A novel DNA primase-helicase pair encoded by SCCmec elements
Figures
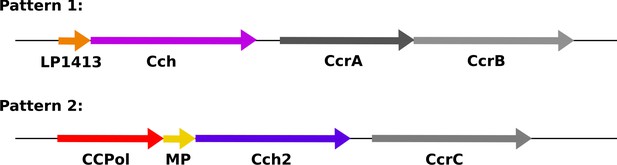
Organization of conserved ORFs in SCC elements.
Pattern 1 elements (including types I-IV, VI, and VIII-XI) carry an operon encoding a short ssDNA-binding protein, LP1413, and a helicase, Cch, whereas Pattern 2 elements (including types V, VII, and XII-XIV, and SCCmer) carry an operon encoding CCPol, MP, and Cch2, which are characterized in this manuscript. Figure 1—figure supplement 1 shows the sequence alignments of CCPol, MP, and Cch2 from various SCC elements. The recombinase genes (CcrAB or CcrC) are found after these operons, followed by three additional small ORFs not shown.
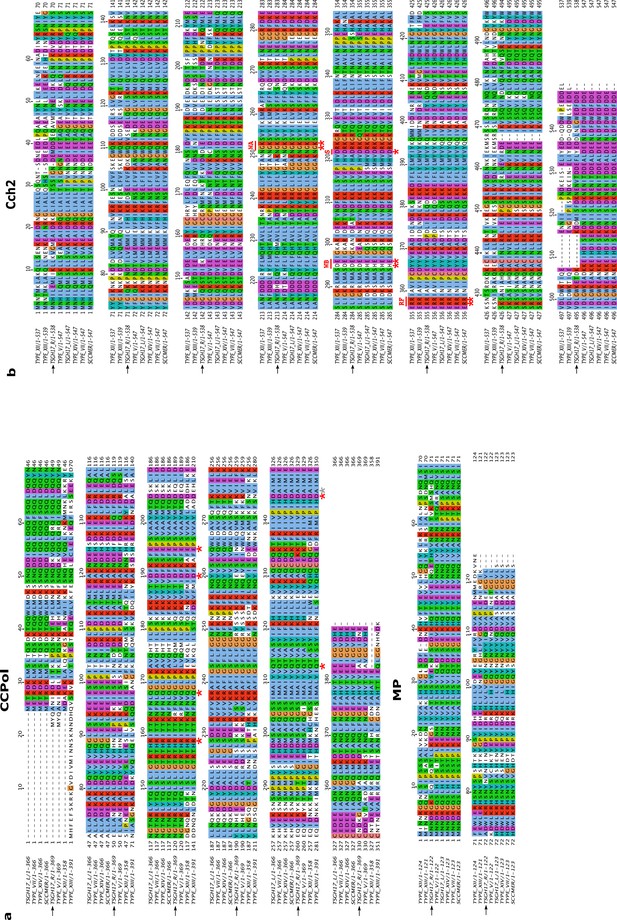
Sequence alignments of Cch2 operon proteins CCPol, MP, and Cch2.
The alignments were prepared in Clustal Omega (Sievers et al., 2011) and visualized in Jalview (with Jalview coloring) (Waterhouse et al., 2009). The following SCC sequences were used for the alignments: the two gene clusters (SCCmer-like L and type V-like R) from a composite SCCmec from TSGH17 strain (SCC accession number AB512767), type V element from WIS strain (AB121219), type VII from JCSC6082 (AB373032), type XII from strain BA01611 (JCSC6082), type XIII from isolate 55-99-44 (MG674089), type XIV from strain SC792 (LC424989), and SSCmer (carrying a mercury resistance cluster) from strain 85/2082 (AB037671). The proteins investigated in this study are marked with arrows. Residues predicted to be important for activity for CCPol (based on similarities to other A-family polymerases Kiefer et al., 1997) and Cch2 (based on similarities to Cch and the SaPI5 Rep protein Mir-Sanchis et al., 2016) are marked with red asterisks (with a grey asterisk marking an alanine residue which in other A-family polymerases is invariably a catalytic glutamic acid). For Cch2 alignment, predicted motifs are: WA – Walker A motif, WB – Walker B motif, S – sensor I, and RF – Arginine finger.
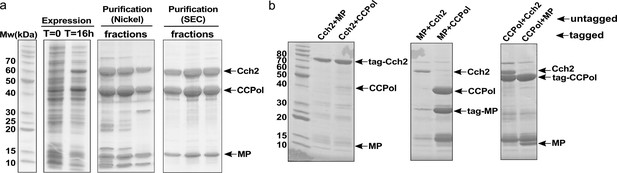
CCPol, MP, and Cch2 interact with one another.
(a) Co-expression and co-purification of His6-CCPol, MP, and Cch2 from the Cch2 operon of a type V SCCmec element; relevant fractions run on SDS-PAGE. Figure 2—figure supplement 2 shows the oligomeric states of individually purified Cch2 and CCPol-MP. (b) Pairwise pulldowns of Cch2 operon proteins. One protein within each pair was tagged with N-terminal His6-SUMO tag and co-expressed with a second, untagged protein. Cleared E. coli lysates were applied to Nickel-sepharose beads and eluted proteins were analyzed by SDS-PAGE. The contaminating band just above MP is most likely prematurely cleaved His6-SUMO tag. See Figure 2—figure supplement 1 for untagged controls.
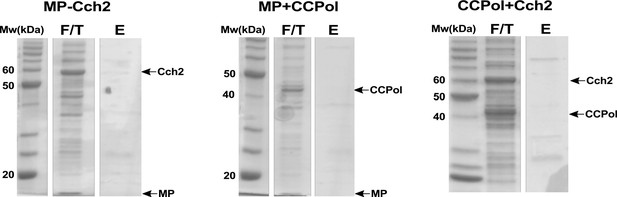
Pairwise pulldowns of Cch2 operon proteins: untagged controls.
In each experiment (performed as those shown in Figure 2b), two proteins were co-expressed with each other. Cleared E. coli lysates were applied to Nickel-sepharose beads and flow-through (F/T) fractions were collected. Proteins were eluted with imidazole (E) and all samples were analyzed on SDS-PAGE.
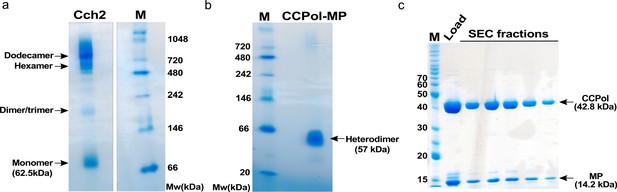
Oligomeric states of purified Cch2 (a) and CCPol-MP (b,c).
(a) Blue native PAGE of Cch2. M denotes a molecular weight marker. Note that under these conditions, the Cch2 bands migrate consistently slower than expected. (b) Blue native PAGE of CCPol-MP. M denotes a molecular weight marker. (c) Comigration of CCPol and MP during size exclusion chromatography on a Superdex 75 gel filtration column, analyzed by SDS-PAGE. M denotes a molecular weight marker.
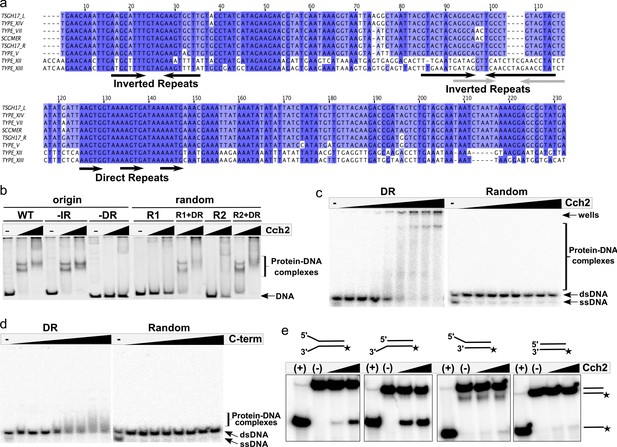
Cch2 binds a specific sequence downstream from its gene and is an active 3’-to-5’ helicase.
(a) Sequence alignment of the intergenic region between the cch2 and ccrC genes, with eight sequences from different S. aureus pattern 2 SCC elements: the two gene clusters (L and R) from a composite SCCmec from TSGH17 strain (SCC accession number AB512767), type V element from WIS strain (AB121219), type VII from JCSC6082 (AB373032), type XII from strain BA01611 (JCSC6082), type XIII from isolate 55-99-44 (MG674089), type XIV from strain SC792 (LC424989), and SSCmer (carrying a mercury resistance cluster) from strain 85/2082 (AB037671). Shading indicates sequence conservation. Inverted repeats (IR) that could potentially form hairpins and direct repeats (DR) that could act as iterons are marked below the sequence; arrows in grey show inverted repeats found in type XII and XIII elements. (b) Cch2 binding to the PCR-amplified putative origin of replication, its derivatives, and synthetic duplexes of random sequence (R1 and R2), without and with (+DR) the direct repeat sequences. Final protein concentrations were 1 and 2 µM in monomers. (c) Cch2 (10–500 nM) binding to 23 bp ds-DNA oligos (5 nM) representing the DR from the putative origin of replication or a random sequence. The experiment was repeated and the results are shown in Figure 3—figure supplement 1a. (d) Binding of the Cch2 C-terminal domain (C-term (10–500 nM)) to 23 bp ds-DNA oligos (5 nM) representing the DR from the putative origin of replication or a random sequence. The experiment was repeated and the results are shown in Figure 3—figure supplement 1b. (e) Helicase assay with four different 60 bp substrates (forked, 3’-overhang, 5’-overhang, and blunt-ended) and Cch2 (2 and 3 µM). The star represents the 32P on the bottom strand. DNA unwinding results in the formation of ssDNA product that can be detected by native PAGE. Figure 3—figure supplement 3 shows additional requirements for Cch2 helicase activity.
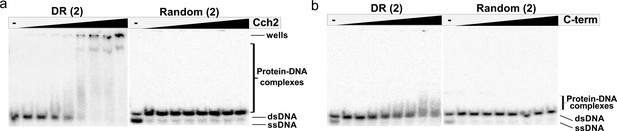
Binding of Cch2 and its C-terminal domain (C-term) to 23 bp ds-DNA oligos representing the DR from the putative origin of replication or a random sequence – repeat of the experiment shown in (a) Figure 3c and (b) Figure 3d.
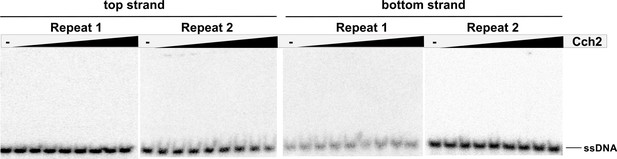
Binding of Cch2 to 23-nt ss-DNA oligos representing the top and bottom strands of the DR from the putative origin of replication.
Two repeats for each top and bottom strand are shown.
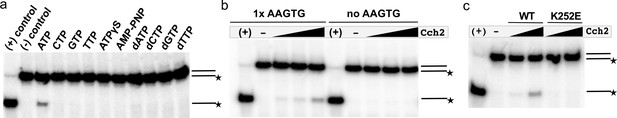
Cch2 helicase activity requires ATP, the AAGTG iteron sequence in the substrate oligo, and catalytic Lys252.
(a) Dependence of Cch2 (3µM) helicase activity on various nucleotide cofactors in the presence of a dsDNA substrate with a 3’ overhang (HELA-5F/HELA-6R; Supplementary file 1 - Table 2). (+) control denotes a sample with no protein added that was subjected to 95 °C heat treatment. (-) control denotes no-protein sample. (b) Dependence of Cch2 (1-3µM) helicase activity on the presence of the iteron sequence AAGTG within the dsDNA substrate containing a 3’ overhang. The AAGTG sequence in the right panel was replaced with unrelated sequence, while the surrounding basepairs remained unchanged (see Supplementary file 1 - Table 2). (+) denotes a positive control sample subjected to 95 °C heat treatment. (c) Cch2 helicase activity assay with wild-type (WT) and mutant (K252E) Cch2 (2-3µM each) in the presence of a dsDNA substrate with a 3’ overhang (HELA-5F/HELA-6R; Supplementary file 1 - Table 2). (+) control denotes a sample with no protein added that was subjected to 95 °C heat treatment. (-) control denotes no-protein sample.
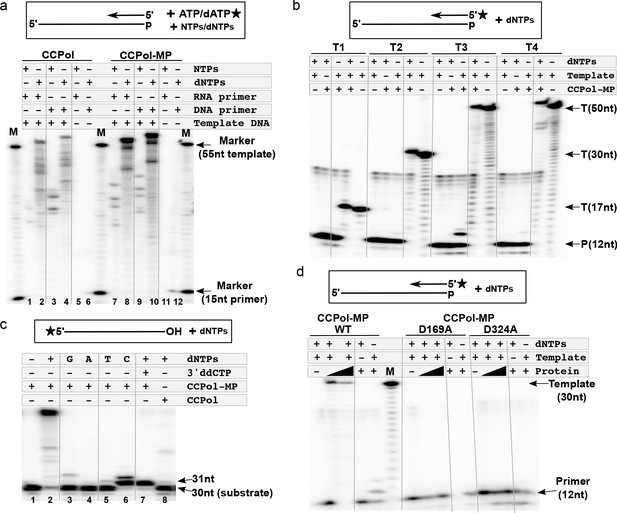
CCPol is an active polymerase.
(a) Primer extension assay in the presence of CCPol-MP or CCPol (5 µM each), with DNA or RNA primers and dNTPs or NTPs. The 15-nt primer was extended in the presence of 55-nt complementary 3’-phosphorylated template and α-32P -labeled dATP/ATP as shown in the diagram; (b) Primer extension of a 32P-labeled DNA primer in the presence of CCPol-MP (5 µM), four different 3’-phosphorylated templates (T1-T4; sequences in Supplementary file 1 under ‘primer extension assays’), and dNTPs as indicated; (c) Terminal deoxynucleotidyl transferase activity of CCPol-MP and CCPol in the presence of individual (‘G’, ‘A’, ‘T’, and ‘C’) or combined (‘+’) dNTPs as indicated. As a control, template with 3’ ddC (preventing addition of dNTPs) was used. (d) Primer extension assay with wild-type CCPol-MP complex and its variants containing predicted catalytic CCPol mutations (5 µM each) extending a 32P-labeled DNA primer in the presence of a 30-nt 3’-phosphorylated template and dNTPs as indicated. Figure 4—figure supplement 1 shows structure-based sequence alignments of CCPol and other bacterial DNA polymerases.

Structure-based sequence alignments of CCPol and the polymerase domains of several bacterial DNA Pol Is.
The alignments were prepared using PROMALS3D multiple sequence and structure alignment server (Pei et al., 2008). The following protein structures were used for the alignments: 6q4u - Thermus aquaticus Taq polymerase residues 423–832; 1l5u - Geobacillus stearothermophilus DNA Pol I residues 469–876; and 1kln - E. coli DNA Pol I residues 520–918. A structural model of CCPol was prepared with I-Tasser (Yang et al., 2015). The polymerase sequences corresponding to the exonuclease domains absent from CCPol were removed from the alignment. The sequences are colored as follows: red – alpha helix, blue – beta strand, gray –E. coli Pol I thumb residues disordered in the structure used. Consensus amino acids are shown in below the structure, with conserved amino acids in bold and uppercase; aliphatic residues (I, V, L): green l; aromatic residues (Y, H, W, F): green @; hydrophobic residues (W, F, Y, M, L, I, V, A, C, T, H): green h; alcohol residues (S, T): o; polar residues (D, E, H, K, N, Q, R, S, T): p; tiny residues (A, G, C, S): t; small residues (A, G, C, S, V, N, D, T, P): s; bulky residues (E, F, I, K, L, M, Q, R, W, Y): b; positively charged residues (K, R, H): blue +; and charged residues (D, E, K, R, H): c. Consensus secondary structures are marked with pink h for alpha helix and blue e for beta strand. Polymerase domains (Thumb, fingers, and palm) are marked underneath the alignment. Predicted mechanistically important residues are marked with red asterisks (with a grey asterisk marking a CCPol alanine residue which in other PolA-like polymerases is invariably a catalytic glutamic acid) above the sequence.
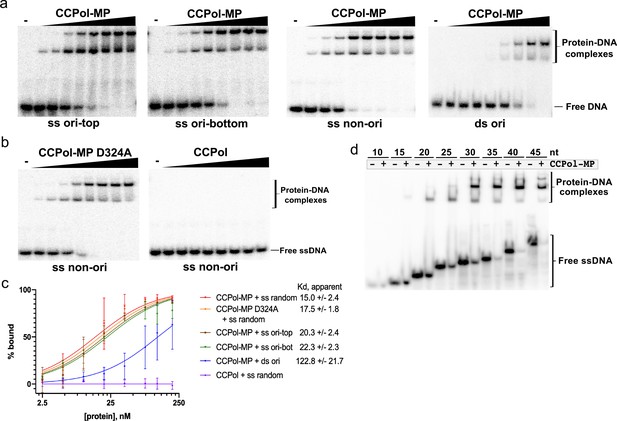
CCPol requires MP for ssDNA binding.
(a) CCPol-MP (0–200 nM) binding to 30-nt ss and dsDNA substrates (2 nM) representing a fragment of the putative origin of replication (ori) or an unrelated sequence (non-ori) as determined by EMSA; (b) Binding to 30-nt ssDNA (2 nM) by CCPol-MP D324A and CCPol (concentrations of 0–200 nM). Figure 5—figure supplement 1 shows the repeats of experiments in (a) and (b); (c) Quantification of DNA-binding assays shown in (a), (b), and Figure 5—figure supplement 1. Two repeats were quantified for each experiment; the mean and standard deviation are shown for each point. The curves show the fit to the data of the equation Y = Bmax*X/(Kd + X), where X equals percent bound, Y equals protein concentration, and Bmax is constrained to 100. For raw data, see Figure 5—source data 1. (d) CCPol-MP (180 nM) binding to ssDNA oligonucleotides ranging in size from 10 to 45 nt.
-
Figure 5—source data 1
DNA binding data for CcPol-MP and CcPol.
- https://cdn.elifesciences.org/articles/55478/elife-55478-fig5-data1-v2.txt
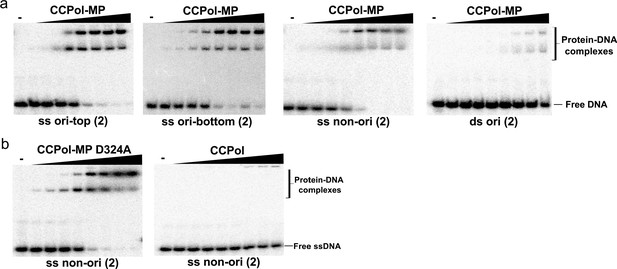
Binding of CCPol, CCPol-MP, and CCPol-MP D324A to 30-nt ss- and ds-DNA oligos representing a fragment of the putative origin of replication (ori) or an unrelated sequence (non-ori) – repeat of the experiments shown in (a) Figure 5a and (b) Figure 5b.
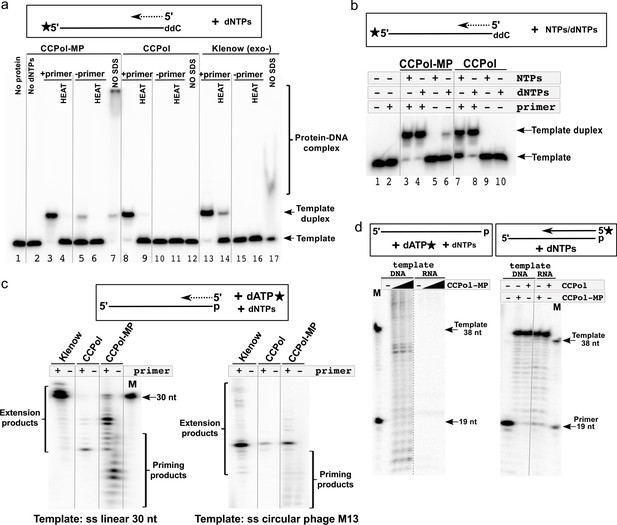
CCPol-MP has primase activity.
(a) Primase assay with CCPol-MP, CCPol, and Klenow (exo-) polymerases. Formation of a template-product duplex was monitored on a non-denaturing gel in the presence or absence of a complementary primer (12 nt). The 3’ end of the template (30 nt) was blocked with ddC. Formation of DNA duplexes was confirmed by denaturation back to ssDNA (‘HEAT’). Samples without added SDS buffer (‘NO SDS’) show native DNA-protein complexes; (b) Primase/primer extension assay on a non-denaturing gel with CCPol-MP and CCPol (5 µM each) in the presence of radiolabeled 30-nt ssDNA template and dNTPs or NTPs as indicated; (c) Priming/primer extension assay with short (30 nt) linear (left) and phage M13 circular ssDNA (right) templates in the presence of α-32P dATP, in the presence or absence of a primer (12 and 24 nt, respectively); (d) Primase (left panel) and primer extension (right panel) assays in the presence of a DNA or RNA template and CCPol-MP or CCPol as indicated. M = markers with sizes corresponding to the primer (19 nt) and template (38 nt). The dashed grey line indicates that irrelevant lanes were removed from the gel; the solid grey lines were added to guide the eye.
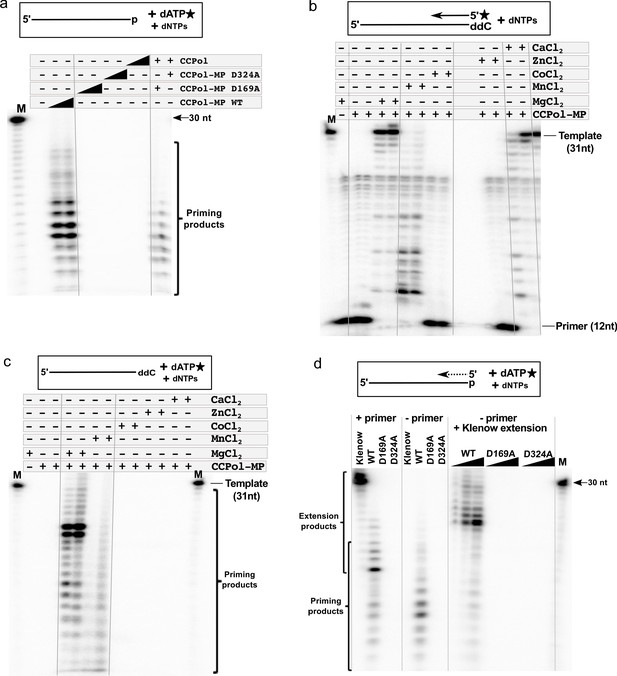
CCPol active site and divalent metal ions are required for primase activity.
(a) Primase assay with CCPol-MP, CCPol-MP with the CCPol active site mutated, and CCPol alone (concentrations: 2 and 5 µM) in the presence of 30-nt ssDNA template and α-32P dATP. The last two lanes show a complementation assay where CCPol-MP mutants (2.5 µM) were supplemented with WT CCPol (2.5 µM); (b) Primer extension of a 32P-labeled DNA primer (12 nt) in the presence of CCPol-MP (1 and 2 µM) and a 30-nt template with the 3’ end blocked with ddC, in the presence or absence of selected divalent metal ions (20 mM); (c) Primase assay with CCPol-MP, a template (30 nt) with the 3’ end blocked with ddC, and α-32P dATP, in the presence or absence of selected divalent metal ions (20 mM); (d) Primase/primer extension with Klenow (exo-), CCPol-MP, and its two mutant variants D169A and D324A (5 µM each in the left panel, 1–5 µM each in the right panel), with a 30-nt template and α-32P dATP in the presence (left panel) or absence (middle panel) of a primer. Some of the priming reactions were then further incubated with Klenow (exo-) to allow elongation of the CCPol-MP – made primers (right panel). Unless otherwise noted, 5 µM CCPol-MP or CCPol and 5U Klenow were used in the experiments shown in this figure.
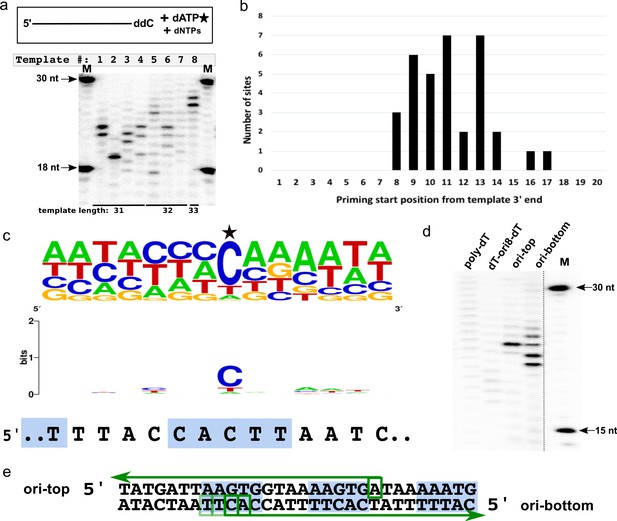
CCPol-MP priming start sequence preference.
(a) CCPol-MP prime-then-elongate assay with eight different templates (31–33 nt including 3’ ddC, as shown) with random sequences in the presence of radiolabeled dATP. Primers synthesized by CCPol-MP were further elongated by the complex upon addition of excess of dNTPs. Priming start sites can be mapped by reading the product size off the gel, then subtracting one nt to account for the terminal nucleotide added by CCPol-MP, and mapping the product length to the template starting at its 5’ end; (b) Positions of 34 CCPol-MP priming start sites (Supplementary file 1) identified as in (a) with respect to the 3’ end of the template; (c) Sequence consensus logo of the priming start sites identified as in (a). 39 unique priming start sites (Supplementary file 1) from 25 different templates are included in the logo, with the start site marked with a star. The sequence of direct repeat region within the putative origin of replication is aligned under the logo, with the 5-nt iterons marked by blue boxes; (d) CCPol-MP prime-then-elongate assay as in (a) with 30-nt templates (each with a 3’ phosphate) as indicated; (e) Mapping of CCPol-MP priming start sites on the templates representing the putative origin of replication (ori-top and ori-bottom) based on the band position on the gel shown in (d). Green arrows represent the elongated DNA products, with green boxes marking the identified start sites. The darkness of the boxes corresponds to the efficiency of priming at that site as judged by the brightness of the bands in (d). Blue boxes mark the iterons within the putative origin of replication.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Staphylococcus aureus) | SCCmec | GenBank | AB512767.1 | Mobile genetic element |
| Gene (Staphylococcus aureus) | CCPol | GenBank | AB512767.1 ORF TS025; BAK57481.1 | |
| Gene (Staphylococcus aureus) | MP | GenBank | AB512767.1 ORF TS025; BAK57482.1 | |
| Gene (Staphylococcus aureus) | Cch2 | GenBank | AB512767.1 ORF TS025; BAK57483.1 | |
| Strain, strain background (Escherichia coli) | DH5alpha | Thermo Fischer | Cat # 18265017 | Cloning strain (chemically competent cells) |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | Novagen (Sigma-Aldrich) | Cat # 69450 | Expression strain (chemically competent cells) |
| Strain, strain background (Escherichia coli) | Rosetta (DE3) pLysS | Novagen (Sigma-Aldrich) | Cat # 70956 | Expression strain (chemically competent cells) |
| Recombinant DNA reagent | pET21a (plasmid) | Novagen (Sigma-Aldrich) | Cat # 69740 | Expression plasmid |
| Recombinant DNA reagent | pET28a (plasmid) | Novagen (Sigma-Aldrich) | Cat # 69864 | Expression plasmid |
| Recombinant DNA reagent | pUC19 (plasmid) | NEB | Cat # N3041 | Cloning plasmid |
| Recombinant DNA reagent | pET28-SUMO | This work | Expression plasmid with N-terminal SUMO tag introduced. Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET21-CCPol | This work | CCPol expression plasmid (C-terminal His6 tag). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-Cch2 | This work | Cch2 expression plasmid (TEV-cleavable N-terminal His6 tag). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-Cch2_operon | This work | CCPol-MP-Cch2 operon expression plasmid (TEV-cleavable N-terminal His6 tag on CCPol). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO-CCPol+MP | This work | CCPol-MP expression plasmid (terminal His6-SUMO tag on CCPol). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO-MP+CCPol | This work | MP-CCPol expression plasmid (terminal His6-SUMO tag on MP). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO-CCPol+Cch2 | This work | CCPol-Cch2 expression plasmid (terminal His6-SUMO tag on CCPol). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO- Cch2+CCPol | This work | Cch2-CCPol expression plasmid (terminal His6-SUMO tag on Cch2). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO- Cch2+MP | This work | Cch2-MP expression plasmid (terminal His6-SUMO tag on Cch2). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO-MP+Cch2 | This work | MP-Cch2 expression plasmid (terminal His6-SUMO tag on MP). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO- MP+CCPol | This work | MP-CCPol expression plasmid (no tags). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO- CCPol+Cch2 | This work | CCPol-Cch2 expression plasmid (no tags). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pET28-SUMO- MP+Cch2 | This work | MP-Cch2 expression plasmid (no tags). Plasmid available upon reasonable request. | |
| Recombinant DNA reagent | pUC19-ori | This work | Plasmid containing the intergenic region between Cch2 and CcrC (putative origin of replication). Plasmid available upon reasonable request. | |
| Sequence-based reagent | Synthesized oligonucleotides | IDT | Primers/oligonucleotides | For sequences, see Supplementary file 1 |
| Commercial assay or kit | QuikChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat # 210519 | |
| Software, algorithm | ImageJ | NIH | https://imagej.nih.gov/ij/ | |
| Software, algorithm | Graphpad Prism | Graphpad Software, Inc, La Jolla, CA | http://www.graphpad.com/ |
Additional files
-
Supplementary file 1
Supplementary tables.
Table 1. Primers used in the study. Table 2. Oligonucleotides used for in vitro assays. Table 3. Primase start sites identified in the study.
- https://cdn.elifesciences.org/articles/55478/elife-55478-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55478/elife-55478-transrepform-v2.docx





