Characterising a healthy adult with a rare HAO1 knockout to support a therapeutic strategy for primary hyperoxaluria
Figures
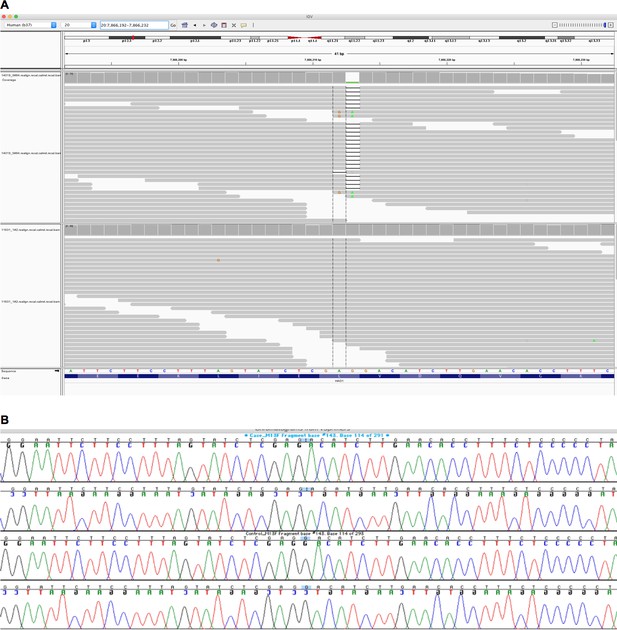
Integrative Genomics Viewer (IGV) image of sequencing alignments around the ENST00000378789.3:c.997delC HAO1 variant (rs1186715161, GRCh37:20;7866212 AG/A).
(A) Exome sequencing performed using Agilent in solution capture and Illumina short read sequencing using blood DNA. Top half of image, individual with homozygous ENST00000378789.3:c.997delC genotype. Bottom half of image, a different individual with homozygous reference sequence genotype. Neither sequencing assay (see also Figure 1B) in the different tissues suggested mosaicism however additional tissues (e.g. skin) for more definitive exclusion could not be obtained. (B) Sanger sequencing around the ENST00000378789.3:c.997delC HAO1 (rs1186715161) variant using saliva DNA taken at a different timepoint to the blood DNA in Figure 1A.
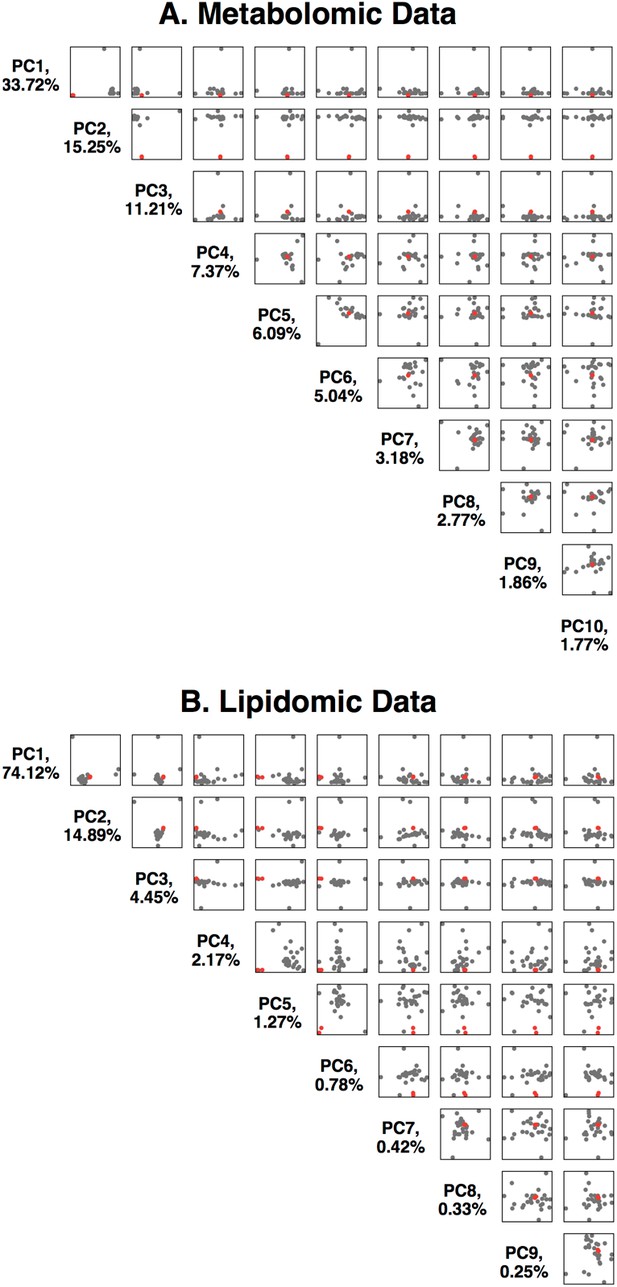
Principal component analysis of the Metabolon metabolomics data.
(A) Data is available in Supplementary file 1. The first 10 principal components are plotted against each other, with %variance explained. The two replicates of the HAO1 knockout volunteer are shown as red dots, control individuals as black dots. (B) Principal component analysis of the Metabolon lipidomics data. Data is available in Supplementary file 2.
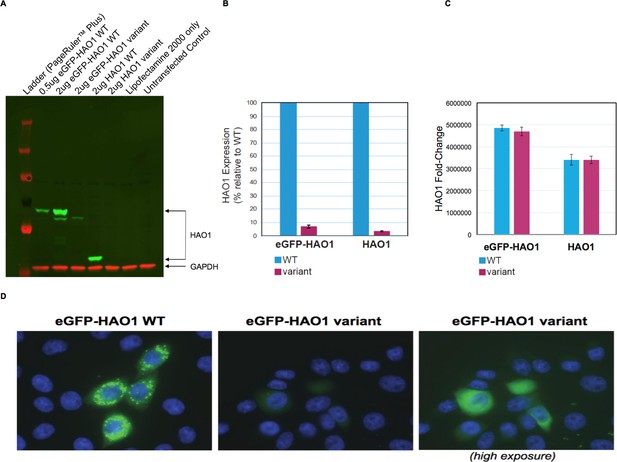
The HAO1 variant p.Leu333SerfsTer4 is expressed at significantly lower levels and mislocalised in cells.
(A) Western blot showing glycolate oxidase (GO, encoded by the HAO1 gene) protein for both tagged and untagged reference sequence HAO1 (wild type, WT) and p.Leu333SerfsTer4. (B) Protein expression quantification of both tagged and untagged wild type and p.Leu333SerfsTer4. (C) mRNA expression quantification of both tagged and untagged wild type and p.Leu333SerfsTer4. (D) Immunofluorescence shows lower expression levels and diffuse signal for p.Leu333SerfsTer4 relative to wild type. Cells imaged at 60X magnification. The right panel was adjusted to be 2.3-fold brighter than the left or middle panel in ImageJ software in order show green fluorescence.
Tables
Blood and Urine biochemical measurements in a healthy woman with a p.Leu333SerfsTer4 HAO1 knockout.
| Sample and assay | Level | Reference Range* |
|---|---|---|
| Venous blood | ||
| Plasma oxalate | <1.0 mcmol/L | <1.6 |
| Plasma glycolate | 171 nmol/mL | ≤14 |
| Plasma glycerate | <1 nmol/mL | ≤28 |
| Urine (single void, same day as blood tests) | ||
| Urine oxalate | 0.16 mmol/L | Not applicable |
| Urine oxalate/creatinine ratio | 20 mg/g | ≤75 |
| Urine glycolate/creatinine ratio | 199 mg/g | ≤50 |
| 24 hr urine (on a different day) | ||
| Urine oxalate (total/24 hr) | 0.27 mmol/24 hr | <0.46 |
| Urine oxalate/creatinine ratio | 23 mg/g | ≤75 |
| Urine glycolate/creatinine ratio | 309 mg/g | ≤50 |
-
*Reference ranges (mean+2sd) derived from studies of 67 healthy adults at the Mayo Clinic.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Chinese Hamster Ovary) | CHO-K1 | ATCC CCL-61 | RRID:CVCL_0214 | |
| Commercial assay or kit | Taqman Assay for RT-qPCR HAO1 FAM | Thermofisher | Assay Id Hs00213909_m1 | |
| Commercial assay or kit | Taqman Assay for RT-qPCR GAPDH VIC | Thermofisher | Assay Id Cg04424038_gH |
Additional files
-
Supplementary file 1
Metabolon HD4 metabolomic data.
- https://cdn.elifesciences.org/articles/54363/elife-54363-supp1-v2.xlsx
-
Supplementary file 2
Metabolon CLP lipidomic data.
- https://cdn.elifesciences.org/articles/54363/elife-54363-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54363/elife-54363-transrepform-v2.docx





