mRNA decapping is an evolutionarily conserved modulator of neuroendocrine signaling that controls development and ageing
Figures
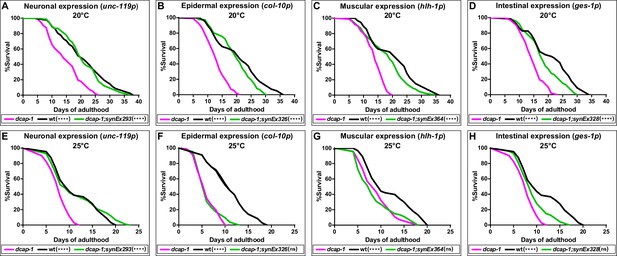
Neuronal restoration of dcap-1 rescues the short lifespan of dcap-1 mutants.
Lifespan of dcap-1 worms overexpressing a dcap-1::gfp fusion in neurons, epidermis, muscles or intestine at (A–D) 20°C and (E–H) 25°C. Statistical significance is calculated in comparison to dcap-1. ****p<0.0001. Log-rank (Mantel-Cox) test.
-
Figure 1—source data 1
Lifespan replicates of dcap-1(tm3163) worms that overexpress a dcap-1::gfp fusion tissue-specifically at 20°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Lifespan replicates of dcap-1(tm3163) worms that overexpress a dcap-1::gfp fusion tissue-specifically at 25°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig1-data2-v1.xlsx
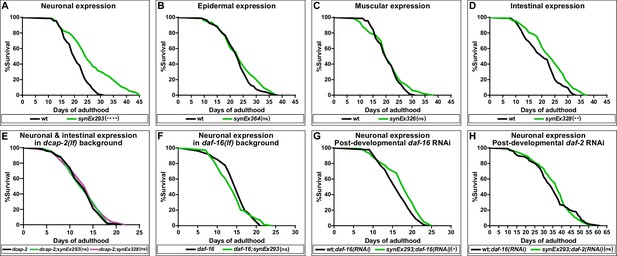
Neuronal overexpression of dcap-1 extends lifespan through mRNA decapping, DAF-16/FOXO and IIS.
Lifespan at 20°C of (A–D) wt worms overexpressing a dcap-1::gfp fusion in neurons, epidermis, muscles or intestine, (E) worms overexpressing neuronal or intestinal dcap-1::gfp in dcap-2 mutant genetic background, (F) worms overexpressing neuronal dcap-1::gfp in daf-16 mutant genetic background, (G) worms overexpressing neuronal dcap-1::gfp under post-developmental daf-16 RNAi knockdown, (H) worms overexpressing neuronal dcap-1::gfp during post-developmental daf-2 RNAi knockdown. *p<0.05, **p<0.01, ****p<0.0001. Log-rank (Mantel-Cox) test.
-
Figure 2—source data 1
Lifespan replicates of worms that overexpress a dcap-1::gfp fusion tissue-specifically in wtgenetic background at 20°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Lifespan replicates of worms depleted of daf-2 or daf-16 at 20°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Lifespan replicates of dcap-2(ok2023) worms that overexpress a dcap-1::gfp fusion tissue-specifically at 20°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig2-data3-v1.xlsx
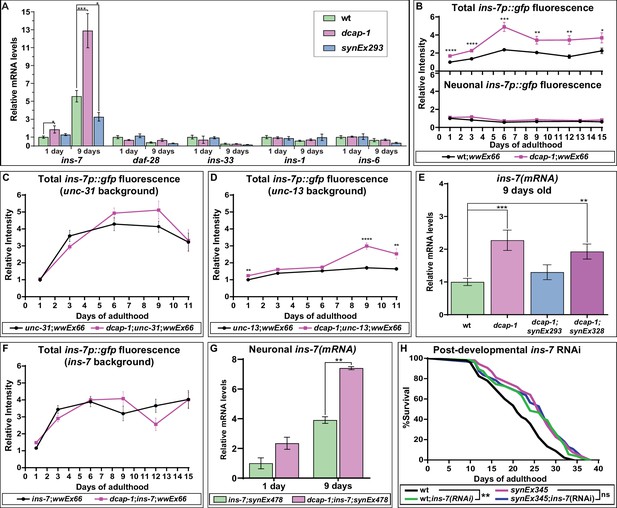
Neuronal dcap-1 regulates ins-7 expression to control longevity.
(A) mRNA levels of lifespan-regulating ILPs in young (1 day old) and mid-aged (9 days old) wt, dcap-1 and neuronal dcap-1 overexpressing worms. (B) Fluorescence intensity of ins-7p::gfp reporter in whole worms or head neurons of wt and dcap-1 animals. (C–D) Total fluorescence intensity of ins-7p::gfp reporter in neurosecretion defective unc-31 or unc-13 mutants. (E) ins-7 mRNA levels in 9 days old worms overexpressing dcap-1::gfp in neurons (synEx293) or intestine (synEx328). (F) Total fluorescence intensity of ins-7p::gfp reporter in ins-7 mutants. (G) ins-7 mature mRNA/pre-mRNA ratio in ins-7 and dcap-1;ins-7 animals that carry a neuronally expressed unc-119p::ins-7 transgene. (H) Lifespan of wt and neuronal dcap-1 overexpressing worms sensitized for neuronal RNAi during post-developmental ins-7 RNAi knockdown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars indicate mean ± SD. Unpaired t-test (A–G); Log-rank (Mantel-Cox) test (F).
-
Figure 3—source data 1
Lifespan replicates of worms exposed to ins-7 RNAi at 20°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Figure 3A, E and G and Figure 3—figure supplement 6.
Numeric values of relative mRNA levels of insulin-like peptides in the indicated genetic backgrounds and ages.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig3-data2-v1.xlsx
-
Figure 3—source data 3
Figure 3B, C, D and F and Figure 3—figure supplement 2A.
Numeric values of relative fluorescence intensity of ins-7p::gfp and ges-1p::gfp reporters in the indicated genetic backgrounds.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig3-data3-v1.xlsx
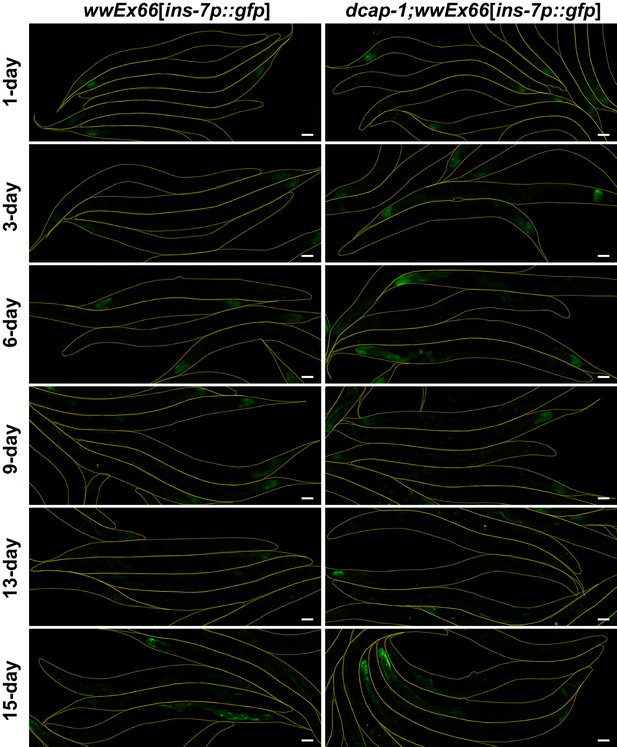
Transcriptional activity of ins-7 promoter is significantly increased in dcap-1 mutants.
Representative confocal images (maximum projections) of wt or dcap-1(tm3163) worms that express gfp under the control of ins-7 promoter (array wwEX66) at various ages. Scale bar = 50 μm.
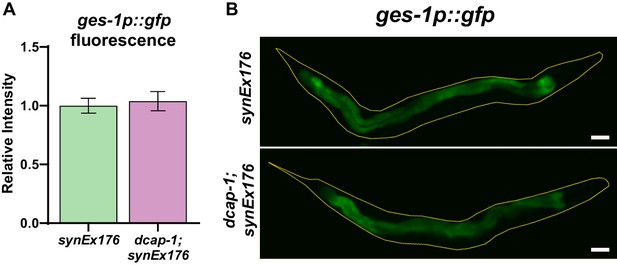
Mutation of dcap-1 does not affect the expression of a ges-1p::gfp transgene.
(A) Fluorescence intensity of ges-1p::GFP reporter in wild type and dcap-1 mutant worms at day 1 of adulthood. (B) Representative fluorescent images of wt and dcap-1 mutant worms expressing a ges-1p::gfp transgene. Scale bar = 50 μm.
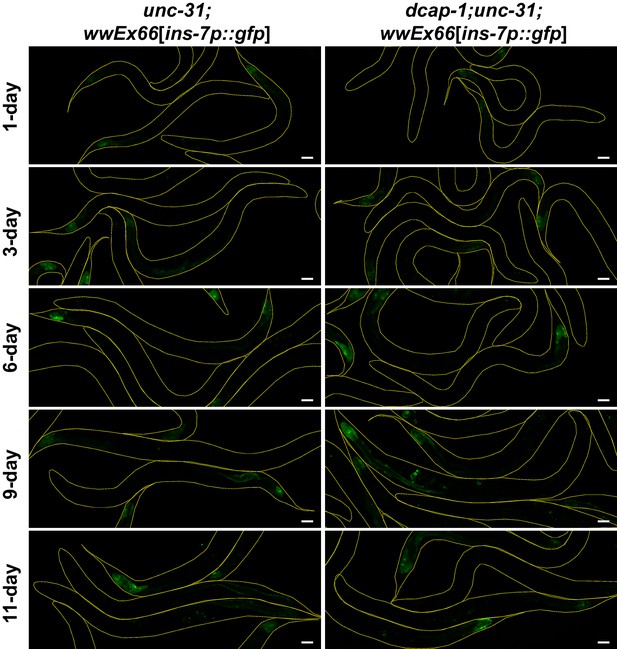
Transcriptional activity of ins-7 promoter is not increased in dcap-1 mutants when unc-31 is missing.
Representative confocal images (maximum projections) of unc-31(e928) or dcap-1(tm3163);unc-31(e928) worms that express gfp under the control of ins-7 promoter (array wwEX66) at various ages. Scale bar = 50 μm.
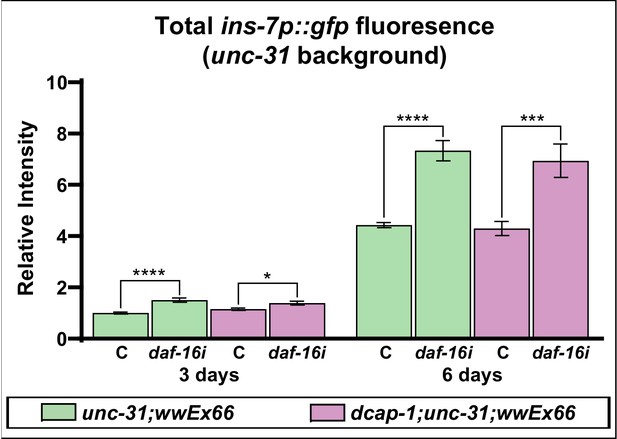
Transcriptional activity of ins-7 promoter in unc-31 mutants is further induced upon daf-16 knockdown.
Total fluorescence intensity of ins-7p::gfp reporter in neurosecretion defective unc-31 and dcap-1;unc-31 1 day and 3-day-old adults under control conditions (C) or daf-16 knockdown (daf-16i).
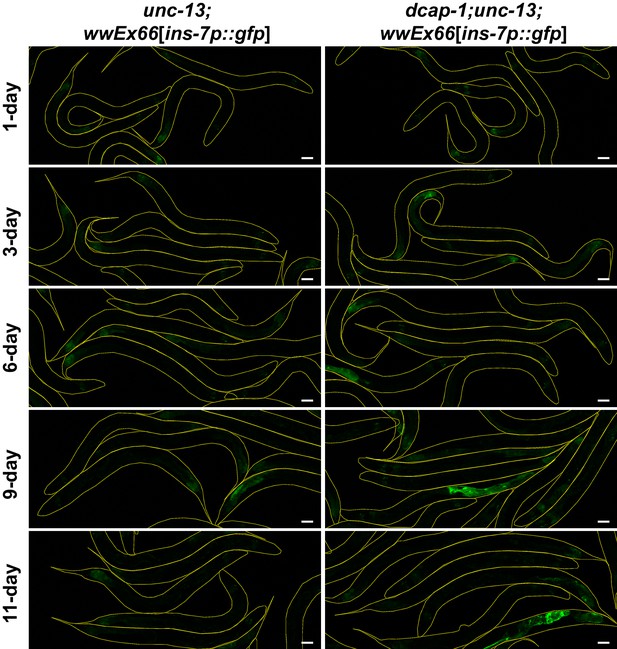
Transcriptional activity of ins-7 promoter is significantly increased in dcap-1 mutants independently of unc-13.
Representative confocal images (maximum projections) of unc-13(e450) or dcap-1(tm3163);unc-13(e450) worms that express gfp under the control of ins-7 promoter (array wwEX66) at various ages. Scale bar = 50 μm.
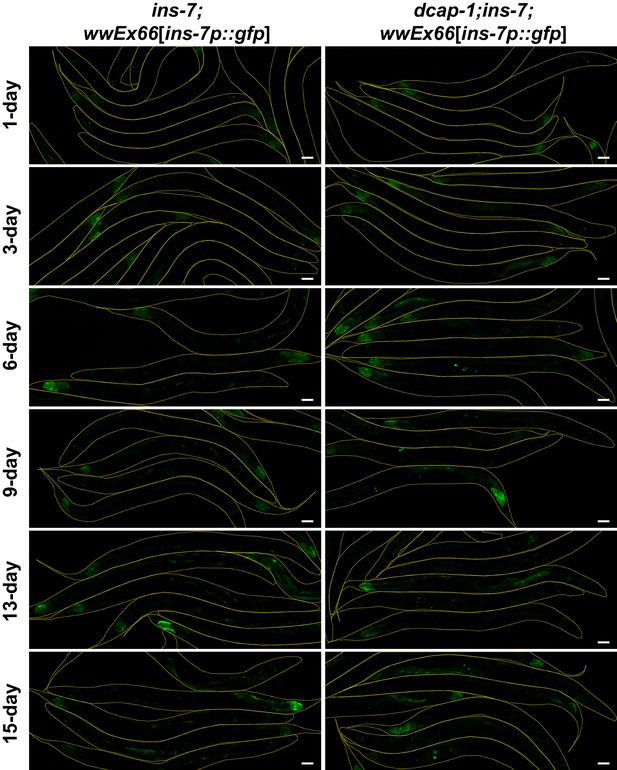
Transcriptional activity of ins-7 promoter is not increased in dcap-1 mutants when ins-7 gene product is missing.
Representative confocal images (maximum projections) of ins-7(ok1573) or dcap-1(tm3163);ins-7(ok1573) worms that express gfp under the control of ins-7 promoter (array wwEX66) at various ages. Scale bar = 50 μm.

The ratio of eft-3 mature mRNA/pre-mRNA is not affected by dcap-1 mutation.
eft-3 (F31E3.5) mature mRNA/pre-mRNA ratio in ins-7 and dcap-1;ins-7 animals that carry a neuronally expressed unc-119p::ins-7 transgene.
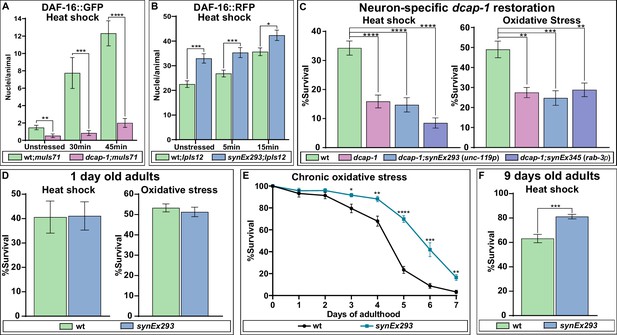
DCAP-1 affects DAF-16 localization and stress resistance.
(A) Quantification of DAF-16a::GFP positive nuclei during early stages of heat shock response in 1-day-old wt and dcap-1 mutant worms. (B) Quantification of DAF-16a::RFP positive nuclei during early stages of heat shock response in 1-day-old wt and neuronal dcap-1::gfp overexpressing worms. (C) Survival of 1-day-old dcap-1 worms that express dcap-1::gfp in their neurons, after acute exposure to high temperature or high concentration of sodium arsenite. (D) Survival of 1-day-old wt worms overexpressing neuronal dcap-1::gfp after acute exposure to high temperature or high concentration of sodium arsenite. (E) Survival of wt worms overexpressing neuronal dcap-1::gfp during sustained exposure to low concentration of sodium arsenite. (F) Survival of 9 days old wt worms overexpressing neuronal dcap-1::gfp after acute exposure to high temperature. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars indicate mean ± SD. Unpaired t-test.
-
Figure 4—source data 1
Numerical values of DAF-16-positive nuclei plotted in panels A and B.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Numerical values of % survival plotted in panels C, D, E and F.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig4-data2-v1.xlsx
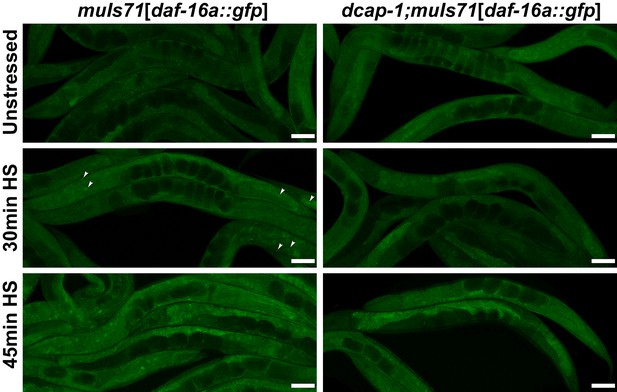
Translocation of DAF-16 to the nucleus is impaired in dcap-1 mutants.
Representative confocal images (maximum projections) of wt and dcap-1(tm3163) worms that express a daf-16a::gfp fusion under the control of the native daf-16 promoter, after mild heat shock at 35°C. Arrowheads point to nuclei. Scale bar = 50 μm.
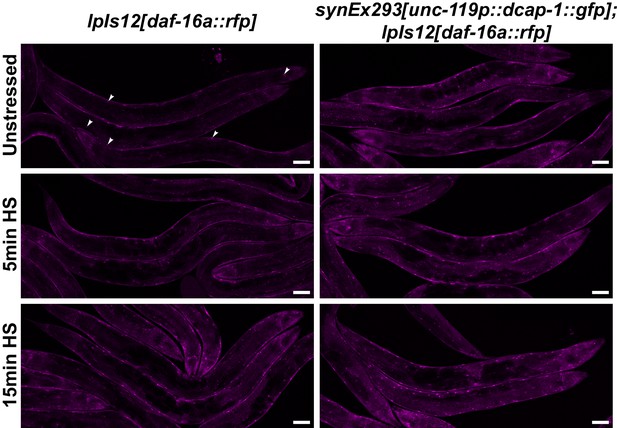
Translocation of DAF-16 to the nucleus is facilitated in neuronal dcap-1 overexpressing worms.
Representative confocal images (maximum projections) of wt and neuronal dcap-1 overexpressing worms (synEx293) that express a daf-16a::rfp fusion under the control of the native daf-16 promoter, after mild heat shock at 35°C. Arrowheads point to nuclei. Scale bar = 50 μm.
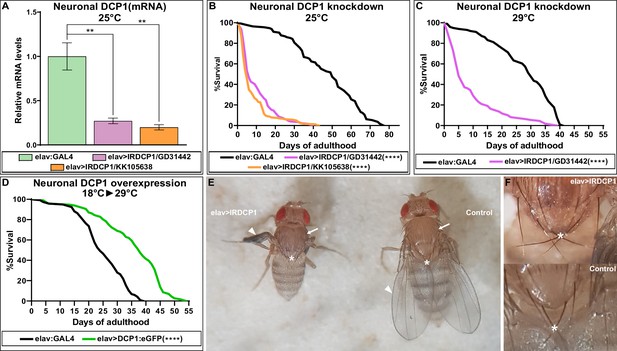
Neuronal DCP1 affects lifespan and adult wing expansion in D. melanogaster.
(A) DCP1 mRNA levels in heads of two fly strains subjected to neuronal DCP1 knockdown. (B, C) Lifespan of flies subjected to neuronal DCP1 knockdown. (D) Lifespan of flies that neuronally overexpress a DCP1:eGFP fusion post-developmentally. (E, F) Malformations of adult flies subjected to neuron-specific DCP1 knockdown during their development (see text). Arrowheads point to wing morphology, arrows to the difference in cuticle tanning and asterisks to the crossing of postscutellar bristles. **p<0.01, ****p<0.0001. Error bars indicate mean ± SD. Unpaired t-test (A); Log-rank (Mantel-Cox) test (B–D).
-
Figure 5—source data 1
Lifespan replicates of flies with neuronal DCP1 knockdown or overexpression.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig5-data1-v1.xlsx
-
Figure 5—source data 2
Numeric values of relative mRNA levels plotted in panel 5A and figure supplement 2.
- https://cdn.elifesciences.org/articles/53757/elife-53757-fig5-data2-v1.xlsx
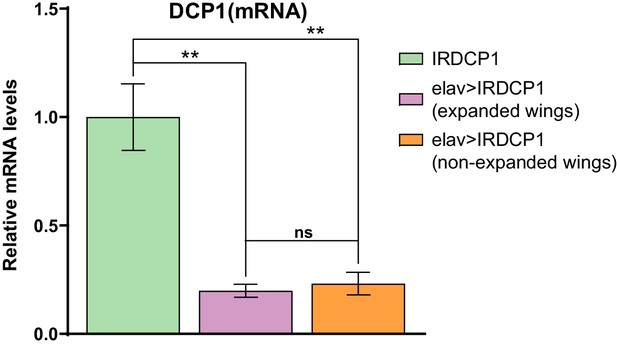
The penetrance of unexpanded wings phenotype does not correlate with the efficiency of DCP1 knockdown.
DCP1 mRNA levels in heads of flies with expanded or non-expanded wings, subjected to neuronal DCP1 knockdown.
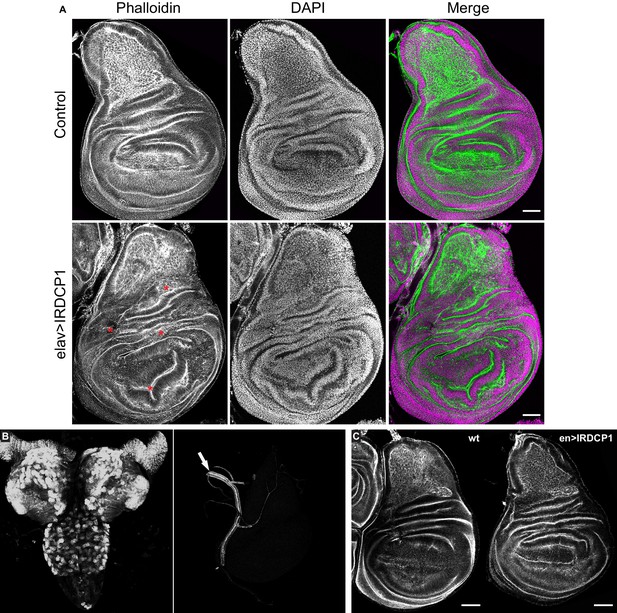
Neuronal DCP1 affects wing disk morphology.
(A) Phalloidin staining of wing imaginal discs derived from animals subjected to neuron-specific DCP1 knockdown. Asterisks indicate abnormalities in epithelial folding. Green: phalloidin, Magenta: DAPI. (B) Fluorescent images of brain (left) and imaginal disc (right) dissected from elav >DCP1:eGFP larvae. Arrow shows autofluorescence in the trachea. (C) Phalloidin staining of wing discs derived from flies subjected to DCP1 knockdown only in the posterior part of wing disc epithelium (using engrailed expression driver). Anterior is to the left and posterior to the right. Scale bar = 50 μm.
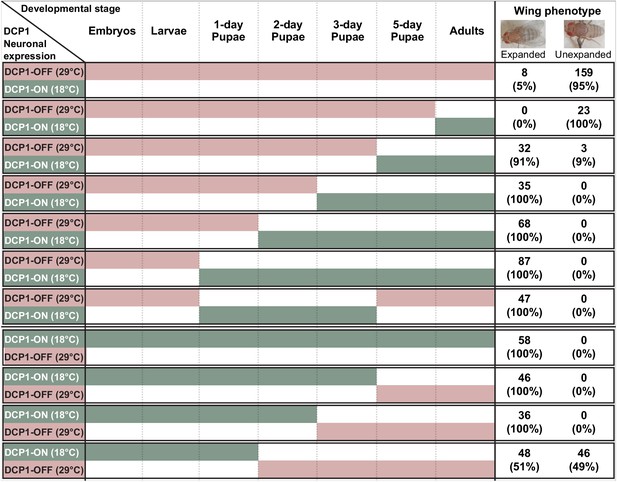
Schematic representation of thermoinducible DCP1 knock down at different developmental stages.
The crucial period where DCP1 activity affects adult wing expansion is traced between the 1-day and the 3-day pupa stages.
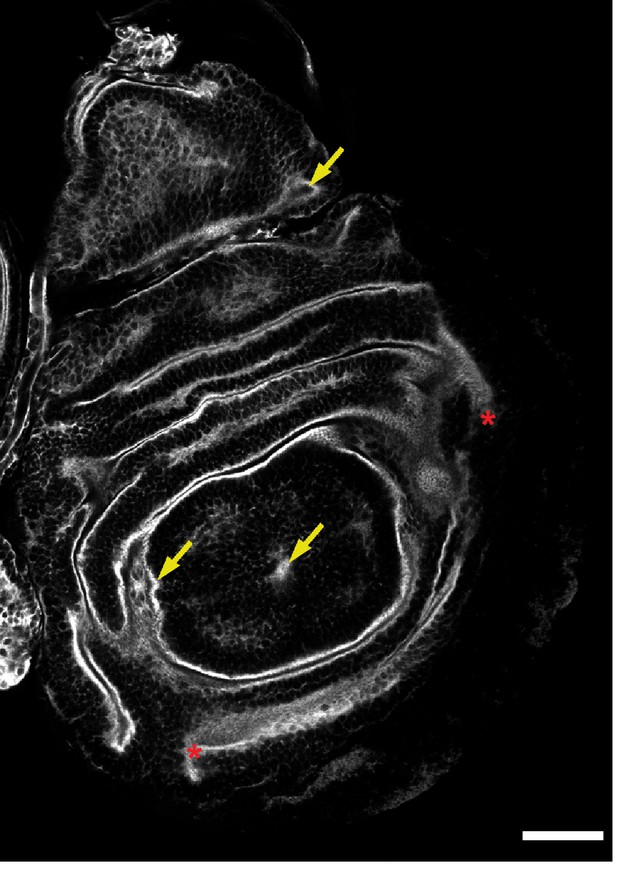
Abnormalities in wing discs of larvae subjected to neuron-specific DCP1 knockdown at 29°C.
Phalloidin staining of wing imaginal discs derived from 3rd instar larvae subjected to neuron-specific DCP1 knockdown at 29°C. Yellow arrows indicate aberrant F-actin accumulation and red asterisks abnormalities in epithelial folding. Scale bar = 50 μm.
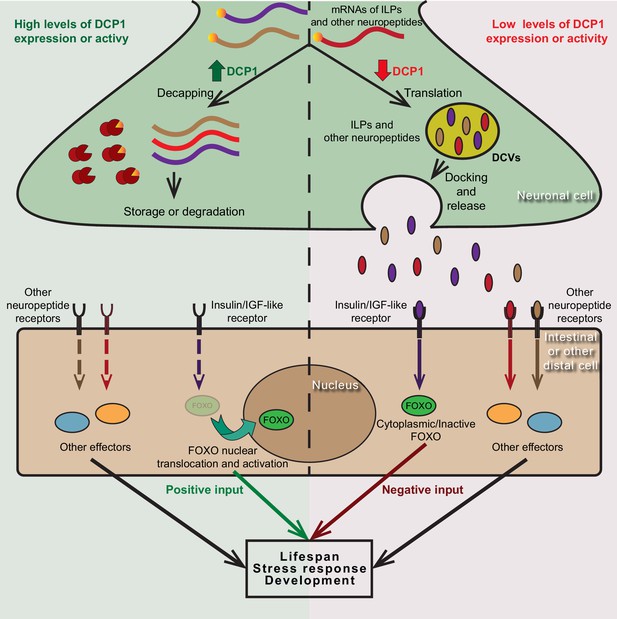
Putative model of DCP1-mediated regulation of neuroendocrine signalling that controls lifespan, stress response and development.
DCP1 activity or availability in neuronal cells controls the equilibrium between translation and storage/degradation of mRNAs encoding for insulin-like peptides (ILPs) or other neuropeptides. High levels of DCP1 promote decapping, storage and degradation, while low levels favour mRNA translation and secretion the resulting ILPs or neuropeptides through DCVs. Secreted ILPs/neuropeptides bind to Insulin/IGF-like receptor or their corresponding receptor in distal cells and control the activity of downstream effectors (like FOXO transcription factor), ultimately affecting lifespan, stress response and developmental events.
Videos
Neuronal overexpression of dcap-1 delays the onset of age-dependent movement impairment. Video of 20 days old worms.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | dcap-1 | www.wormbase.org | CELE_Y55F3AM.12 | WormBase ID: WBGene00021929 |
| Gene (Caenorhabditis elegans) | dcap-2 | www.wormbase.org | CELE_F52G2.1 | WormBase ID: WBGene00003582 |
| Gene (Caenorhabditis elegans) | daf-16 | www.wormbase.org | CELE_R13H8.1 | WormBase ID: WBGene00000912 |
| Gene (Caenorhabditis elegans) | ins-7 | www.wormbase.org | CELE_ZK1251.2 | WormBase ID: WBGene00002090 |
| Gene (Drosophila melanogaster) | DCP1 | www.flybase.org | CG11183 | FlyBase ID: FBgn0034921 |
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center (CGC) | OP50 | For standard NGM plates |
| Strain, strain background (Escherichia coli) | HT115 | Caenorhabditis Genetics Center (CGC) | HT115(DE3) | For RNAi plates |
| Strain, strain background (Caenorhabditis elegans) | C. elegans strains used in this study | This study | Supplementary file 3 | |
| Strain, strain background (Drosophila melanogaster) | D. melanogaster strains used in this study | This study | Supplementary file 4 | |
| Recombinant DNA reagent | Primers used in this study | This study | Supplementary file 5 | |
| Recombinant DNA reagent | promoterless dcap-1::gfp | Borbolis et al., 2017 doi: 10.1098/rsob.160313 | PS#302 | pBluescript KS(+);4181 bp dcap-1::gfp |
| Recombinant DNA reagent | unc-119p::dcap-1::gfp | Borbolis et al., 2017 doi: 10.1098/rsob.160313 | PS#293 | pBluescript KS(+);2200 bp unc-119p;4181 bp dcap-1::gfp |
| Recombinant DNA reagent | rab-3p::dcap-1::gfp | This study | PS#345 | pBluescript KS(+); 1200 bp rab-3p;4181 bp dcap-1::gfp |
| Recombinant DNA reagent | ges-1p::dcap-1::gfp | Borbolis et al., 2017 doi: 10.1098/rsob.160313 | PS#328 | pBluescript KS(+);1541 bp ges-1p;4181 bp dcap-1::gfp |
| Recombinant DNA reagent | hlh-1p::dcap-1::gfp | Borbolis et al., 2017 doi: 10.1098/rsob.160313 | PS#326 | pBluescript KS(+);3100 bp hlh-1p; 4181 bp dcap-1::gfp |
| Recombinant DNA reagent | col-10p::dcap-1::gfp | Borbolis et al., 2017 doi: 10.1098/rsob.160313 | PS#364 | pBluescript KS(+);2000 bp col-10p; 4181 bp dcap-1::gfp |
| Recombinant DNA reagent | ges-1p::gfp | This study | PS#176 | pPD95.77;1541 bp ges-1p |
| Recombinant DNA reagent | unc-119p::ins-7 | This study | PS#478 | pBluescript SK II; 2200 bp unc-119p; 1082 bp ins-7 |
| Recombinant DNA reagent | UAS:DCP1:eGFP | This study | PS#394 | pUASTattB; 1116 bp dcp1; 924 bp egfp |
| Recombinant DNA reagent | daf-16 RNAi | This study | PS#48 | pPD129.36(L4440);1721 bp genomic DNA |
| Recombinant DNA reagent | daf-2 RNAi | This study | PS#36 | pPD129.36(L4440);1393 bp genomic DNA |
| Recombinant DNA reagent | ins-7 RNAi | This study | PS#452 | pPD129.36(L4440);743 bp genomic DNA |
| Commercial assay or kit | QIAprep spin Miniprep Kit (50) | QIAGEN | QIA.27104 | |
| Commercial assay or kit | Qiaquick PCR purification kit | QIAGEN | QIA.28104 | |
| Commercial assay or kit | Nucleospin RNA XS | Macherey-Nagel | 740902.50 | |
| Commercial assay or kit | FIREScipt RT cDNA Synthesis kit | SOLIS BIODYNE | 06-15-00200 | |
| Commercial assay or kit | Maxima H Minus First Strand cDNA Synthesis Kit | ThermoFisher Scientific | K1682 | |
| Commercial assay or kit | KAPA SYBR FAST 2X MasterMix, Universal | Kapa Biosystems | KK4602 | |
| Commercial assay or kit | DNA oligos synthesis | IDT | ||
| Chemical compound | Tri Reagent | Sigma-Aldrich | T9424 | |
| Chemical compound | FUdR | Sigma-Aldrich | F0503 | |
| Software, algorithm | Prism 8 | Graphpad Software | N/A | www.graphpad.com/scientific-software/prism/ |
| Software, algorithm | Fiji | Image J | N/A | imagej.net/Fiji |
| Software, algorithm | Illustrator CS6 | Adobe | www.adobe.com/products/illustrator.html | |
| Software, algorithm | Photoshop CS6 | Adobe | www.adobe.com/products/photoshop.html | |
| Software, algorithm | HCI Imaging | HAMAMATSU | hcimage.com/ | |
| Software, algorithm | Leica Application Suite Advanced Fluorescence | Leica |
Additional files
-
Supplementary file 1
Neuronal overexpression of a dcap-1::gfp fusion results in its aggregation to P-body like structures.
Representative fluorescent images of wild type and dcap-2(ok3032) worms that express a dcap-1::gfp fusion under the control of the pan-neuronal unc-119 promoter. Arrowheads point to P-body like structures. Scale bar = 20 μm.
- https://cdn.elifesciences.org/articles/53757/elife-53757-supp1-v1.jpg
-
Supplementary file 2
mRNA levels of bursicon and rickets.
Relative mRNA levels of bursicon and its receptor rickets in heads dissected form newly eclosed adults subjected to neuron-specific DCP1 knockdown at 29°C.
- https://cdn.elifesciences.org/articles/53757/elife-53757-supp2-v1.jpg
-
Supplementary file 3
List of C. elegans strains used in this study.
- https://cdn.elifesciences.org/articles/53757/elife-53757-supp3-v1.xlsx
-
Supplementary file 4
List of D. melanogaster strains used in this study.
- https://cdn.elifesciences.org/articles/53757/elife-53757-supp4-v1.xlsx
-
Supplementary file 5
List of primers used in this study.
- https://cdn.elifesciences.org/articles/53757/elife-53757-supp5-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53757/elife-53757-transrepform-v1.docx





