parameter | Model | Contrast | T value | P value |
|---|---|---|---|---|
| Shoot Biomass | LM | EV/EV: S(n = 5) - P(n = 5) | −7.823 | <0.0001† |
| EV/irMPK4: S(n = 4) - P(n = 6) | −6.232 | <0.0001† | ||
| irMPK4/irMPK4: S(n = 6) - P(n = 6) | −4.442 | <0.0001† | ||
| TFC | LM | EV/EV: S(n = 9) - P(n = 11) | −2.637 | 0.0106‡ |
| EV/irMPK4: S(n = 13) - P(n = 12) | −3.620 | 0.0006† | ||
| irMPK4/irMPK4: S(n = 13) - P(n = 10) | −0.024 | 0.9813 |
-
*extracted from linear (LM) or generalized least squares (GLS) models with significant ANOVA results TFC: Total Fitness Correlates; S: Singles; P: Paired.
†p value < 0.001; ‡p value < 0.05.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (N. attenuata) | A-04-266-3 | Bubner et al., 2006 DOI:10.1007/s00299-005-0111-4 | Empty vector control | |
| Genetic reagent (N. attenuata) | A-7–163 | Hettenhausen et al., 2012 DOI:10.1086/342519 | Stably silenced in MPK4 via RNAi | |
| Genetic reagent (N. attenuata) | A-09-1212-1-4 | Groten et al., 2015 DOI:10.1111/pce.12561 | Stably silenced in CCaMK via RNAi | |
| Software | R version 3.4.2 | R Development Core Team, 2017 | ||
| Software | RStudio version 1.0.153 | Rstudio Team, 2016 |
Additional files
-
Supplementary file 1
Table of emmeans contrasts, within genotypes from Figure 3—figure supplement 6α.
- https://cdn.elifesciences.org/articles/53517/elife-53517-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53517/elife-53517-transrepform-v2.docx





