Gradients in the biophysical properties of neonatal auditory neurons align with synaptic contact position and the intensity coding map of inner hair cells
Figures
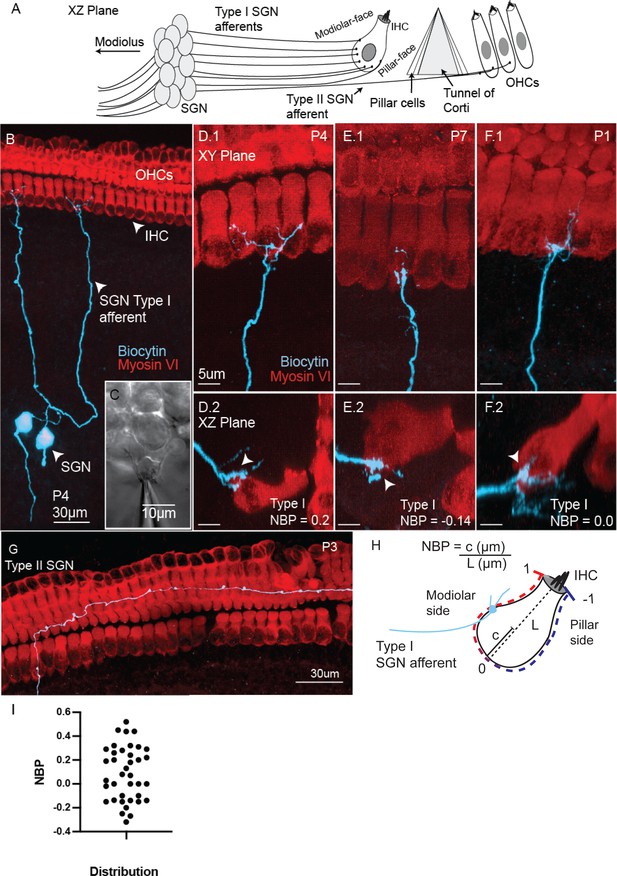
Semi-intact preparation combines SGN biophysics with identification of pattern of connectivity with hair cells.
(A) Schematic of the innervation at auditory hair cell along with key anatomical structures provided with the semi-intact preparation presented in the XZ plane. (B) Confocal image of two type I spiral ganglion neurons (SGN) injected with biocytin in the acute semi-intact preparation of rat middle-turn cochlea. Hair-cells and biocytin-labeled fibers are visualized by immunolabeling for Myosin VI (red; hair cells) and streptavidin conjugated to Alexa Flour 488 (green, fiber). (C) Myelinating Schwann cells are mechanically removed by suction pipettes to make SGN somata accessible to recording electrodes. D.1-F.1. Confocal images in the XY plane show type I SGN afferent fibers making exclusive connections with inner hair cells. D.2-F.2 The synaptic position of the afferent fiber onto the inner hair cell is assessed by examining the cross-section of the hair cell in XZ plane. (G) Type II SGN projecting radially and turn within the outer hair cell region. (H) A schematic of an individual inner hair cell with guides showing how the normalized basal position (NBP) is measured for each type I SGN. NBP is measured by the synaptic position along the radial axis of the hair cell,c, divided by the maximum length of the inner hair cell, L. Positive NBP values indicate that the synaptic position is on the modiolar-side of the inner hair cell, while negative NBP values indicate that the synaptic position is on the pillar-side of the inner hair cell. (I) The distribution of type I SGN contacts as a function of NBP. NBP values averaged at 0.11 +/- 0.23 with a range from −0.27 to +0.52 (n = 38).
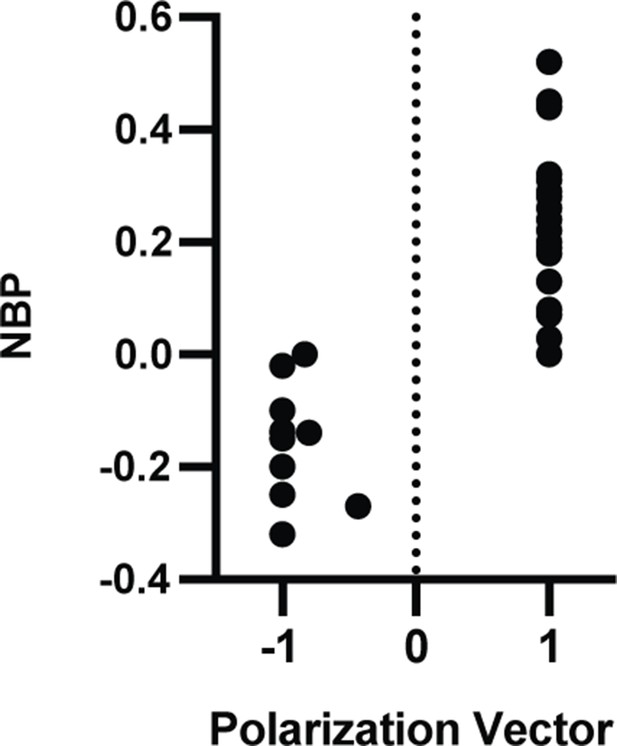
Polarization vector shows that branching fibers have a preferred polarization to either modiolar or pillar face.
The net polarization of the fiber was computed by taking the sum of the individual polarizations divided by the total number of branches (Figure 1).
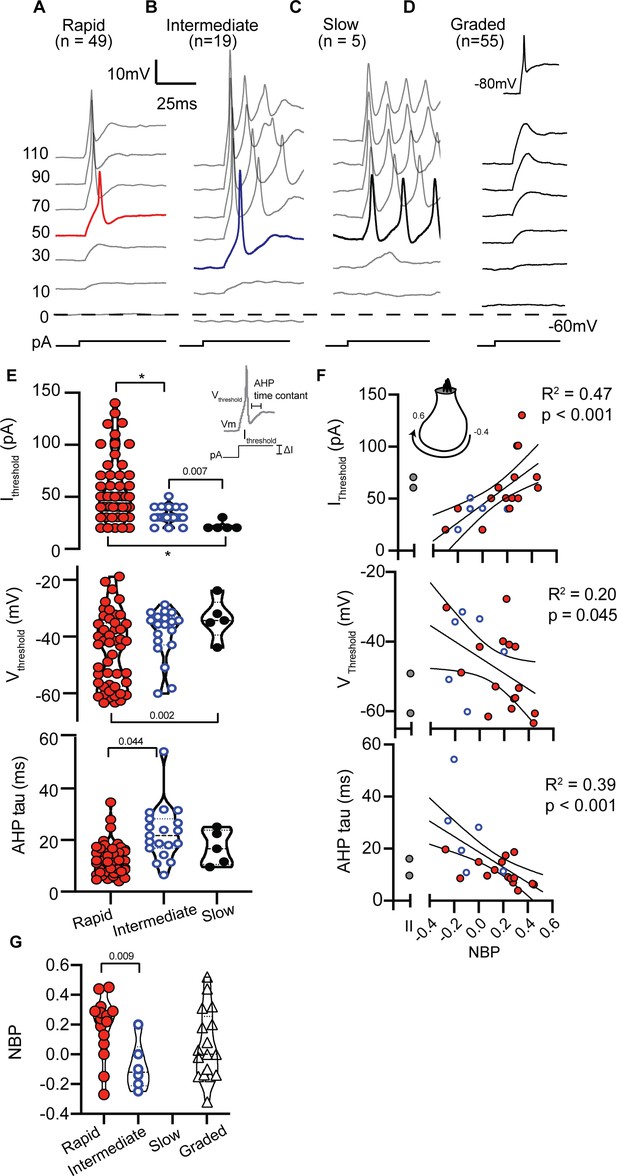
SGN can be qualitatively grouped by firing patterns in response to step current injections.
(A) Rapidly-accommodating neurons fire a single action potential. (B) Intermediately-accommodating neurons fire more than one action potential and eventually reach a steady-state potential. (C) Slowly-accommodating neurons fire multiple actions and do not reach a steady-state potential. (D) Graded-response neurons do not produce an action potential, but produce graded-depolarization in response to incrementing current steps. These neurons are capable of firing action potentials after their membrane potential are held at −80 mV (inset). (E) Action potential features including current threshold (Ithreshold), voltage threshold (Vthreshold), and after hyperpolarization potential time constant (AHP tau) vary among spiking neurons. The p-values indicate when two means were assessed to be significantly different (see Materials and methods); asterisks indicate p<0.001. (F) Action potential features were significantly correlated with normalized basal position (NBP). As NBP values increased, current thresholds increased (r(20) = 0.68, p=0.0007), voltage thresholds hyperpolarized (r(20) = −0.45, p=0.045), and AHP time constants became faster (r(20) = 0.62, p<0.0001). Best fit lines with 95% confidence intervals are plotted to show the gradients along the normalized basal position axis. (G) The distribution of firing pattern subgroups as a function of NBP shows a significant relationship between the firing-pattern subgroup and NBP (p=0.0097, t-test), with rapidly-accommodating neurons found primarily on the modiolar-face and intermediately-accommodating neurons found on the pillar-face of the inner hair cell. Graded-firing neurons are found throughout the NBP scale on both the modiolar and pillar faces of the hair cell.
-
Figure 2—source data 1
Current-clamp features versus normalized basal position data set for Figure 2F.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig2-data1-v1.xlsx
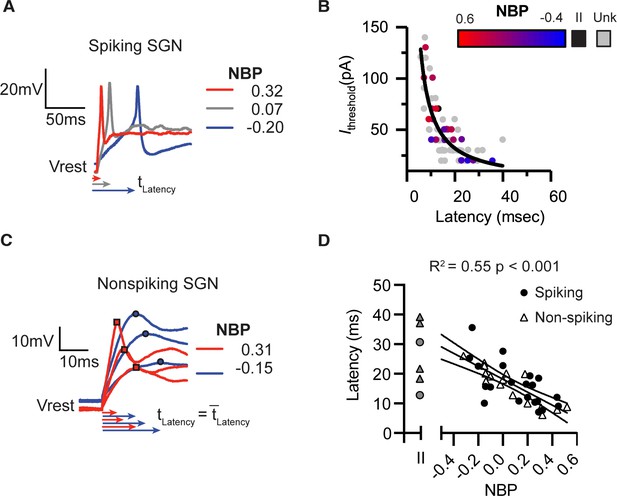
Response latencies are alternate predictors for normalized basal position.
There is a significant inverse relationship between first-spike latency and current thresholds. Black line shows a fit Ithresh = constant/latency. Color transition from red to blue indicates position on NBP scale. Type II SGN are colored black. Unlabeled and therefore unclassified SGN are in gray. (A) First spike latencies compared between three spiking SGN with different NBP values. Latencies are faster for more positive NBP (B) In non-spiking neurons, response latency is computed by averaging the time from the onset of the stimulus until peak depolarization potential over a series of current steps. Similar to the dependence of first-spike latency, average response latency is faster for modiolar-contacting SGN (NBP >0, red) than for pillar-contacting SGN (NBP <0, blue). (C) Response latencies for all spiking (black dot) and non-spiking (triangle) neurons plotted against NBP. In both spiking- and non-spiking SGN, response latencies become faster as NBP values increase (R2 = 0.55, p<0.001). Latencies are highly variable across type II fibers (gray symbols).
-
Figure 3—source data 1
Current threshold versus latency dataset for Figure 3B.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Latency versus normalized basal position dataset for Figure 3D.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig3-data2-v1.xlsx
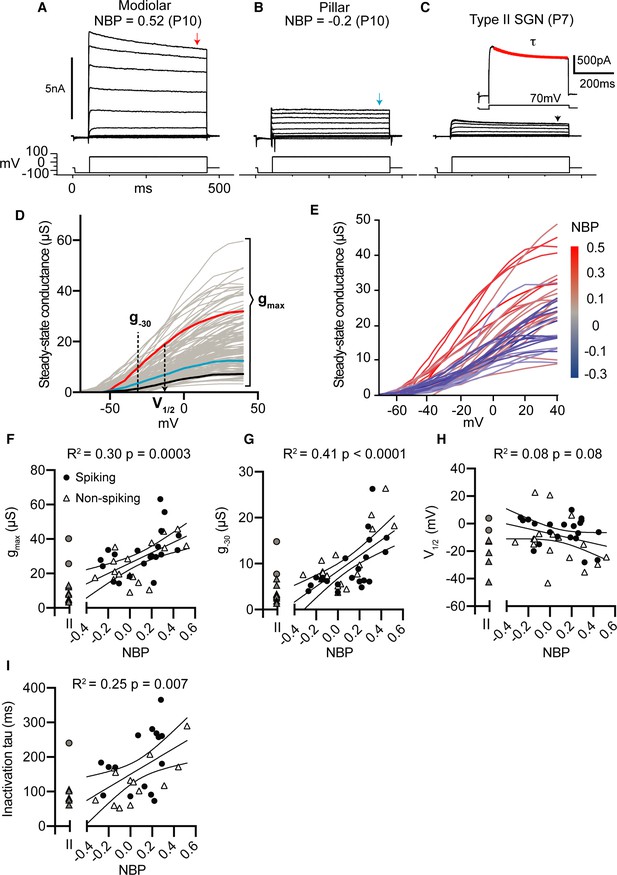
Net whole-cell conductances increase as SGN synaptic position moves from pillar to modiolar-face of the inner hair cell.
(A–C) Voltage-clamp responses of a modiolar-contacting (A), pillar-contacting type I (B), and a type II SGN (C). For each cell, we held the potential at −60 mV, followed by a series of voltage 400 ms steps from −120 mV to 70 mV, and then finally back to the holding potential. For each cell, we measured current-voltage relationships at approximate steady-state (400 ms indicated by the arrow). We measured the time course for outward current inactivation by fitting a single exponential curve to the current evoked by a +70 mV voltage step (inset). (D) Steady-state conductance in microSiemens as a function of command voltage of all SGN recorded with three measurements: the magnitude of conductance at −30 mV (g-30), the maximum conductance (gmax), and the half-activation potential (V1/2) via Boltzmann function. (E) Steady-state conductances of identified type I SGN as a function of normalized basal position. (F–I) Voltage clamp properties as a function of normalized basal position (NBP) (Spiking, black dot; non-spiking, triangle). Linear regression models were fitted to each parameter with statistical significance test of p<0.05. All parameters were significant except for V1/2.
-
Figure 4—source data 1
Voltage-clamp features versus normalized basal position for Figure 4F through 4I.
Conductance values are shown in units of micro Siemens.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig4-data1-v1.xlsx
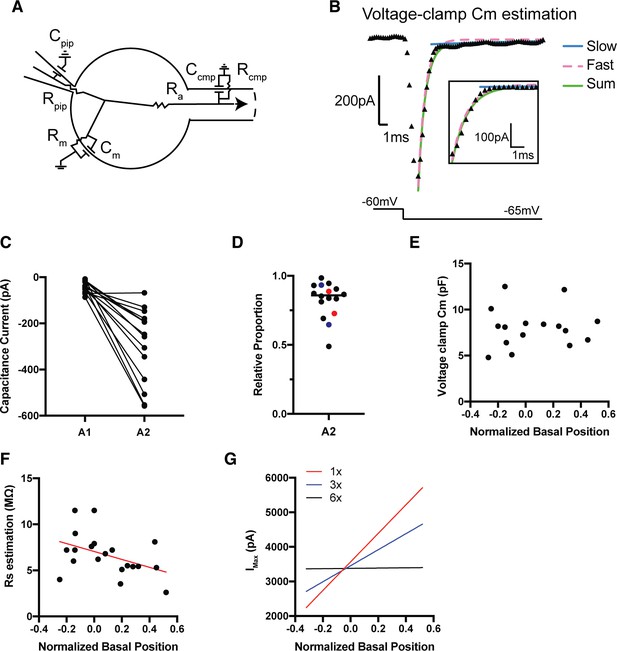
Considering the influence of space-clamp and series resistance errors.
(A) Circuit diagram of an acute SGN in the semi-intact preparation clamped in whole-cell configuration. This diagram shows the series resistance (Rpip), membrane resistance (Rm), axial resistance (Ra), and a secondary compartment resistance from the neurite (Rcomp) with corresponding capacitors (C). (B) Voltage-clamp response of the transient capacitance current of an SGN in the semi-intact preparation fitted with a bi-exponential function (green trace). The fast (pink trace) and slow (blue trace) components of the bi-exponential function are plotted to show the contribution of the current coming for the somata (fast/pink) and neurite (slow/blue). (C) The relative size of the magnitude of the fast (A2) and slow (A1) components of the trace, indicating that the fast component dominates the fit and currents are measured primarily from the soma. (D) The fast component of the bi-exponential (A2) relative proportion to the sum of the bi-exponential function (A1+A2). A2 relative proportion dominates in both modiolar(red) and pillar (blue) contacting SGN. (E) Cell capacitance, measured as the integral of the capacitance current, is not correlated with basal position (r = 0.05, p=0.85). (F) Series resistance as a function of normalized basal with the mean trend line is shown in red. (G) After performing series resistance error corrections per cell, we see that the position correlation of maximum outward current (Imax) and NBP does not collapse, and remains correlated (R2 = 0.23, p=0.0003). If we simulate increasing levels of systematic series resistance errors by three (blue trace) and six (black trace) times the amount we measured, trends along NBP do not collapse.
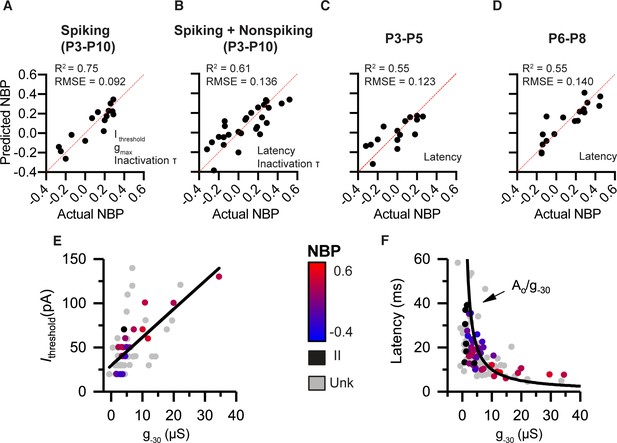
Combining voltage-clamp and current-clamp features in multiple-variable regression models to predict normalized basal position (NBP) based on biophysics.
(A) Prediction model for spiking neurons (P3–P10) relies on Ithresh, gmax and tinact. (B) Prediction model that pools spiking and non-spiking neurons (P3–P10) is successful by using latency in place of current threshold (C,D), respectively. Biophysical gradients are still successful at predicting spatial position even after filtering data into narrow age bins. P3-P5 in C and P6-P8 in D. Predictor variables used in each model are presented at the bottom right of the predicted vs. actual plot. (E) Current threshold increase with increases in g-30. (F) Response latency as a function of g-30. Fit is a function of the form latency = constant/g-30.
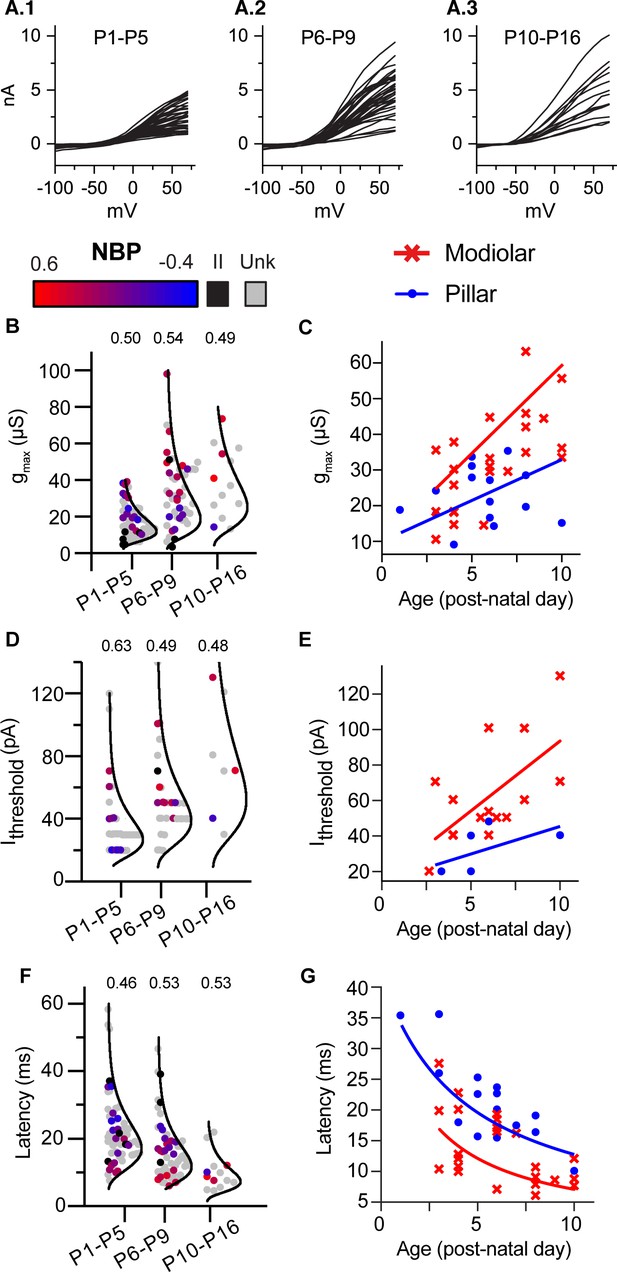
Biophysical properties of SGN change with age but remain diverse throughout 3 weeks of post-natal development.
(A) 1-A.3 Steady-state current/voltage curves show that net outward currents grow in three age bins (P1-P5; P6-9; and P10-P16). Small and large currents are evident in all age ranges. (B,D and F) Log-normal plots showing the distribution of maximum conductance (gmax), current threshold (Ithreshold) and latency in three age bins, respectively. Points are color coded to indicate normalized basal position (red to blue gradient), unlabeled cells (gray) and type II neurons (black). Within each age bin, biophysical properties of type I SGN change as a gradient along the NBP scale. The relative dispersion in each distribution is quantified by the CV (coefficient of variation indicated above each distribution) and is not systematically dependent on age. C,E and F Plots of same three features, gmax, current threshold, and latency plotted with age as a continuous variable but with spatial position binned as ‘modiolar-contacting’ (NBP >0; red symbols) and ‘pillar-contacting’ (NBP <= 0, blue symbols). Solid lines in C and E show the best regression fits through modiolar-contacting and pillar-contacting groups . Solid lines in F plot curves defined by the inverting the regression plots in C; .
-
Figure 6—source data 1
Maximum conductance versus age for Figure 6C.
Conductance values are in microSiemens.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Current threshold versus age dataset for Figure 6E.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Latency versus age dataset for Figure 6G.
- https://cdn.elifesciences.org/articles/55378/elife-55378-fig6-data3-v1.xlsx
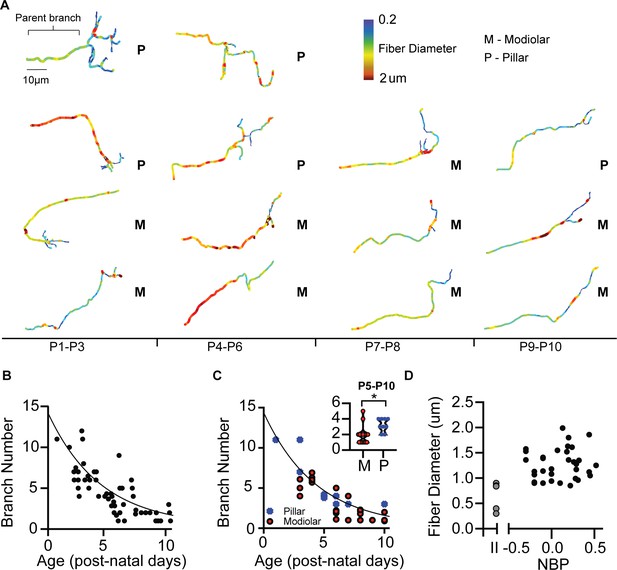
Morphological development of spiral ganglion terminal branches.
(A) Three-dimensional reconstructions of SGN terminals are made to quantify the number of branches at each terminal and the approximate diameter of the fibers. (B) The number of branches exponentially decreases over the course of development. One-to-one connections with the inner hair cell can be seen at P5. In some cases, terminals have multiple branches at P10. (C) Pillar-contacting SGN are not observed to have the mature one-connection morphology. (D) Fiber diameters do not differ between type I SGN along the NBP scale. Type II SGN have significantly smaller fiber diameters than type I SGN.
Tables
Analysis of covariance.
| Figure | Source | d.f. | Sum of squares | F-score | p-value |
|---|---|---|---|---|---|
| 3D Latency | Spike (Spike/Non-Spike) | 1 | 0.158 | 0.0052 | 0.94 |
| NBP | 1 | 967.79 | 32.04 | <0.0001 | |
| Spike * NBP | 1 | 4.14 | 0.13 | 0.71 | |
| 4F gmax | Spike | 1 | 150.99 | 1.37 | 0.25 |
| NBP | 1 | 1578.89 | 14.29 | 0.0006 | |
| Spike * NBP | 1 | 0.038 | 0.0004 | 0.98 | |
| 4 G g-30 | Spike | 1 | 41.27 | 1.97 | 0.17 |
| NBP | 1 | 568.15 | 27.16 | <0.0001 | |
| Spike * NBP | 1 | 12.14 | 0.58 | 0.45 | |
| 4 HV1/2 | Spike | 1 | 632.64 | 3.53 | 0.068 |
| NBP | 1 | 743.44 | 4.15 | 0.0495 | |
| Spike * NBP | 1 | 107.86 | 0.60 | 0.44 | |
| 4I | Spike | 1 | 18715.3 | 3.64 | 0.068 |
| NBP | 1 | 42318.4 | 8.24 | 0.0084 | |
| Spike * NBP | 1 | 9.85 | 0.0019 | 0.96 | |
| 6C. gmax | Class (Modiolar/Pillar) | 1 | 1508.38 | 7.32 | 0.0107 |
| Age | 1 | 1963.45 | 9.53 | 0.0041 | |
| Class* Age | 1 | 1232.78 | 5.98 | 0.0199 | |
| 6E Ithreshold | Class (M/P) | 1 | 3827.39 | 8.87 | 0.0087 |
| Age | 1 | 2333.04 | 5.35 | 0.0335 | |
| Class * Age | 1 | 280.75 | 0.64 | 0.4334 | |
| 6G Latency | Class (M/P) | 1 | 577.732 | 36.97 | <0.0001 |
| Age | 1 | 483.887 | 30.96 | <0.0001 | |
| Class * Age | 1 | 131.247 | 8.4 | 0.0066 |
Correlations with normalized basal position.
| Spiking neurons P3-P10 | Non-spiking P3-P10 | Spiking plus non-spiking P3-P5 | Spiking plus non-spiking P6-P8 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R | p-value | R | p-value | R | p-value | R | p-value | ||
| Current-clamp features | *IThreshold | 0.68 | 0.0007 | 0.81 | 0.0080 | 0.44 | 0.0772 | ||
| *First-spike Latency | −0.65 | 0.0013 | −0.76 | 0.0168 | −0.45 | 0.22 | |||
| *Voltage Threshold | −0.39 | 0.0169 | −0.61 | 0.0837 | 0.15 | 0.7390 | |||
| *AHP Tau | −0.61 | 0.0031 | −0.65 | 0.0217 | −0.21 | 0.5616 | |||
| *AP Height | 0.31 | 0.17 | 0.03 | 0.5634 | 0.63 | 0.0069 | |||
| Average Response Latency | −0.89 | <0.0001 | −0.76 | 0.0010 | −0.77 | 0.0003 | |||
| Resting Potential | −0.54 | 0.0112 | −0.31 | 0.229 | −0.07 | 0.8048 | −0.48 | 0.0487 | |
| Voltage-clamp features | gmax | 0.52 | 0.0166 | 0.58 | 0.0177 | 0.78 | 0.8799 | 0.62 | 0.0083 |
| V1/2 | −0.27 | 0.233 | −0.39 | 0.137 | −0.10 | 0.71 | −0.09 | 0.74 | |
| g-30 | 0.59 | 0.0045 | 0.74 | 0.0010 | 0.44 | 0.10 | 0.59 | 0.0122 | |
| 0.5 | 0.0066 | 0.67 | 0.0179 | 0.48 | 0.10 | 0.45 | 0.1459 | ||
Multiple variable regression models.
| Figure | Variable | ß | SE | Df | /t/ | VIF | RMSE | p-value |
|---|---|---|---|---|---|---|---|---|
| 5A | Ithreshold | 0.0099 | 0.0016 | 11 | 6.102 | 2.420 | 0.090 | 0.0004 |
| gmax | −0.0182 | 0.0047 | 3.858 | 3.858 | ||||
| 0.0012 | 0.0005 | 2.625 | 2.165 | |||||
| 5B | Latency | −0.0231 | 0.0045 | 24 | 5.158 | 1.241 | 0.136 | <0.0001 |
| 0.0005 | 0.0004 | 1.404 | 1.241 | |||||
| 5C | Latency | −0.0192 | 0.00454 | 14 | 4.23 | 1.00 | 0.123 | 0.0010 |
| 5D | Latency | −0.0310 | 0.0067 | 16 | 4.62 | 1.00 | 0.140 | 0.0003 |
Table of regression slopes.
| Figure | Term | Estimate | Standard error | T | p-value |
|---|---|---|---|---|---|
| 3D Latency | Slope | −22.8 | 4.02 | −5.66 | <0.0001 |
| Spike | −2.98 | 8.05 | −0.37 | 0.71 | |
| Non-Spike | 2.98 | 8.05 | 0.37 | 0.71 | |
| 4F gmax | Slope | 27.87 | 2.55 | 10.89 | <0.0001 |
| Spike | −0.29 | 15.4 | −0.02 | 0.98 | |
| Non-Spike | 0.29 | 15.4 | 0.02 | 0.98 | |
| 4 Gg-30 | Slope | 17.46 | 0.80 | 10.6 | <0.0001 |
| Spike | 5.1 | 6.7 | 0.76 | 0.45 | |
| Non-Spike | −5.1 | 6.7 | −0.76 | 0.45 | |
| 4 HV1/2 | Slope | −19.98 | 9.8 | −2.04 | 0.0495 |
| Spike | −15.22 | 19.62 | −0.78 | 0.44 | |
| Non-Spike | 15.22 | 19.62 | 0.78 | 0.44 | |
| 4I | Slope | 178.9 | 62.3 | 2.87 | 0.0084 |
| Spike | −5.46 | 124.63 | −0.04 | 0.97 | |
| Non-Spike | 5.46 | 124.63 | 0.04 | 0.97 | |
| 6C gmax | Slope | 2.80 | 1.06 | 2.63 | 0.0127 |
| Pillar | −2.60 | 1.06 | −2.45 | 0.0199 | |
| Modiolar | 2.60 | 1.06 | 2.45 | 0.0199 | |
| 6E Ithreshold | Slope | 4.68 | 2.26 | 2.07 | 0.054 |
| Pillar | −1.81 | 2.26 | −0.8 | 0.4334 | |
| Modiolar | 1.81 | 2.26 | 0.8 | 0.4334 | |
| 6G Latency | Slope | −1.752 | 0.29 | −5.97 | <0.0001 |
| Pillar | −0.85 | 0.29 | −2.9 | 0.0066 | |
| Modiolar | 0.85 | 0.29 | 2.9 | 0.0066 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (species) | Long-Evans (rat) | USC Vivarium and Charles River | RRID:RGD_2308852 | Freshly isolated cochlear middle turns |
| Antibody | anti-myosin-VI (rabbit polyclonal) | Proteus Biosciences | Cat# 25–6791, RRID:AB_10013626 | IF(1:1000) |
| Antibody | anti-peripherin (rabbit polyclonal) | Millipore | Cat# 183 AB1530, RRID:AB_90725 | IF(1:500) |
| Antibody | streptavidin Alexa Fluor 488 conjugate | Molecular Probes | Cat# S32354, 182 RRID:AB_2315383 | IF(1:200) |
| Antibody | Alexa Fluor 594 (goat anti-rabbit, polyclonal) | Thermo Fisher Scientific | Cat# A-11080, 185 RRID:AB_2534124 | IF(1:200) |
| Other | Vecta-shield | Vector Laboratories | Cat# H-191 1000, RRID:AB_2336789 | |
| Software, algorithm | JMP | SAS Institute | RRID:SCR_002865 | |
| Software, algorithm | Matlab | Mathworks | RRID:SCR_001622 | |
| Software, algorithm | pClamp | Molecular Devices | RRID:SCR_011323 | |
| Software, algorithm | OriginPro | Origin Labs | RRID:SCR_015636 | |
| Software, algorithm | Imaris | Oxford Instruments | RRID:SCR_007370 | |
| Software, algorithm | Prism 8 | Graph Pad | RRID:SCR_002798 | |
| Software, algorithm | FilamentTracer in Imaris | Oxford Instruments | RRID:SCR_007366 |





