A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate
Figures
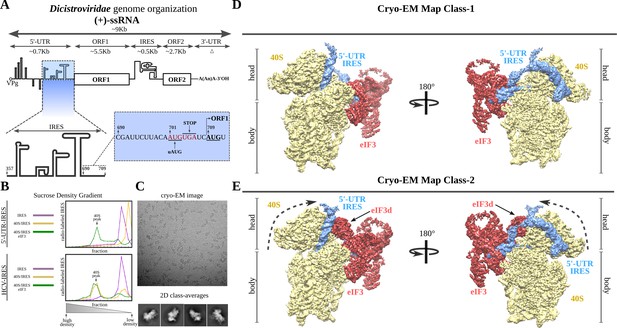
Dicistroviridae genome organization, in vitro complex formation and cryo-EM maps.
(A) Top, schematic representation of the genome organization of Dicistroviruses. The approximate genomic lengths of the different components are indicated by the arrows. Bottom, detailed view of the region described as harboring the IRES activity of the 5'-UTR of the CrPV. On the right, the sequence adjacent to the initiation AUG codon of ORF1, located at nucleotide 709 and preceded by a ‘start-stop uORF’ indicated in red. (B) Sucrose-gradient analysis showed that the 5'-UTR-IRES is dependent on eIF3 in order to form a stable complex with the 40S. 5'-UTR-IRES co-migrates with the 40S only in the presence of eIF3 (top). By contrast, HCV IRES does not require eIF3 for 40S binding (bottom). (C) Representative cryo-EM micrograph of the 40S–5'-UTR-IRES–eIF3 complex. Bottom, representative reference-free 2D class averages used for further image processing. (D, E) After 3D classifications, two classes showing density for 40S (yellow), eIF3 (red) and 5'-UTR-IRES (blue) could be identified in the dataset. Class-1 (D) presents a non-swiveled configuration of the 40S head and the density for eIF3d is absent. Class-2 (E) shows a swiveled configuration of the 40S head (arrows) with eIF3d (indicated) contacting eIF3's core subunits.
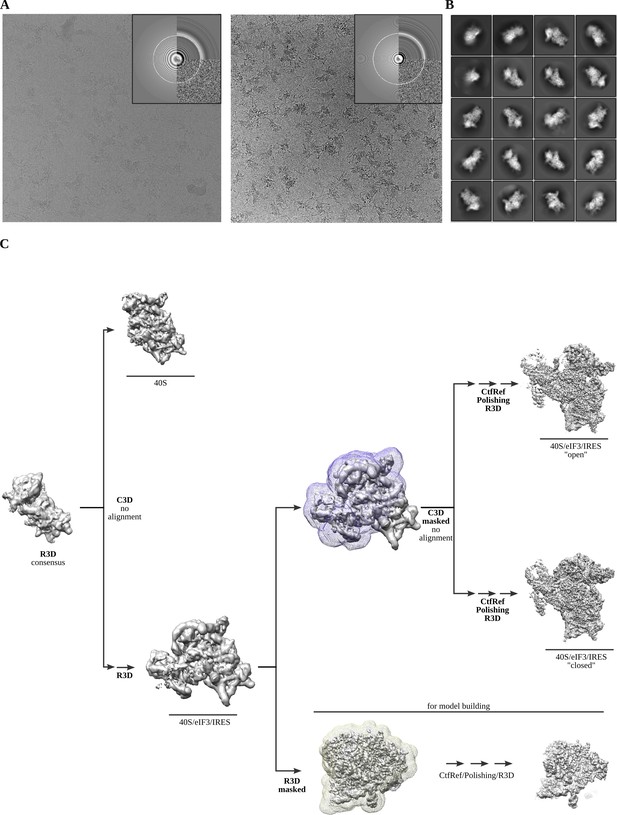
Cryo-EM representative images and classification workflow.
(A) Two examples of aligned micrographs used for image processing. Data collection in thick ice was instrumental to avoid complex disassembly and preferential orientation. (B) Representative reference-free 2D averages. (C) The classification scheme followed to identify the two described complex classes.
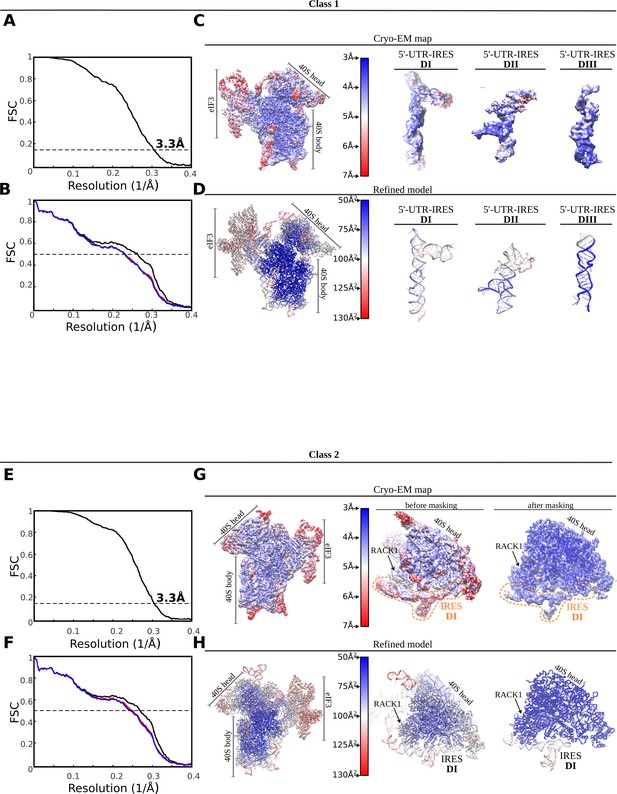
FSC correlation curves, local resolution and model validation.
Top, class-1. (A) Gold-standard Fourier Shell Correlation (FSC) curves between half-maps independently refined in Relion3. Global resolution by the 0.143 cutoff criterium was estimated to be 3.3 Å. (B) FSC between the final refined model and the final unsharpened map (black curve). The absence of model overfitting is demonstrated by the overlapping of FSC curves between half-map 1 (included in the refinement, blue) and the model and half-map 2 (not included in the refinement, red). (C) Unsharpened map colored according to local resolution as computed by ResMap. On the right, detailed views for the three 5'-UTR-IRES domains. (D) Refined model colored according to estimated B-factors computed by Refmac. Bottom, class-2. (E) Gold-standard FSC curves between half-maps independently refined in Relion3. Global resolution by the 0.143 cutoff criterium was estimated to be 3.3 Å. (F) FSC between the final refined model and the final unsharpened map (black curve). The absence of model overfitting is demonstrated by the overlapping of FSC curves between half-map 1 (included in the refinement, blue) and the model and half-map 2 (not included in the refinement, red). (G) Unsharpened map colored according to local resolution as computed by ResMap. On the right, detailed views for the 40S head before and after masked refinements. (H) Refined model colored according to estimated B-factors computed by Refmac.
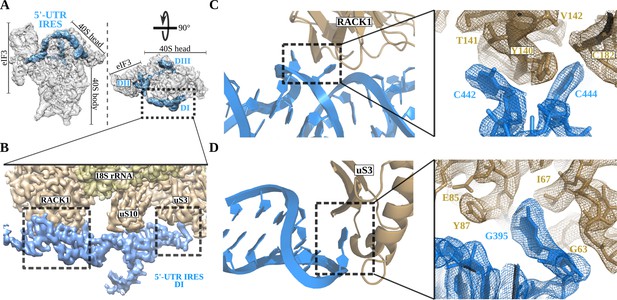
The 5'-UTR-IRES domain I engages ribosomal proteins RACK1 and uS3.
(A) Overview of the 40S–5'-UTR-IRES–eIF3 map, with 40S and eIF3 depicted gray and 5'-UTR-IRES in blue. (B) Detailed view of the cryo-EM density of the 40S–5'-UTR-IRES–eIF3 map centered around 5'-UTR-IRES domain I (DI). Ribosomal proteins are colored brown, 18S rRNA yellow and 5'-UTR-IRES blue. Contacts between 5'-UTR-IRES domain I and ribosomal proteins RACK1 (C) and uS3 (D) could be defined thanks to well-resolved local cryo-EM densities.
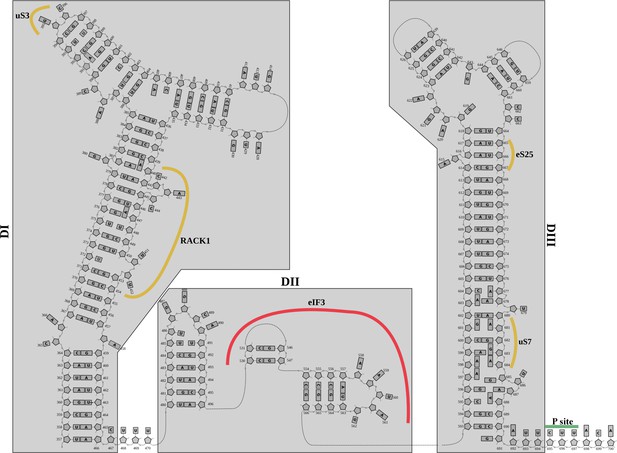
Structurally derived secondary structure diagram for the 5'-UTR-IRES.
Secondary structure diagram for the 5'-UTR-CrPV IRES derived from the cryo-EM structure.
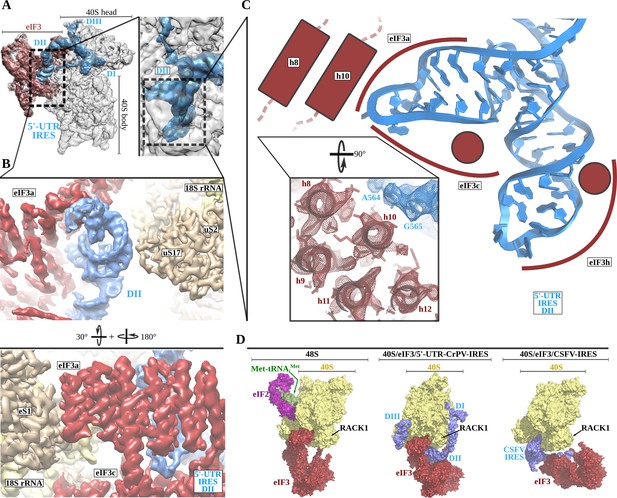
The 5'-UTR-IRES domain II is formed by a dual hairpin that mediates eIF3 recruitment.
(A) Overview of the 40S–5'-UTR-IRES–eIF3 cryo-EM map with 40S colored gray, eIF3 red and 5'-UTR-IRES blue. On the right, a zoomed view centers around 5'-UTR-IRES domain II. (B) Detailed view of the cryo-EM map for the region occupied by 5'-UTR-IRES domain II, with 40S components colored gold, eIF3 red and 5'-UTR-IRES blue. Domain II is sandwiched between ribosomal protein uS17 (located at the back of the 40S body) and eIF3 core subunits a and c. (C) 5'-UTR-IRES domain II is formed by a dual hairpin that establishes interactions with α-helices 8 and 10 from eIF3a. These contacts are mediated mainly by basic residues of eIF3 and the phosphate backbone of the IRES. (D) superposition of the 40S–5'-UTR-IRES–eIF3 complex with the canonical 48S complex (left, PDB ID 6FEC) and with the CSFV-IRES–40S complex (right, PDB ID 4c4q). The 5'-UTR-IRES binds to the 40S with a conformation that is compatible with the canonical position described for eIF3 in the 48S complex.
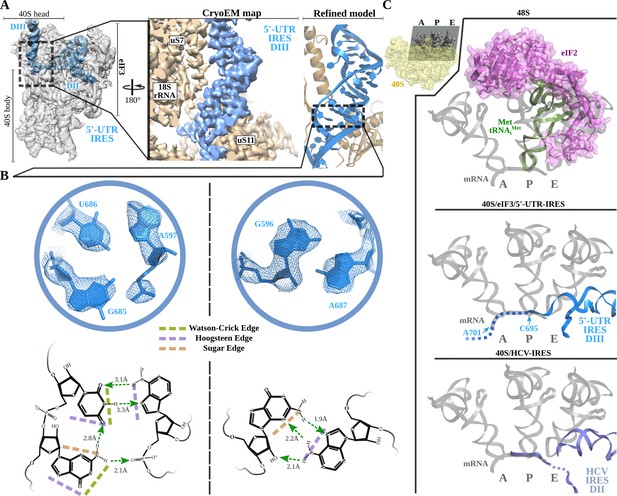
Non-canonical base pairing in 5'-UTR-IRES domain III assists on P-site access.
A) Overview of the 40S–5'-UTR-IRES–eIF3 cryo-EM map with 40S and eIF3 colored gray and 5'-UTR-IRES blue. On the right, a detailed view of the E-site, where 5'-UTR-IRES domain III is placed, shows the cryo-EM map with the 5'-UTR-IRES colored blue and 40S components brown. On the far right, the final refined model is colored following the same color scheme. Ribosomal proteins uS7 and uS11 as well as several 18S rRNA bases contact 5'-UTR-IRES domain III. (B) Non-canonical base pairs found in 5'-UTR-IRES domain III induce a distortion of the double helix near the E-site. At the top, two examples are shown with the refined model inserted in the experimental cryo-EM density. The corresponding chemical diagrams below show the base edges and the hydrogen bonds involved in interactions. (C) Superpositions of the canonical 48S complex (PDB ID 6FEC, top) with the HCV-IRES/40S complex (PDB ID 5A2Q, bottom) and the 40S/5'-UTR-IRES/eIF3 (middle) models, focused on the tRNA A, P and E binding sites. 5'-UTR-IRES domain III and HCV-IRES occupies a space on the E-site that overlaps with the position described for eIF2 in the canonical 48S complex. In the middle, the last residue of 5'-UTR-IRES is indicated (C695), as is the putative path along the mRNA binding channel followed by the IRES (dashed line).
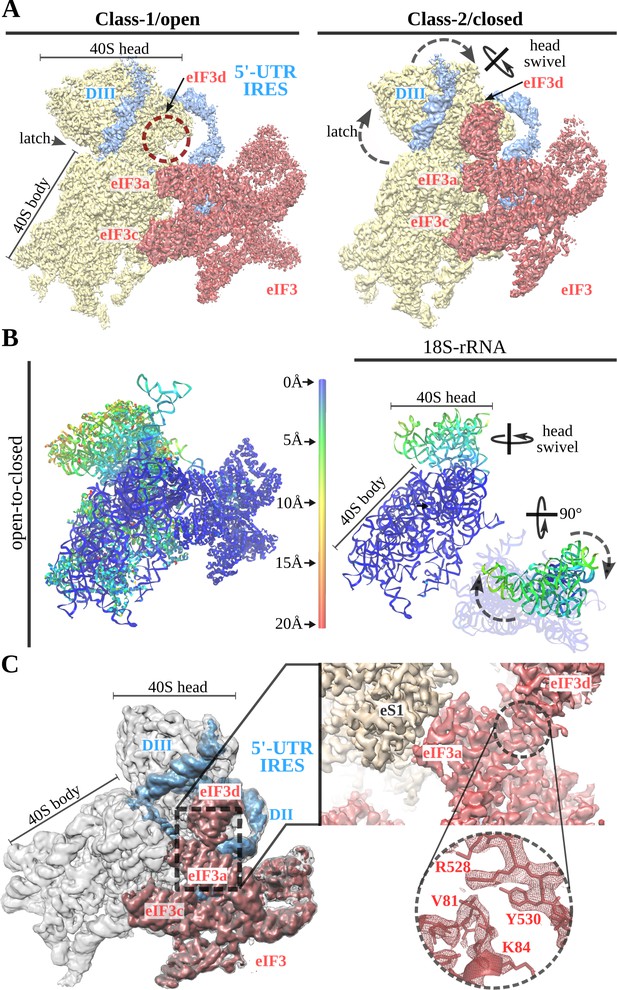
A 40S head swiveling movement ‘locks’ 5'-UTR-IRES on the 40S, inducing a compact eIF3 configuration.
(A) Cryo-EM maps obtained for the two classes present in the 40S–5'-UTR-IRES–eIF3 dataset with 40S colored gold, eIF3 red and 5'-UTR-IRES blue. The positions of the latch, eIF3d and the swiveling rotation axis are indicated. A 40S head swiveling movement in class-2 brings eIF3d closer to the core subunit of eIF3 (i.e. eIF3a), establishing interactions that stabilize its conformation. (B) Left, ribbon diagram colored according to pairwise root mean square deviation (r.m.s.d.) displacements for the open-to-closed transition, with displacements scale at the center. On the right, a simplified diagram shows only the 18S rRNA colored according to the used on the left. Two orthogonal views are shown, in which it can be appreciated that the main displacement is localized at the 40S head. (C) Overview of the closed class with 40S colored gray, eIF3 red and 5'-UTR-IRES blue. Inset, detail of the experimental density obtained for the eIF3a–eIF3d interface for this class. Clear information on side chains was present in the maps, allowing proper model building and refinement.
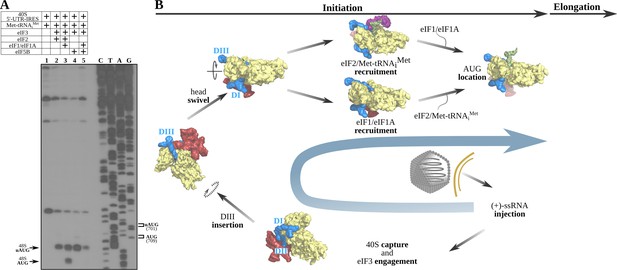
The 5'-UTR-IRES requires TC, eIF1 or eIF1A to assemble a functional initiation complex via an uAUG intermediate.
(A) Toe-print analysis of 48S initiation complexes assembled on 5'-UTR-IRES in an in vitro reconstituted system. eIF2 delivers Met-tRNAiMet to the uAUG (lane 2) and requires the presence of eIF1 or eIF1A to transition to the bona fide AUG codon of ORF1 (lane 3). Under some conditions, eIF5B can substitute for eIF2 in Met-tRNAiMet delivery (Terenin et al., 2008). In the absence of eIF1 or eIF1A, a robust toe-print signal is detected in the presence of eIF5B (lane 4), however, Met-tRNAiMet is delivered to the uAUG, and thus eIF5B is unable to find the annotated AUG even in the presence of eIF1 or eIF1A (lane 5). (B) A model for 5'-UTR-IRES-mediated translation initiation. From the bottom right: injection of the genomic (+)-ssRNA of the CrPV into the cytosol allows the 5'-UTR-IRES to capture free 40S and eIF3, which is recruited to the complex with an initial canonical conformation of the 40S head without tilt or swivel. Insertion of 5'-UTR-IRES DIII in the vicinity of the E-site and a swiveling movement of the 40S head induces a ‘locked’ conformation of the complex, with the uAUG at 701 in the vicinity of the P-site. Delivery of Met-tRNAiMet and location of ORF1 AUG is achieved by the concerted action of eIF2, eIF1 and eIF1A. Large subunit recruitment mediated by eIF5B will allow transitioning into elongation.
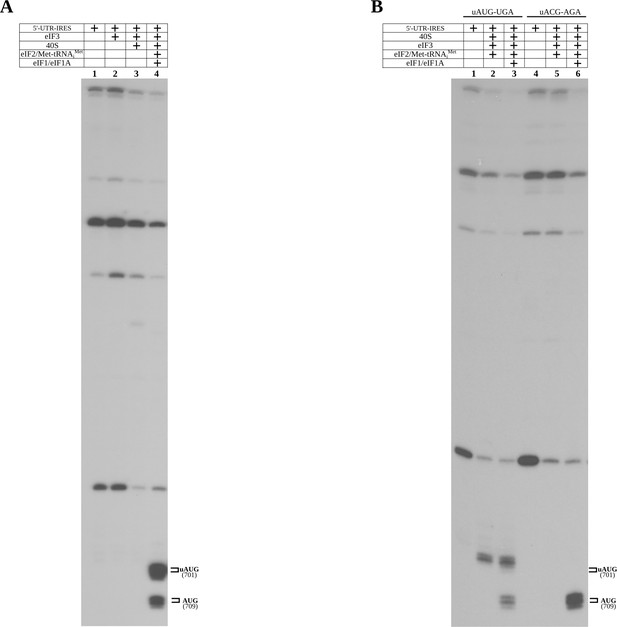
Control toe-print experiments.
(A) Toe-print analysis for 5'-UTR-IRES, 5'-UTR-IRES–eIF3 and 40S/5'-UTR-IRES–eIF3 complexes (lanes 1, 2 and 3, respectively). No toe-print signal was detected that is ascribable to uAUG or annotated AUG in any of these reactions. Robust toe-print signal could be detected 15–17 nucleotides downstream of uAUG and AUG only for 40S–5'-UTR-IRES–eIF3, and in the presence of TC (eIF2–Met-tRNAiMet–GTP) and eIF1 or eIF1A. (B) The uAUG toe-print signal can be abolished by mutation of the uAUG to ACG, redirecting the Met-tRNAiMet loading event exclusively to the annotated AUG in the presence of TC and eIF1 or eIF1A.





