JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes
Figures
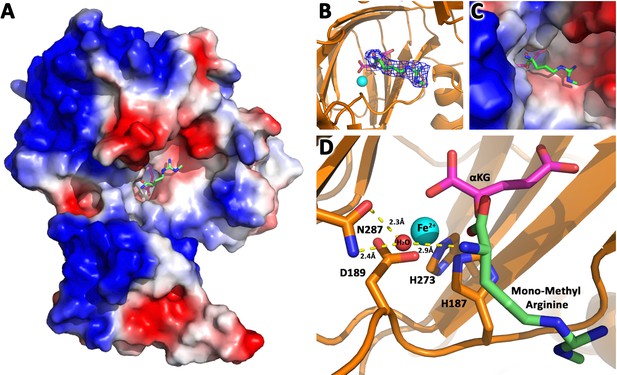
JMJD6 binds to monomethyl arginine (1-of-4).
(A) Complex structure of JMJD6 (1–343) and monomethyl arginine (MM-Arg). Surface charges were generated using PyMOL (Action > generate > vacuum electrostatics > protein contact potential; https://pymol.org/2/). Red represents negatively-charged surface, Gray represents neutral-charged surface, and Blue represents positively-charged surface. (B) Omit map 2Fo-Fc electron density of MM-Arg. (C) Magnified view of MM-Arg in the catalytic center of JMJD6 (D) Coordination of elements at catalytic center.
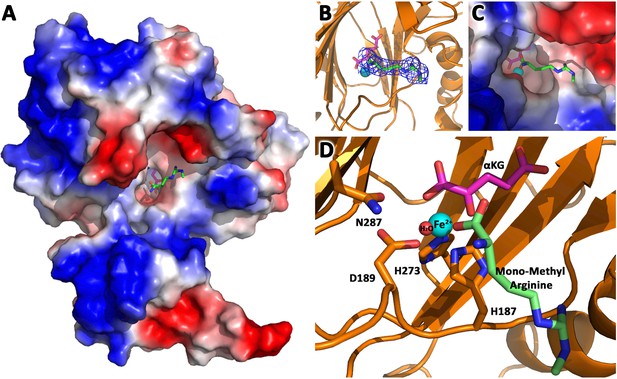
JMJD6 binds to monomethyl arginine (2-of-4).
(A) Complex structure of JMJD6 (1–343) and monomethyl arginine (MM-Arg). Surface charges were generated using PyMOL (Action > generate > vacuum electrostatics > protein contact potential; https://pymol.org/2/). Red represents negatively-charged surface, Gray represents neutral-charged surface, and Blue represents positively-charged surface. (B) Omit map 2Fo-Fc electron density of MM-Arg. (C) Magnified view of MM-Arg in the catalytic center of JMJD6 (D) Coordination of elements at catalytic center.
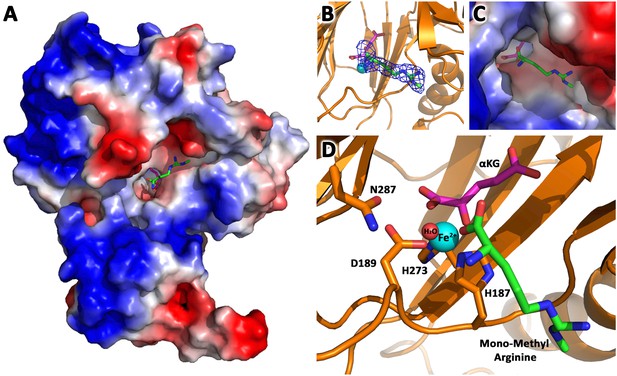
JMJD6 binds to monomethyl arginine (3-of-4).
(A) Complex structure of JMJD6 (1–343) and monomethyl arginine (MM-Arg). Surface charges were generated using PyMOL (Action > generate > vacuum electrostatics > protein contact potential; https://pymol.org/2/). Red represents negatively-charged surface, Gray represents neutral-charged surface, and Blue represents positively-charged surface. (B) Omit map 2Fo-Fc electron density of MM-Arg. (C) Magnified view of MM-Arg in the catalytic center of JMJD6 (D) Coordination of elements at catalytic center.
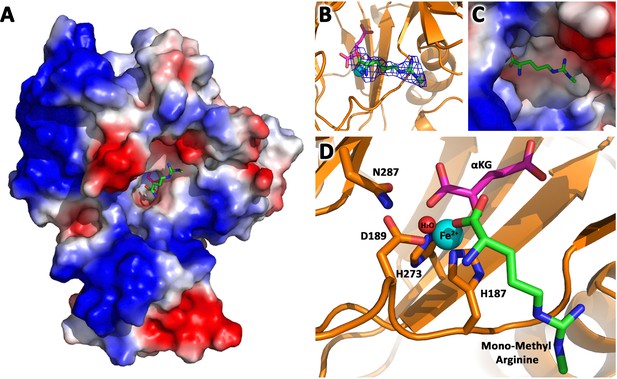
JMJD6 binds to monomethyl arginine (4-of-4).
(A) Complex structure of JMJD6 (1–343) and monomethyl arginine (MM-Arg). Surface charges were generated using PyMOL (Action > generate > vacuum electrostatics > protein contact potential; https://pymol.org/2/). Red represents negatively-charged surface, Gray represents neutral-charged surface, and Blue represents positively-charged surface. (B) Omit map 2Fo-Fc electron density of MM-Arg. (C) Magnified view of MM-Arg in the catalytic center of JMJD6 (D) Coordination of elements at catalytic center.
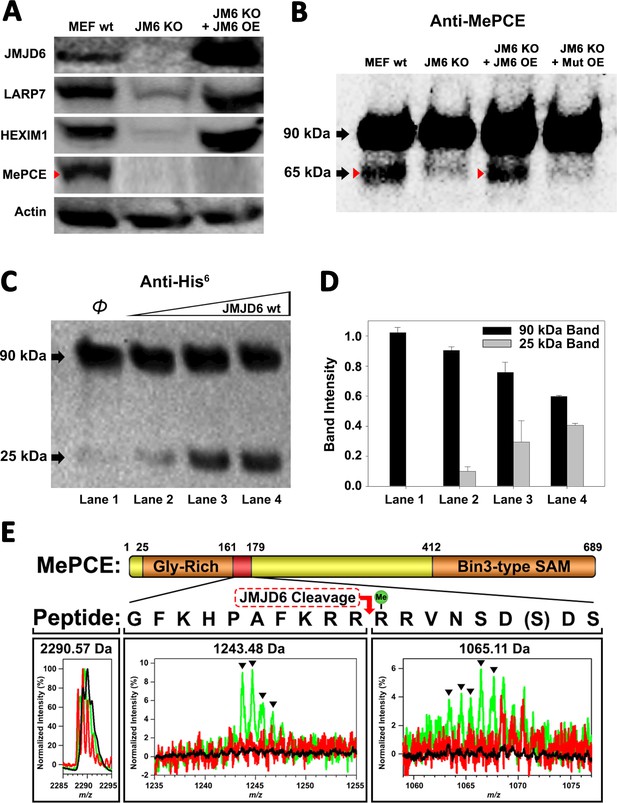
JMJD6 targets MePCE for proteolysis.
(A) Western blot of wild-type MEF, Jmjd6 knockout MEF, and JMJD6 overexpression in Jmjd6 knockout MEF probed with antibodies specific for JMJD6, Actin, and components of the 7SK snRNP complex; LARP7, HEXIM1, and MePCE. (B) Western blot of MePCE overexpressed respectively in wild-type MEF, Jmjd6 knockout MEF, wild-type JMJD6 overexpression in Jmjd6 knockout MEF, and inactive mutant JMJD6 overexpression in Jmjd6 knockout MEF; probed with antibody specific for MePCE. (C) Wild-type JMJD6 titrated into full-length MePCE with N-terminal His6-tag. Enzymatic activity of JMJD6 is probed with anti-His6 antibody. (D) Quantification of c. (E) The endopeptidase activity of JMJD6 on synthesized MePCE (161-179) R171-me2s/C177S peptide. The mass spectrum is normalized to the intensity of the undigested peptide input. The peptide is assayed with wild-type JMJD6 (green), inactive mutant JMJD6 (red), or peptide alone (black). The MePCE (161-179) peptide with symmetric dimethylation on R171 and C177S mutation has a molecular weight of 2,290.57 Da. After wild-type JMJD6 cleavage between R170 and R171, the major peaks* (black triangles) with the molecular weight of 1,243.48 Da corresponds to the N-terminal product and the molecular weight of 1,065.11 Da corresponds to the C-terminal product respectively. *The multiple peaks are isotopic distributions, which are characteristic of MALDI-TOF.
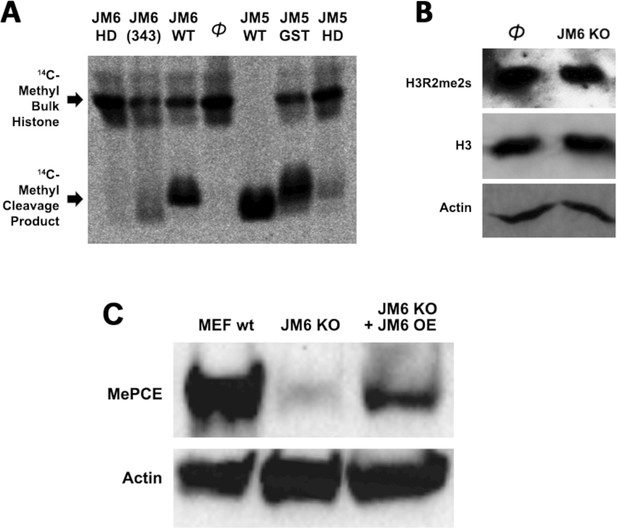
Histone tails are not the cognate substrate of JMJD6.
(A) 14C-labeled bulk histone reaction and resultant cleavage product when treated with JMJD6 and JMJD5 in vitro. (B) JMJD6 knockout has no effect in the content of both arginine methylated H3 and total H3 in MEF cells in vivo. (C) Western blot of wild-type MEF, Jmjd6 knockout MEF, and JMJD6 overexpression in Jmjd6 knockout MEF probed with antibodies specific for MePCE and Actin.
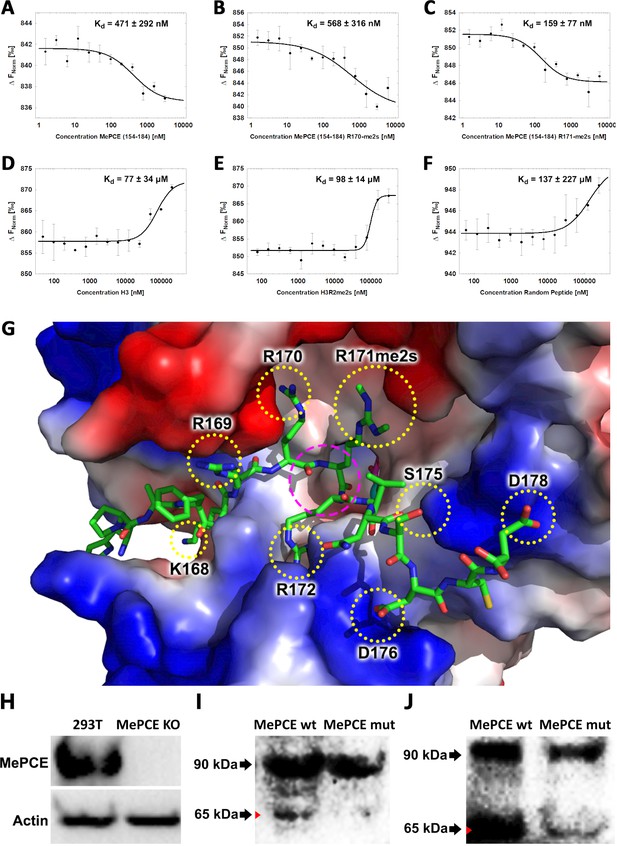
JMJD6 specifically binds to MePCE site containing residues 164–178 (determined via MST).
(A) The binding of His-JMJD6 (1–343) to unmodified MePCE (154-184). (B) The binding of His-JMJD6 (1–343) to MePCE (154-184) R170-me2s. (C) The binding of His-JMJD6 (1–343) to MePCE (154-184) R171-me2s. (D) The binding of His-JMJD6 (1–343) to unmodified Histone 3 (1-21). (E) The binding of His-JMJD6 (1–343) to Histone 3 (1-16) R2-me2s. (F) The binding of His-JMJD6 (1–343) to C-peptide (57-87). (G) Electrostatic interactions between JMJD6 (1–343) and MePCE (164-178) determined by YASARA Energy Minimization server are highlighted in yellow dotted circles and catalytic center is highlighted in magenta dashed circle. (H) Western blot of wild-type 293 T cells (left lane) and MePCE knockout 293 T cells (right lane) probed with antibody specific for MePCE. (I) Western blot of MePCE knockout 293 T cells overexpressing C-terminal His6-tagged wild-type MePCE (left lane) or C-terminal His6-tagged mutant MePCE (right lane), respectively, probed with antibody specific for His6-tag. (J) Western blot of wild-type JMJD6 overexpression in Jmjd6 knockout MEF cells overexpressing C-terminal His6-tagged wild-type MePCE (left lane) or C-terminal His6-tagged mutant MePCE (right lane), respectively, probed with antibody specific for His6-tag.
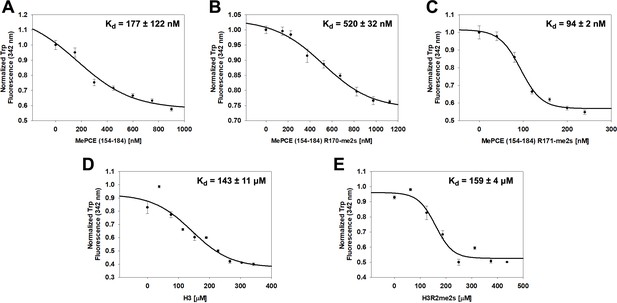
JMJD6 specifically binds to MePCE site containing residues 164–178 (determined via fluorescence polarization).
(A) The binding of His-JMJD6 (1–343) to unmodified MePCE (154-184). (B) The binding of His-JMJD6 (1–343) to MePCE (154-184) R170-me2s. (C) The binding of His-JMJD6 (1–343) to MePCE (154-184) R171-me2s. (D) The binding of His-JMJD6 (1–343) to unmodified Histone 3 (1-21). (E) The binding of His-JMJD6 (1–343) to Histone 3 (1-16) R2-me2s.
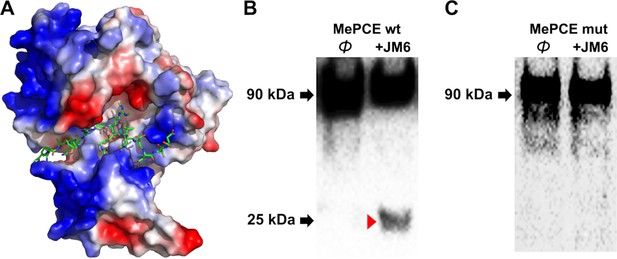
Computational complex structure model of JMJD6 (1–343) and MePCE (164-178) derived from YASARA Energy Minimization server and verification.
JMJD6 activity against wild-type MePCE or mutant MePCE in vitro. (A) Minor spatial adjustments were made on R171me2s of computational model to better overlap with experimental model. (B) Activity of JMJD6 against wild-type MePCE in vitro. (C) Activity of JMJD6 against mutant MePCE with –DNDN- replacing –RRRR- at the cleavage site.
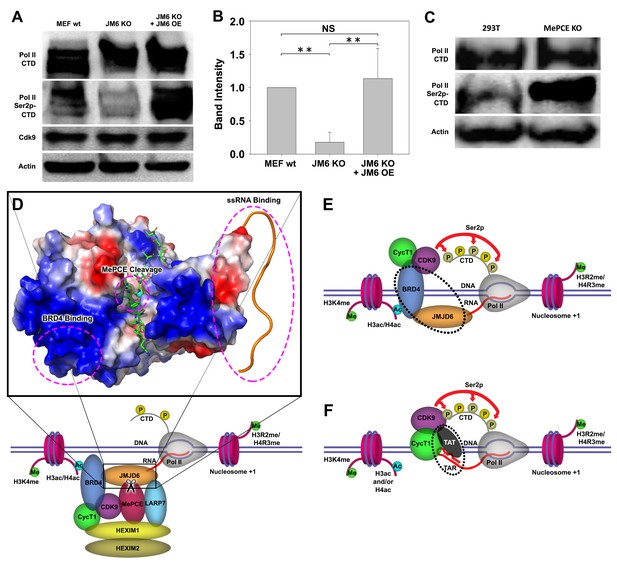
JMJD6 regulates Pol II Ser2-CTD phosphorylation.
(A) Western blot of wild-type MEF, Jmjd6 knockout MEF, and JMJD6 overexpression in Jmjd6 knockout MEF probed with antibodies specific for Pol II CTD, Pol II ser2p-CTD, CDK9, and Actin. (B) Quantification of Pol II Ser2p-CTD on a. (**, p<0.01; NS, not significant) (C) Western blot of wild-type 293 T cells and MePCE knockout 293 T cells probed with antibodies specific for for Pol II CTD, Pol II ser2p-CTD, and Actin. (D) Model of JMJD6 cleavage of MePCE within the 7SK snRNP complex. A representative surface charge model of JMJD6 with unstructured C-terminal tail highlights respective JMJD6 interaction sites with MePCE, BRD4, and ssRNA in magenta dashed circles. (E) ssRNA-bound JMJD6 and acetylated H3/H4-bound BRD4 in conjunction (black dotted circle) bridges P-TEFb to paused Pol II. (F) TAR-bound TAT (black dotted circle) bridges P-TEFb to paused Pol II.
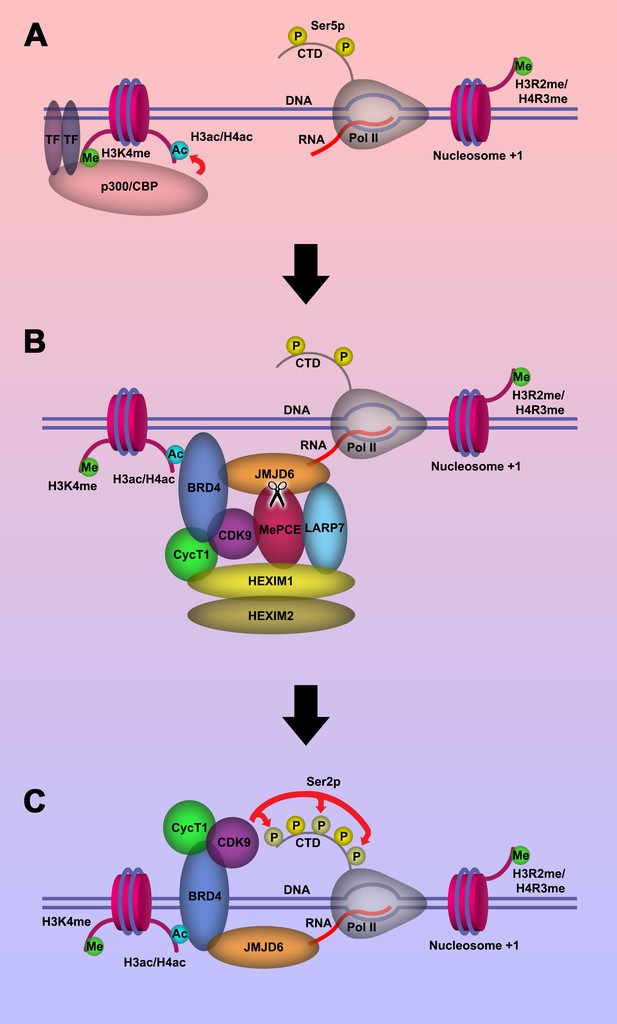
Model of P-TEFb release from 7SK snRNP complex.
(a.) Pol II is initiated at the TSS and remains in the paused state until further stimulation. Transcription factors and H3K4me recruits p300/CBP, which acetylates and generates H3ac and/or H4ac. (b) H3ac and/or H4ac recruits BRD4, which associates with 7SK snRNP/P-TEFb complex and JMJD6. JMJD6 associates with ssRNA from Pol II. JMJD6 digests MePCE to disrupt the 7SK snRNP complex to release P-TEFb (CDK9). (c) BRD4/JMJD6 brings CDK9 in close proximity of CTD of Pol II. CDK9 phosphorylates Ser2 motifs on CTD of Pol II.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (M. musculus) | JMJD6 KO cell on MEF background | Böse et al., 2004 | Cell line from Dr. Andreas Lengeling lab | |
| Cell line (M. musculus) | JMJD6 Re-expression cell line on the JMJD6 KO background | This paper | MSCV-JMJD6 transduced JMJD6 KO cell | |
| Cell line (Homo-sapiens) | 27–13 MEPCE KO cell on the HEK293 background | This paper | Cell line from Drs. Yuhua Xue lab | |
| Cell line (Homo-sapiens) | MEPCE Re-expression cell line on the MEPCE KO cell line 27–13 background | This paper | MSCV-MEPCE with c-terminal 6*His tag transduced MEPCE KO cell | |
| Antibody | Rabbit polyclonal Phospho-Serine 2 CTD Pol II | Nat Struct Mol Biol 15, 71–78 (2008) | Dr. David Bentley Lab | WB (1:2000) |
| Antibody | Rabbit polyclonal CTD Pol II antibody | Nat Struct Mol Biol 15, 71–78 (2008) | Dr. David Bentley Lab | WB (1:2000) |
| Antibody | Mouse monoclonal JMJD6 antibody | Santa Cruz Biotechnology | sc-28348 | WB (1 µg/ml) |
| Antibody | Rabbit polyclonal MEPCE antibody | Bethyl Laboratories Inc | A304-184A | WB (1:2000) |
| Antibody | Rabbit polyclonal MEPCE antibody | Novus | NBP2-34858 | WB (1:2000) |
| Antibody | Rabbit polyclonal LARP7 antibody | Abcam | ab-134757 | WB (1:2000) |
| Antibody | Mouse monoclonal HEXIM1 antibody | Santa Cruz Biotechnology | sc-390059 | WB (1 µg/ml) |
| Antibody | Mouse monoclonal actin antibody | Santa Cruz Biotechnology | sc-8432 | WB (1 µg/ml) |
| Antibody | Mouse monoclonal CDK9 antibody | Santa Cruz Biotechnology | sc-13130 | WB (1 µg/ml) |
| Antibody | Mouse monoclonal His-probe antibody | Santa Cruz Biotechnology | sc-53073 | WB (1 µg/ml) |
Additional files
-
Supplementary file 1
Crystallographic statistics of complex structure of JMJD6 and a methylated arginine.
- https://cdn.elifesciences.org/articles/53930/elife-53930-supp1-v2.txt
-
Supplementary file 2
Protein Composition Analysis via Mass Spectrometry.
Bacteria expressed and purified JMJD6 are subjected to mass spectrum analysis, all potential contaminated trace protein candidates are listed. No known protease candidate is identified from the list.
- https://cdn.elifesciences.org/articles/53930/elife-53930-supp2-v2.xlsx
-
Supplementary file 3
Total RNA-seq reads of wild-type MEF, JMJD6 knockout MEF, and JMJD6 overexpressed in JMJD6 KO background MEF.
- https://cdn.elifesciences.org/articles/53930/elife-53930-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/53930/elife-53930-transrepform-v2.pdf





