A non-canonical role for the EDC4 decapping factor in regulating MARF1-mediated mRNA decay
Figures
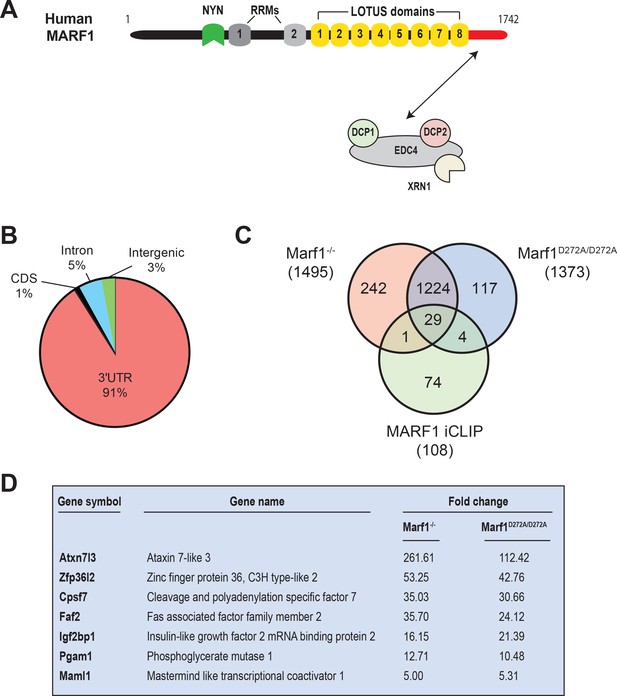
Identification of human MARF1 target mRNAs.
(A) Schematic diagram of full-length MARF1. (B) Distribution of crosslinked sequence reads. (C) Venn diagram illustrating the relationship of MARF1 target mRNAs identified by iCLIP in HEK293 cells with transcripts that were upregulated in Marf1-/- and Marf1D272A/D272A and germinal vesicle-stage mouse oocytes as compared to wild-type. (D) A partial list of mRNAs identified by iCLIP that were upregulated in both Marf1-/- and Marf1D272A/D272A and germinal vesicle-stage mouse oocytes as compared to wild-type.
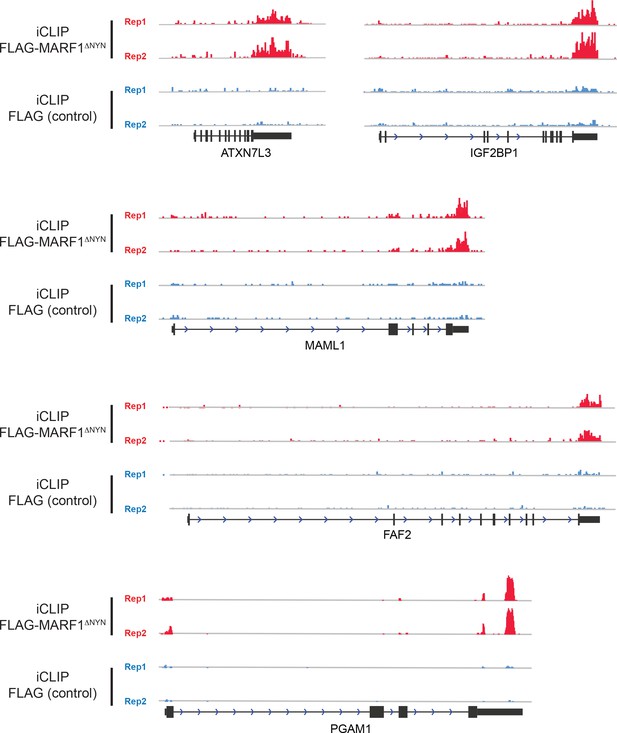
Representative FLAG-MARF1ΔNYN iCLIP coverage at the loci of ATXN7L3, IGFP2BP1, MAML1, FAF2 and PGAM2 loci shows specificity for the 3′UTR.
The negative control iCLIP from HEK293 cells not expressing FLAG-MARF1ΔNYN shows negligible background.
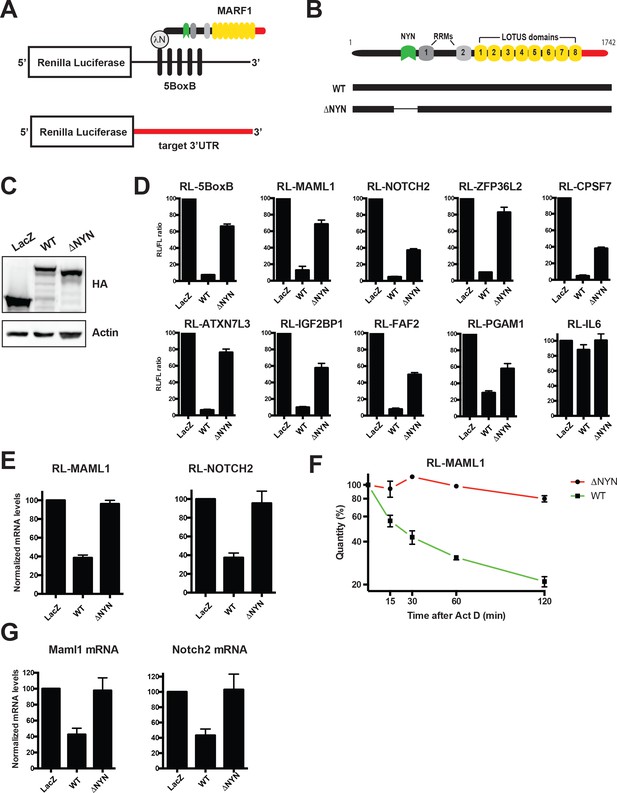
MARF1 represses target mRNAs via their 3’UTRs.
(A) Schematic diagram of the basic Renilla luciferase (RL)-encoding mRNA reporter, containing five 19-nt BoxB hairpins, interacting with λN-HA-MARF1, as well as a RL reporter with a 3’UTR from endogenous mRNAs. (B) Schematic diagram of full-length MARF1 and MARF1 fragments used in tethering assays. (C) Western blot analysis of HEK293 cells expressing indicated proteins. (D) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were cotransfected with constructs expressing the RL-5BoxB reporter, FL, and indicated fusion proteins. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-LacZ were set as 100. (E) RL-MAML1 (left panel) and RL-NOTCH2 (right panel) mRNA levels detected in extracts from HEK293 cells expressing the indicated proteins. Histograms represented mean values of RL-MAML1 or RL-NOTCH2 mRNAs normalized to FL mRNA from a minimum of three experiments. mRNA levels values seen in the presence of λNHA-LacZ were set as 100. (F) The stability of RL-MAML1 was assessed by using actinomycin D (5 μg/ml) for the indicated amount of time. Total RNA was isolated, reverse transcribed and RL-MAML1 RNA was quantified by qPCR. RL-MAML1 mRNA decay rates were normalized to FL mRNA levels with the zero time point set at 100. (G) Endogenous MAML1 and NOTCH2 mRNA levels detected in extracts from HEK293 cells expressing the indicated proteins. mRNA levels were normalized to GAPDH mRNA levels for a minimum of three experiments. mRNA levels in the presence of λNHA-LacZ were set as 100. Error bars represent the SEM of multiple independent experiments.
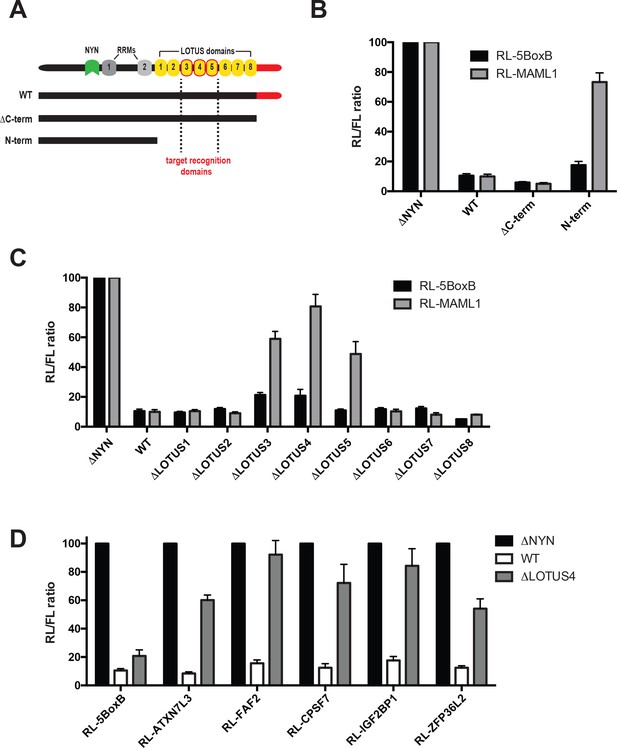
Central MARF1 LOTUS domains are required to silence mRNAs containing 3’UTRs of MARF1 target mRNAs.
(A) Schematic diagram of wild-type MARF1 protein, as well C-terminal deletion mutants. Core LOTUS domains required for MARF1-mediated repression are bordered in red. (B–D) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were cotransfected with constructs expressing the RL-MAML1 reporter or RL-5BoxB reporter, along with FL, and indicated fusion proteins. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN were set as 100.
-
Figure 3—source data 1
Comparative sequence analysis of MARF1 LOTUS domains.
(A) Sequence alignment of conserved amino acids within the MARF1 LOTUS domains 1 through 6 of human (Hs), Drosophila melanogaster (Dm), Xenopus tropicalis (Xt) and Danio rerio (Dr). LOTUS domain amino acids are in bold font. (B) Sequence alignment of human LOTUS domains 3 and 5. Identical amino acids are denoted with an asterisk and conservative amino acids substitutions are denoted with a colon.
- https://cdn.elifesciences.org/articles/54995/elife-54995-fig3-data1-v1.pdf
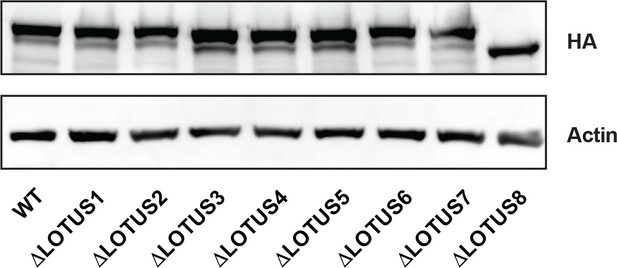
Western blot analysis of HEK293 cells expressing λNHA-MARF1 mutants lacking the indicated LOTUS domains.
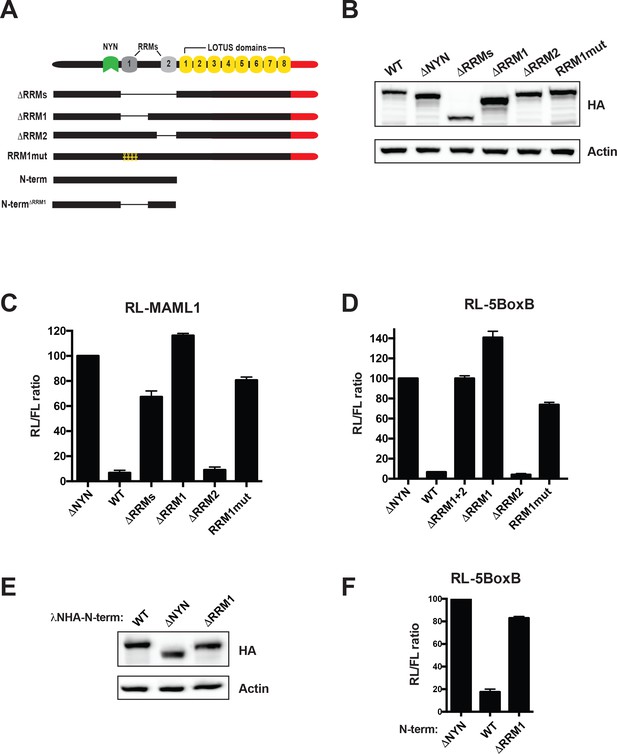
MARF1 RRM1 is required to silence target mRNAs.
(A) Schematic diagram of wild-type MARF1 protein, as well as RRM mutants. (B and E) Western blot analysis HEK293 cells transfected with plasmids expressing full-length (B) or N-terminal fragments (E). (C, D and F) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were cotransfected with constructs expressing the RL-MAML1 reporter (C) or the RL-5BoxB reporter (D and F), FL, and indicated fusion proteins. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN were set as 100. (E).
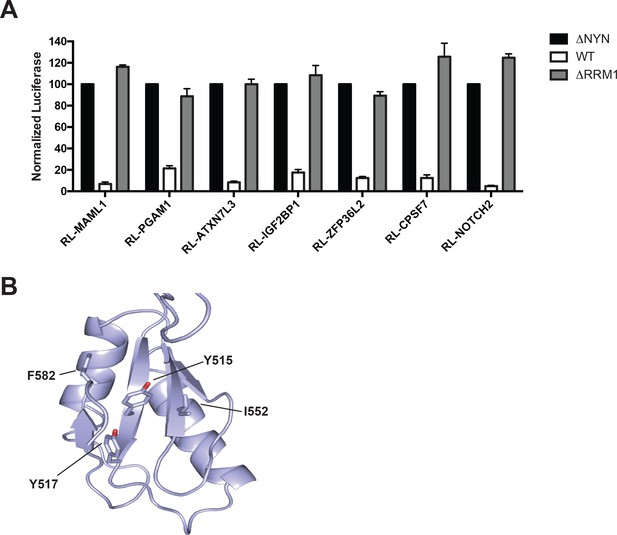
MARF1 RRM1 is required to silence mRNAs containing 3’UTRs of MARF1 target mRNAs.
(A) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were cotransfected with constructs expressing the RL reporter mRNAs containing the 3’UTRs of MARF1 target mRNAs along with FL, and indicated fusion proteins. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN were set as 100. (B) Structural model of MARF1 RRM1. The model was generated using the Phyre2 structure prediction server (Kelley et al., 2015) and is shown in ribbon format. Conserved surface residues on the putative RNA binding surface are shown in stick format. These residues were substituted with alanines in the RRM1mut protein.
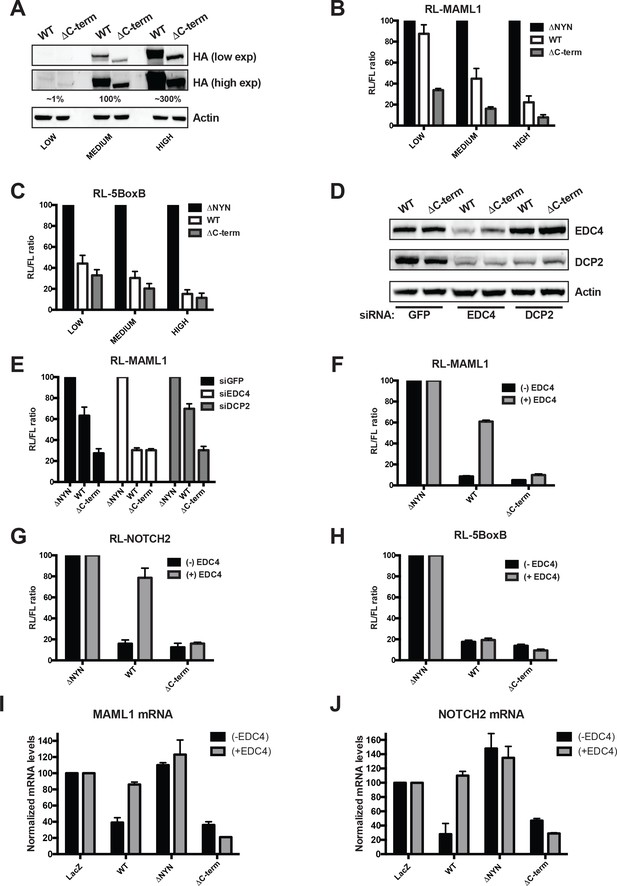
EDC4 impairs MARF1 silencing of endogenous target mRNAs.
(A) Western blot analysis HEK293 cells transfected with different amounts [low (0.1 μg), medium (1.0 μg) and high (3.0 μg)] of wild-type λNHA-MARF1 and λNHA-MARF1ΔC-term plasmids. HA Western blot signals are quantified relative to actin and marked below each lane, with ‘medium’ HA signal set to 100%. (B and C) RL activity detected in extracts from HEK293 cells. Cells were cotransfected with constructs expressing the RL-MAML1 reporter (B) or RL-5BoxB reporter (C), along with FL and different amounts (low, medium and high) of wild-type λNHA-MARF1 and λNHA-MARF1ΔC-term plasmids. (D) Western blot analysis HEK293 cells depleted of EDC4 or DCP2 by siRNA-mediated knockdown. siGFP represents a negative control. (E) siRNA-treated cells were subsequently cotransfected with constructs expressing the RL-MAML1 reporter, FL, and low levels of λNHA-MARF1 or λNHA-MARF1ΔC-term plasmids, and RL activity detected in extracts. (F through J) RL activity and mRNA levels detected in extracts from HEK293 cells normalized to FL activity and mRNA levels. Cells were cotransfected with constructs expressing the RL-MAML1 reporter (F and I), RL-NOTCH2 reporter (G and J) or RL-5BoxB reporter (H), along with FL, V5-tagged EDC4 and high amounts of wild-type λNHA-MARF1 and λNHA-MARF1ΔC-term plasmids. All Histograms represented normalized mean values of RL activity (F through H) or mRNA levels (G and I) from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN and mRNA levels seen in the presence of λNHA-LacZ were set as 100.
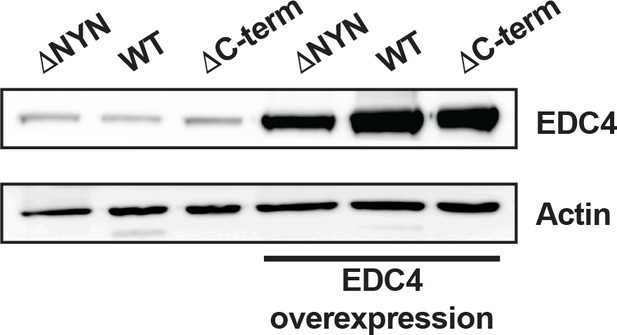
MARF1 C-terminus is sufficient to localize with EDC4 in P-bodies.
Western blot analysis of endogenous EDC4 expression versus ectopic overexpression of V5-tagged EDC4.
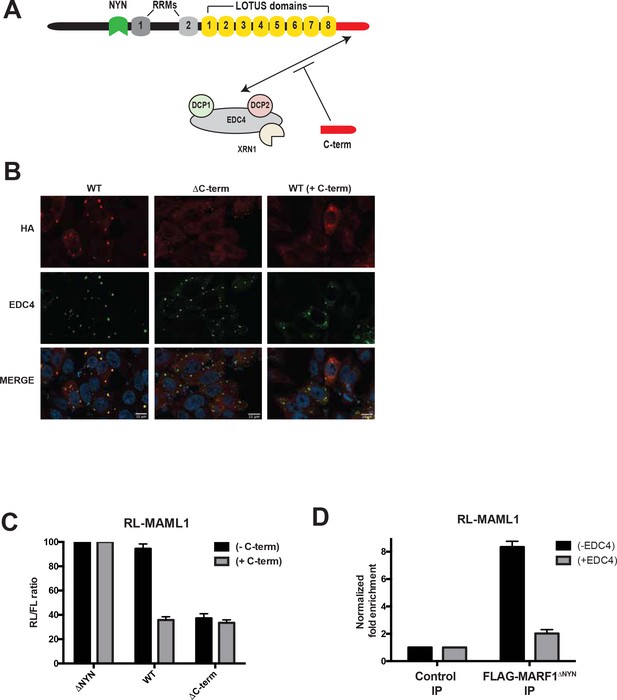
EDC4-MARF1 interaction localizes MARF1 to P-bodies and impairs MARF1 silencing.
(A) Schematic model of MARF1 C-terminus competition assay. MARF1 contains a C-terminal motif (red) that interacts with the mRNA decapping machinery, including EDC4, DCP1, DCP2 and XRN1. Co-transfecting a plasmid encoding the MARF1 C-terminal motif may compete with full-length MARF1 in binding to the decapping machinery. (B) Confocal fluorescence micrographs of fixed HeLa cells expressing wild-type λNHA-MARF1 (with or without FLAG-tagged MARF1C-term) and λNHA-MARF1ΔC-term, along with EDC4. Cells were stained with anti‐HA (red) and anti‐EDC4 (green) antibodies. The merged images show the HA signal in red and the EDC4 signal in green. (C) RL activity detected in extracts from HEK293 cells expressing the indicated proteins. Cells were co-transfected with constructs expressing the RL-MAML1 reporter, FL, low amounts of indicated λNHA-MARF1 constructs, along with/without a plasmid coding for MARF1 C-terminal motif. Histograms represented normalized mean values of RL activity from a minimum of three experiments. RL activity values seen in the presence of λNHA-MARF1ΔNYN were set as 100. (D) RNA immunoprecipitation of RL-MAML1 reporter by FLAG-tagged MARF1ΔNYN in HEK293 cells plus/minus EDC4 overexpression.
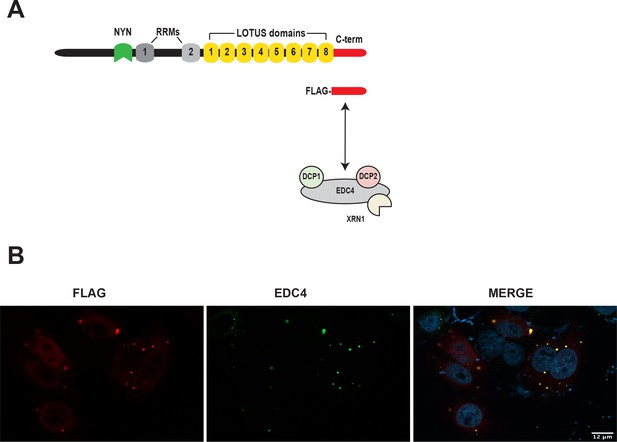
MARF1 C-terminus interfaces with the mRNA decapping machinery and localizes to P-bodies.
(A) MARF1 C-terminus interfaces with the mRNA decapping machinery and localizes to P-bodies. Schematic diagram of FLAG-tagged MARF1Cterminteracting with the mRNA decapping machinery, including EDC4, DCP1, DCP2 and XRN1 . (B) Confocal fluorescence micrographs of fixed HeLa cells expressing FLAG-tagged MARF1C-term and EDC4. Cells were stained with anti‐FLAG (red) and anti‐EDC4 (green) antibodies. The merged image shows the FLAG signal in red and the EDC4 signal in green.
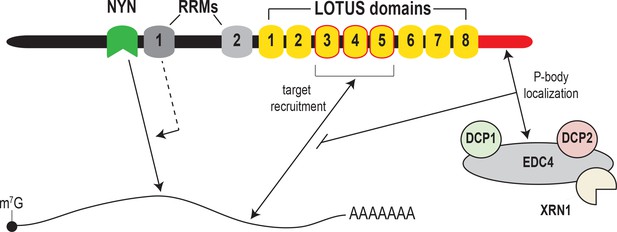
Model for MARF1-mediated mRNA decay.
MARF1 recognizes target mRNAs via LOTUS domains 3 through 5. Subsequently, RRM1 enhances NYN-mediated cleavage of target mRNAs, potentially by assisting in positioning the NYN domain on target. EDC4 regulates MARF1-mediated repression by interacting with the MARF1 C-terminus, segregating MARF1 to P-bodies and preventing MARF1 from binding target mRNAs, potentially by interfering with LOTUS domain-RNA interactions.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | MARF1 | HGNC:HGNC:29562 | ||
| Gene (Homo sapiens) | EDC4 | HGNC:HGNC:17157 | ||
| Cell line (Homo sapiens) | 293T | ATCC | CRL-3216 | Cell line maintained in DMEM + 10% FBS,50 U/mL penicillin, and50 ug/mL streptomycin |
| Cell line (Homo sapiens) | HeLa | ATCC | CCL-2 | Cell line maintained in DMEM + 10% FBS,50 U/mL penicillin, and50 ug/mL streptomycin |
| Cell line (Homo sapiens) | Flp-In T-REx 293 | Thermo Fisher | R78007 | Cell line maintained in DMEM + 10% FBS,50 U/mL penicillin, and50 ug/mL streptomycin |
| Antibody | Anti-HA mouse monoclonal | Covance | 901533 | WB(1:1000); IF(1:200) |
| Antibody | Anti-FLAG M2 mouse monoclonal | Sigma-Aldrich | F1804 | WB(1:1000); IF(1:500) |
| Antibody | Anti-V5 Rabbit monoclonal | Cell Signaling | 13202 | WB(1:1000); IF(1:500) |
| Antibody | Anti-human/mouse EDC4 Rabbit monoclonal | Bethyl | A300-745A | WB(1:1000); IF(1:500) |
| Antibody | Anti-human/mouse DCP2 Rabbit monoclonal | Bethyl | A302-597A | WB(1:1000) |
| Antibody | Anti-human Actin Mouse monoclonal | Cell Signaling | 3700 | WB(1:30000) |
| Antibody | Alexa Fluor 488 anti-rabbit | Invitrogen | A32731 | IF(1:500) |
| Antibody | Alexa Fluor 594 anti-mouse | Invitrogen | A32742 | IF(1:500) |
| Recombinant DNA reagent | PLKO.1-Puro (plasmid) | Sigma-Aldrich | RRID :Addgene10878 | shRNA backbone used for selecting transfected cells with puromycin |
| Recombinant DNA reagent | pCI-neo | Promega | E1731 | For all constructs labelled ‘LNHA’ and ‘RL’ |
| Recombinant DNA reagent | pBABE-3xFLAG-MARF1Cterm | Nishimura et al., 2018 | Construct expressing a fragment of MARF1 previously validated by our group to be sufficient to physically interact with the mRNA decapping machinery. | |
| Recombinant DNA reagent | pcDNA-DEST40 | Thermo Fisher | 12274015 | For expression of gateway cloned V5-tagged EDC4 |
| Sequence-based reagent | MAML1 3’UTR cloning primers | This paper | PCR Primer | FWD: CGGCGCCGCTCTAGAGGTGTTGGGACAGCAGGATA REV: CGGCGCCGCGCGGCCGCCATAGCTCCCCCAAAACACAC |
| Sequence-based reagent | NOTCH2 3’UTR cloning primers | This paper | PCR Primer | FWD: CGGCGCCGCGTCAGCGAGAGTCCACCTCCAGTGTAGAG REV: CGGCGCCGCGCGGCCGCCATGTTCAAATATCTCACTGAC |
| Sequence-based reagent | ZFP36L2 3'UTR cloning primers | This paper | PCR Primer | FWD: GCAGTAATTCTAGAGGCAAGAGGGCGCCAGTGAGGAGGA REV: GCAGTAATGCGGCCGCCCCAAAAATTTTATTGGGGGAAAAC |
| Sequence-based reagent | ATXN7L3 3'UTR cloning primers | This paper | PCR Primer | FWD: GCAGTAATTCTAGACTTGGGTGCAAGGGATAGCCTTTGG REV: GCAGTAATGCGGCCGCCCAACGGGAGATGCAGTTTATTTAC |
| Sequence-based reagent | FAF2 3'UTR cloning primers | This paper | PCR Primer | FWD: GCAGTAATGCTAGCCCTCCTACCCCAGTCCCTAAAAGAA REV: GCAGTAATGCGGCCGCCTGAAACTCTTTGCTTGGCCTTGGC |
| Sequence-based reagent | CPSF7 3'UTR cloning primers | This paper | PCR Primer | FWD: GCAGTAATTCTAGAGGAGTCTGGTTGGAAGCAAATGTTT REV: GCAGTAATGCGGCCGCTCACCGACAACAGGGGGACGGGACC |
| Sequence-based reagent | IGF2BP1 3'UTR cloning primers | This paper | PCR Primer | FWD: GCAGTAATGCTAGCGGAGAACAGGCCTGGTGGGAAAGGC REV: GCAGTAATGCGGCCGCGTAGTTACTAGCACTGCTGGTTCCC |
| Sequence-based reagent | PGAM1 3’UTR cloning primers | This paper | PCR Primer | FWD: GGCGCCGCGGCTAGCCCCACCTGCACATGTCACACTGACCAC REV: GGCGCCGCGCGGCCGCATACTGATATGGAAAAAGGATTTAGTACAG |
| Sequence-based reagent | MARF1ΔNYNcloning primers | This paper | PCR Primer | FWD: GTGCTAGAAAACTTACCCTTCATTTCCGACTTGCCCCCCAGGTTACCAC REV: GGCAAGTCGGAAATGAAGGGTAAGTTTTCTAGCACCTGTCCAGCTACTGC |
| Sequence-based reagent | MARF1ΔRRM1cloning primers | This paper | PCR Primer | FWD: AAAAATGCCACAGACTCCAAAAAATAGAGAACTCTGTG REV: TTCTCTATTTTTTGGAGTCTGTGGCATTTTTAGTGGTAACCTG |
| Sequence-based reagent | MARF1ΔRRM2cloning primers | This paper | PCR Primer | FWD: TGCCCAGACCCACTCTCTTTACTGAGTGCAGAAACAATG REV: CACTCAGTAAAGAGAGTGGGTCTGGGCAGTCGGCTTCGCTG |
| Sequence-based reagent | MARF1Y513A/Y515Acloning primers | This paper | PCR Primer | FWD: GCTCGCTGTTGCTAACCTACCAGCAAATAAGGATGGC REV: GTAGGTTAGCAACAGCGAGCAGAGTGTGGCACTGTGGC |
| Sequence-based reagent | MARF1I552A cloning primers | This paper | PCR Primer | FWD: CTGCAGTGCAGCTCTCCGCTTCATAAACCAAGATAGTG REV: GAAGCGGAGAGCTGCACTGCAGCCTGTGATACTCAGCAC |
| Sequence-based reagent | MARF1F582A cloning primers | This paper | PCR Primer | FWD: TTGTGTCAGCTACTCCAAAAAATAGAGAACTCTGTGAAAC REV: TTTGGAGTAGCTGACACAATGATCCTATTACCAAAGACATC |
| Sequence-based reagent | MARF1ΔLOTUS1cloning primers | This paper | PCR Primer | FWD: TGGTCTCACTTGCCACCGGGGCTGCCAGCAAATCACTACCCAGCAGTCAGGCCCGCCAGA REV: GGGGCTCTGGCGGGCCTGACTGCTGGGTAGTGATTTGCTGGCAGCCCCGGTGGCA |
| Sequence-based reagent | MARF1ΔLOTUS2cloning primers | This paper | PCR Primer | FWD: TCTTACAAGATTCCTTTTGTGATTCTTTCTATTCACAACAAGCCCCCGCC REV: AGTGTTGGGAGGCGGGGGCTTGTTGTGAATAGAAAGAATCACAAAAGGAA |
| Sequence-based reagent | MARF1ΔLOTUS3 cloning primers | This paper | PCR Primer | FWD: CGTTCGAAGAGTCCTGTAGGTAACCCCCAGCACAGGGCCCAGGTGAAGCGCTTTA REV: CTGAGTAAAGCGCTTCACCTGGGCCCTGTGCTGGGGGTTACCTACAGGACTCTTC |
| Sequence-based reagent | MARF1ΔLOTUS4 cloning primers | This paper | PCR Primer | FWD: CGTCTGCTGACCCTTACCCACAGGGCCCAGCCCAAAAGAGAACGCACTCAGGATG REV: TATTTCATCCTGAGTGCGTTCTCTTTTGGGCTGGGCCCTGTGGGTAAGGGTCAGC |
| Sequence-based reagent | MARF1ΔLOTUS5 cloning primers | This paper | PCR Primer | FWD: AGAGAACGCACTCAGGATGAAATAGAAAGGCTTTTCTTCGAGCGGTTCAAAGCTC REV: AGCTAGAGCTTTGAACCGCTCGAAGAAAAGCCTTTCTATTTCATCCTGAGTGCGT |
| Sequence-based reagent | MARF1ΔLOTUS6 cloning primers | This paper | PCR Primer | FWD: TGTCAGAGTAAGGATCTTTTCTTCGAGCGGATCAACCGAAAGTCTCTGCGATCTC REV: AGTGAGAGATCGCAGAGACTTTCGGTTGATCCGCTCGAAGAAAAGATCCTTACTC |
| Sequence-based reagent | MARF1ΔLOTUS7 cloning primers | This paper | PCR Primer | FWD: AGACAGATTCAGCTGATCAACCGAAAGTCTACAAGTCTGTATTTGTTTGC REV: CACATTCTTTGCAAACAAATACAGACTTGTAGACTTTCGGTTGATCAGCT |
| Sequence-based reagent | Full length MARF1 cloning primers | This paper | PCR Primer | FWD: GGACGATCTGCAATTGGAAGGAAACGGAACTGAGAACTCCTGC REV: GCGCCGCGCGGCCGCTTAAAGCTTGGTTATAGGTGCTAAGGAAAAG |
| Sequence-based reagent | MARF1ΔCtermcloning primers | This paper | PCR Primer | FWD: GGACGATCTGCAATTGGAAGGAAACGGAACTGAGAACTCCTGC REV: GCGCCGCGCGGCCGCTTAGAGACTGAGTGAACTCAAACGAC |
| Sequence-based reagent | MARF1N-termcloning primers | This paper | PCR Primer | FWD: GGACGATCTGCAATTGGAAGGAAACGGAACTGAGAACTCCTGC REV: GCGCCGCGCGGCCGCTTACCCGGTGGCAAGTGAGACCAGG |
| Sequence-based reagent | EDC4 Gateway cloning primers | This paper | PCR Primer | FWD: GGGGACAAGTTTGTACAAAAAAGCAGGCTACCATGGCCTCCTGCGCGAGCATCGACATCG REV: GGGGACCACTTTGTACAAGAAAGCTGGGTCAGGGAGGCTGGGGGTCACGA |
| Sequence-based reagent | FL qPCR primers | This paper | FWD: CCTTCGATAGGGACAAGACAA REV: AATCTCACGCAGGCAGTTCT | |
| Sequence-based reagent | RL qPCR primers | This paper | FWD: GAGTTCGCTGCCTACCTGGAGCCAT REV: GGATCTCGCGAGGCCAGGAGAG | |
| S1equence-based reagent | GAPDH qPCR primers | This paper | FWD: GTGGAGATTGTTGCCATCAACGA REV: CCCATTCTCGGCCTTGACTGT | |
| Sequence-based reagent | MAML1 qPCR primers | This paper | FWD: GACTCTCTCAACAAAAAGCGTCT REV: AGGAAATGACTCACTGGGGTTA | |
| Sequence-based reagent | NOTCH2 qPCR primers | This paper | FWD: CTCCAGGAGAGGTGTGCTTG REV: TGATGTCTCCCTCACAACGC | |
| Sequence-based reagent | GFP siRNA | Dharmacon | D-001940-01-05 | Accell eGFP control siRNA |
| Sequence-based reagent | EDC4 siRNA | Dharmacon | L-016635-00-0005 | SMARTpool |
| Sequence-based reagent | DCP2 siRNA | Dharmacon | L-008425-01-0005 | SMARTpool |
| Peptide, recombinant protein | Actinomycin D | Sigma-Aldrich | A1410 | |
| Commercial assay or kit | GoTaq qPCR Master Mix | Promega | A6001 | Reagent for all qPCR assays |
| Commercial assay or kit | Dual-Luciferase Assay | Promega | E1910 | Reagent for all luciferase assays |
| Other | DAPI stain | Invitrogen | D1306 | (1 µg/mL) |
Additional files
-
Supplementary file 1
CLIP mapping and cluster statistics.
- https://cdn.elifesciences.org/articles/54995/elife-54995-supp1-v1.xlsx
-
Supplementary file 2
List of MARF1 target RNAs identified by iCLIP that overlap with upregulated gene expression in Marf1GT/GT and Marf1D272A/D272A germinal vesicles (Yao et al., 2018).
- https://cdn.elifesciences.org/articles/54995/elife-54995-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/54995/elife-54995-transrepform-v1.pdf





