Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing
Figures
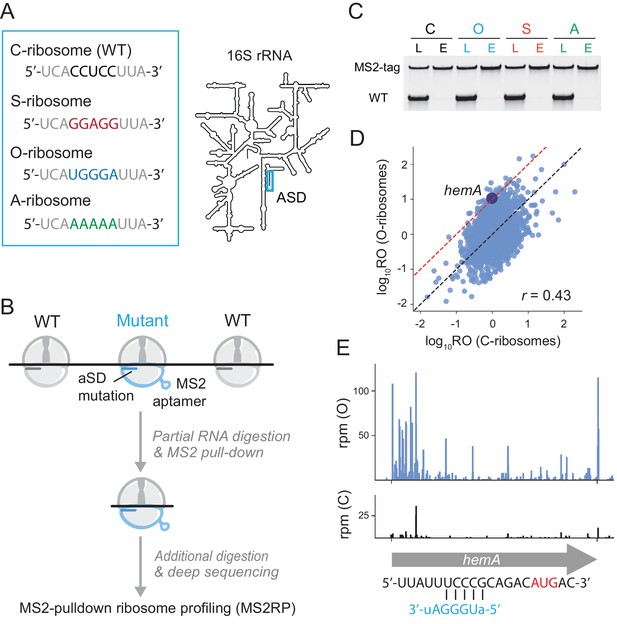
Capturing the role of SD motifs by MS2RP.
(A) ASD mutations at the 3’-end of 16S rRNA are highlighted in color. (B) Schematic of MS2RP: polysomes are collapsed to monosomes by RNase T1 digestion, MS2-tagged monosomes are pulled down with the MS2 coat-protein, and mRNA is fully digested to yield ribosome footprints that are subjected to deep sequencing. (C) RT-PCR of 16S rRNA from cell lysates (L) and the eluate (E) from the MS2 coat-protein column. (D) Scatter plot of ribosome occupancy (RO), the ratio of ribosome profiling to RNA-seq reads, from MS2RP of O-ribosomes vs. C-ribosomes. The red line indicates a 10-fold enrichment and the Pearson correlation is given. (E) Ribosome footprints (in reads per million mapped reads) from MS2RP of O-ribosomes and C-ribosomes on the hemA gene. The sequence upstream of the start codon is predicted to pair with the ASD of O-ribosomes.
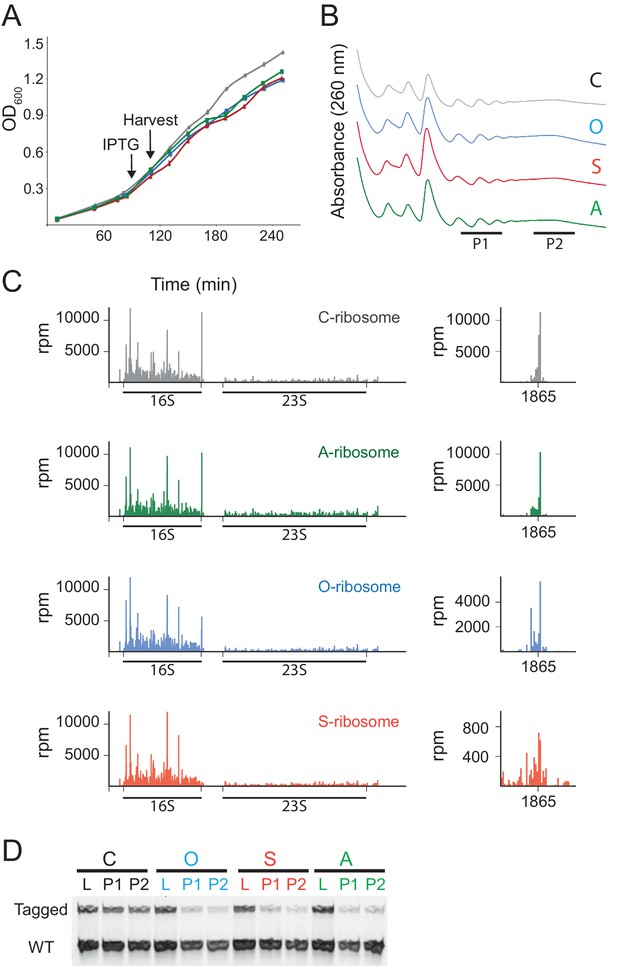
Details of the expression of MS2-tagged mutant ribosomes.
(A) Growth of E. coli MG1655 cells expressing MS2-tagged 16S rRNA from a promoter induced by IPTG. (B) Polysome profiles after 25 min of induction of MS2-tagged ribosomes. (C) RNA-seq analysis of rRNA affinity-purified by the MS2 protein in the absence of nuclease treatment. This procedure should capture all MS2-containing rRNA including processing intermediates. The 16S rRNA ends at 1895 in the mutants as well as in the wild-type, suggesting that there are not gross defects in the processing of the 3’-end of 16S rRNA. (D) position of MS2-tagged ribosomes across the sucrose gradient in cell lysates (L) and light (P1) and heavy (P2) polysomes as detected by RT-PCR of the 16S rRNA.
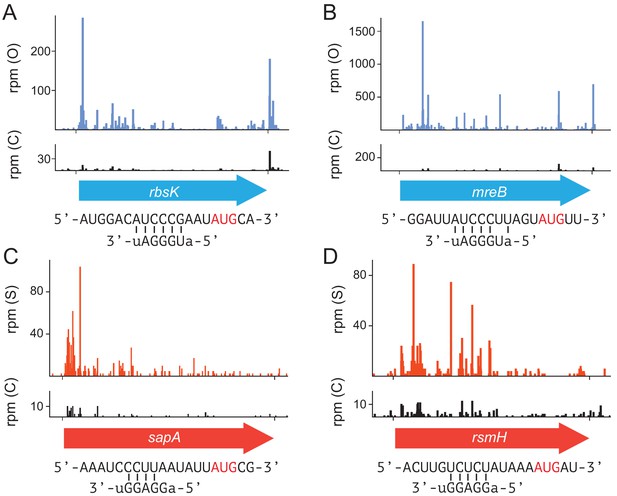
Enhanced translation of genes with ribosome-binding sites complementary to ASD-mutant ribosomes.
(A,B) Ribosome density (in reads per million mapped reads) from MS2RP of O- and C-ribosomes on the rbsK and mreB genes. (C,D) Ribosome density from MS2RP of S- and C-ribosomes on the sapA and rsmH genes.
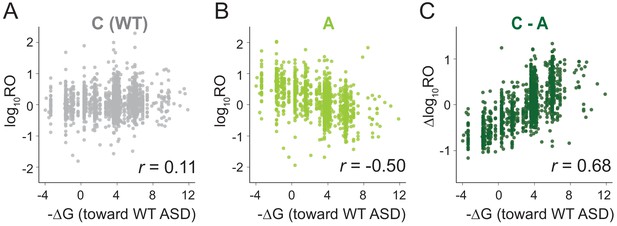
MS2RP reveals that SD motifs enhance translation genome-wide.
Ribosome occupancy (RO) is the ratio of ribosome profiling to RNA-seq reads per gene. Log10RO values are plotted against the SD strength (-∆G of pairing to the wild-type ASD) for each gene with MS2RP data for C-ribosomes (A) and A-ribosomes (B). (C) Scatter plot of ∆logRO (C-ribosomes minus A-ribosomes) and -∆G where r values indicate Pearson correlations.
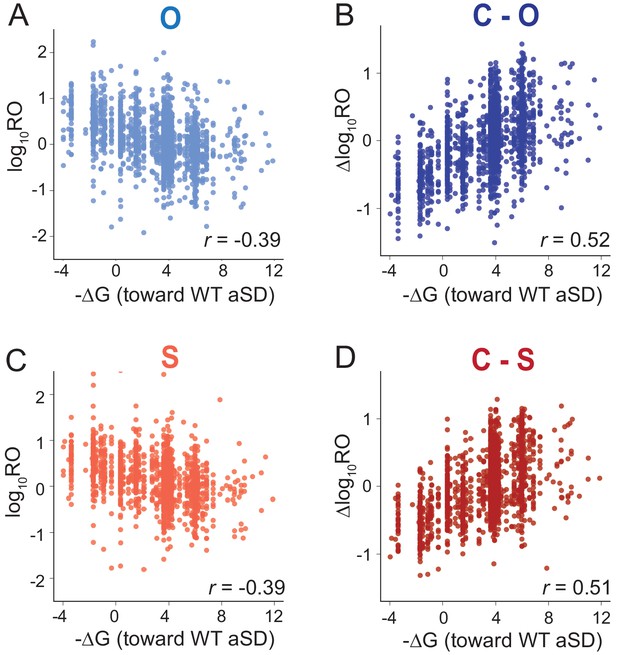
MS2RP reveals that SD motifs enhance ribosome occupancy.
Scatter plots of log10RO and SD strength (-∆G of pairing to the wild-type ASD) for O-ribosomes (A) and S-ribosomes (B). (B,D) Scatter plot of ∆logRO (C-ribosomes minus O- or S-ribosomes) and -∆G where the r values indicate Pearson correlations.
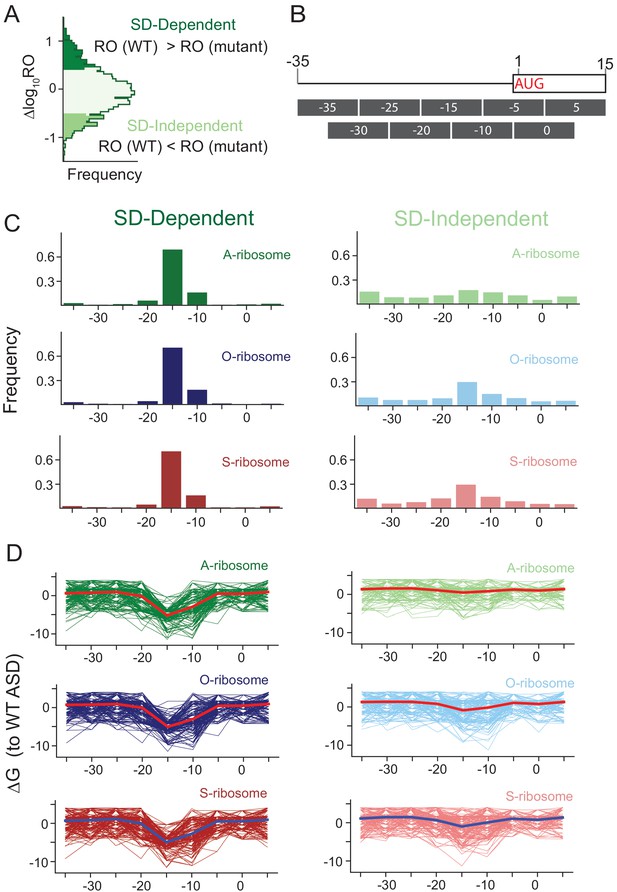
Genes translated better by wild-type ribosomes than by ASD mutants have sequences between −15 and −6 nt upstream of the start codon with high affinity to the ASD.
(A) Genes with the top 15% of ∆logRO values are translated better by the wild-type ribosomes than by ASD mutants and are therefore SD dependent. In contrast, those in the bottom 15% are SD independent. (B) We computed the free energy of base pairing to the wild-type ASD for a tiled set of nine 10 nt windows. (C) Histograms showing the window with the strongest affinity to the wild-type ASD for each mRNA in the set of SD-dependent (left) and SD-independent genes (right) for each of the three ASD mutant datasets. (D) Plots of ∆G of pairing to the WT ASD for individual genes; the thick line indicates the mean of the group. Taken together, these results confirm that we captured the role of SD motifs by MS2RP and justify the use of pairing energies from the −15 to −6 window in this report.
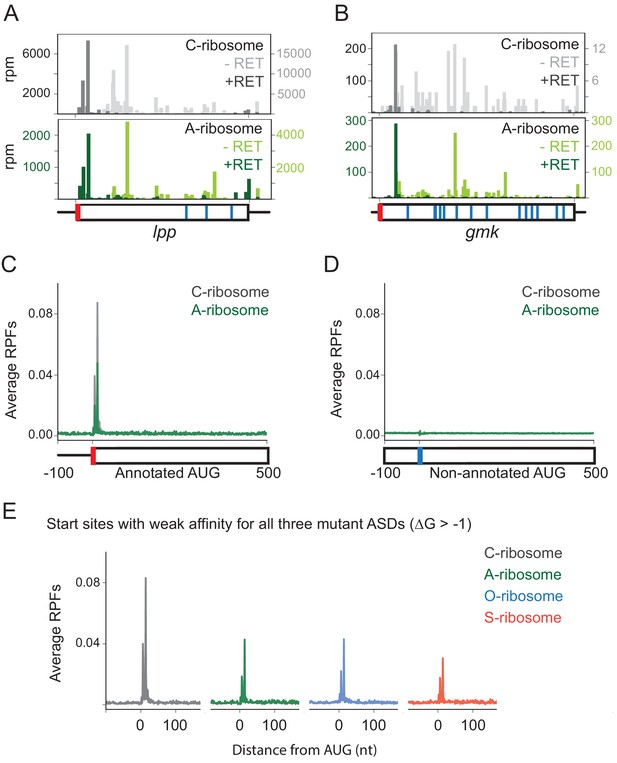
Loss of SD-ASD pairing has little effect on start codon selection.
(A,B) Ribosome footprints on lpp and gmk from MS2RP data obtained with and without retapamulin, an antibiotic that traps ribosomes at start codons. Annotated AUGs are indicated by a red bar, non-annotated AUGs are indicated by blue bars. (C, D) Average ribosome protected fragments (RPFs) at annotated AUGs and non-annotated AUGs (where AUG starts at 1). (E) Average RPFs at the start codon of genes whose ribosome-binding sites have little or no affinity to all three mutant ASD sequences.
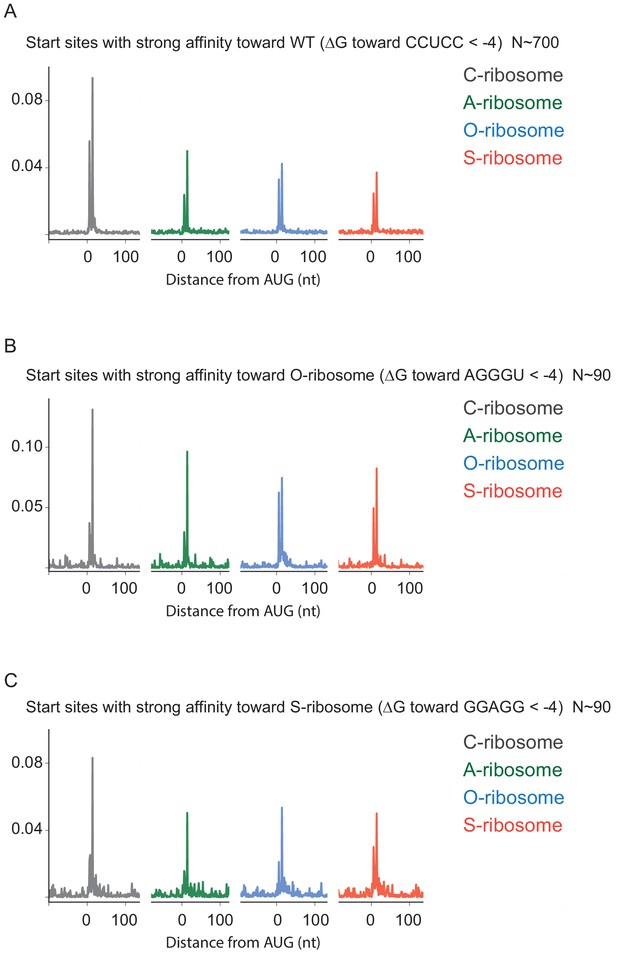
Ribosome density at annotated start sites does not depend on SD-ASD base pairing.
Average ribosome footprints aligned at annotated start sites that have high affinity to the wild-type (A), O-ribosomes (B), and S-ribosomes (C). There were not enough start sites with high affinity to the A-ribosomes for a similar analysis.
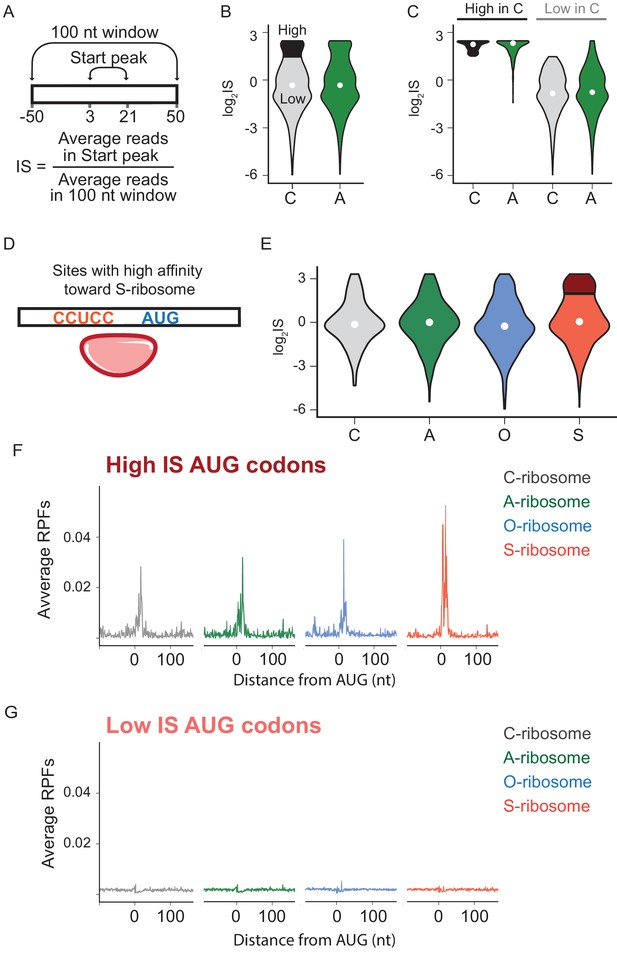
The effects of SD-ASD pairing on initiation at non-canonical sites.
(A) Evaluation of initiation score, IS. (B) Initiation scores on non-annotated AUG triplets. For C-ribosomes, the fraction with IS >1.5 is colored black. (C) Initiation scores for C- and A-ribosomes for the set of sites with IS >1.5 for C-ribosomes (High, colored black in B) and those with IS <1.5 (Low). (D,E) IS values for all four ribosome types on the subset of sites with high affinity for the ASD of the S-ribosome (CCUCC). Average RPFs at the AUG triplets with high IS scores (F) or low IS scores (G) from the S-ribosome data.
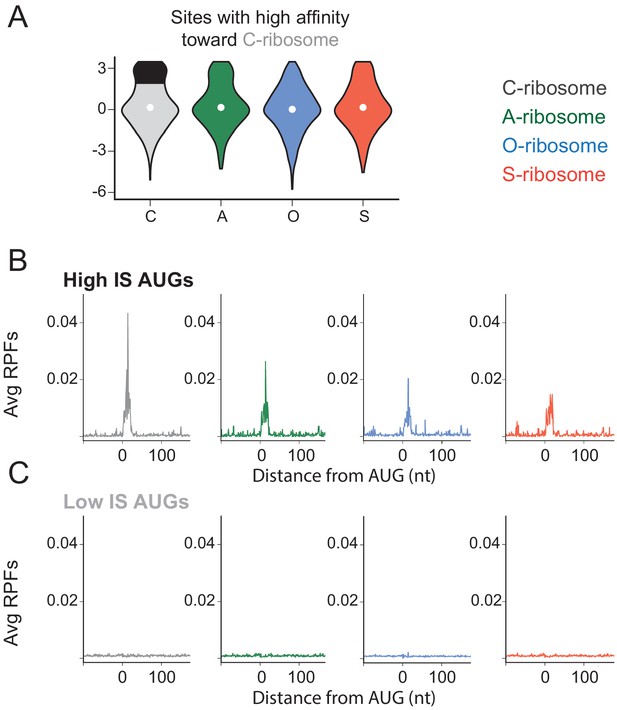
The effects of SD-ASD pairing on initiation at non-canonical sites.
(A) Initiation score (IS) values for all four ribosome types on the subset of non-annotated AUGs in the transcriptome with high affinity for wild-type ASD. (B,C) Average RPFs at AUGs with high IS scores or low IS scores at sites with high affinity for the wild-type ASD (black and grey in panel A, respectively).
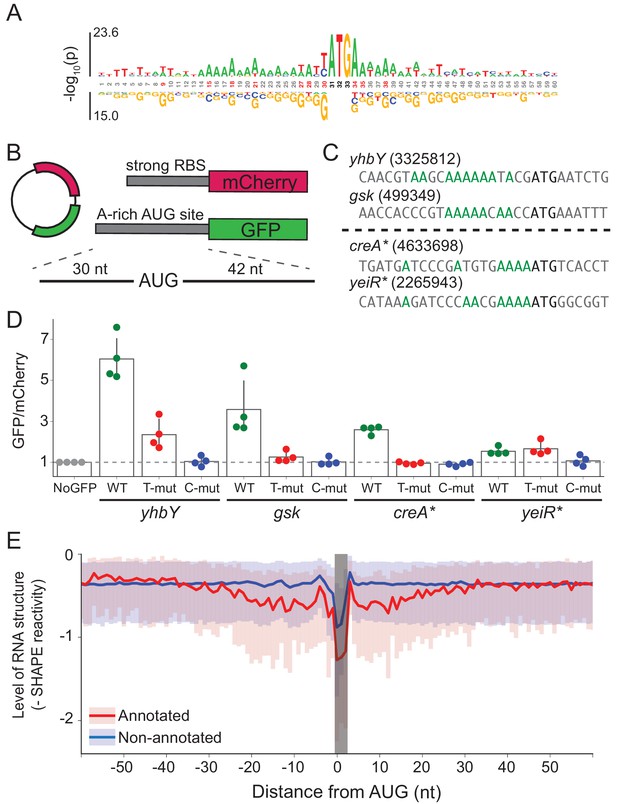
A-rich sequences as a signal for start codon selection.
(A) Probability logo of the region surrounding annotated AUGs with low affinity to the wild-type ASD sequence (∆G > 0) as compared with all non-annotated AUGs in the transcriptome. Enriched nucleotides are shown above the axis and depleted nucleotides below the axis. The height of the letter represents the binomial P-value. (B) Design of the reporter assay. The reporter plasmid encodes mCherry with a strong ribosome binding site (RBS) and separately GFP downstream of a region containing a start site of interest (30 nt upstream of AUG and 42 nt downstream). (C) Initiation sites used in the reporter assay; the number indicates the genomic position of AUG. In the T- and C-mutants, the A’s upstream of AUG (highlighted in green) were substituted by T or C. (D) Results of the reporter assay. Each dot is the median of GFP/mCherry from an independent run of flow cytometry. The bar graph indicates the mean and SD from four independent tests. NoGFP (a plasmid that encodes mCherry but not GFP) serves as a control showing the baseline signal from cellular autofluorescence; the other data are normalized to this ratio. (E) Median (solid line) and interquartile range (shaded) of mRNA structure in SHAPE-MaPseq data for 365 annotated start sites (red) and 7310 non-annotated AUGs within coding sequences (blue).