A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat
Figures
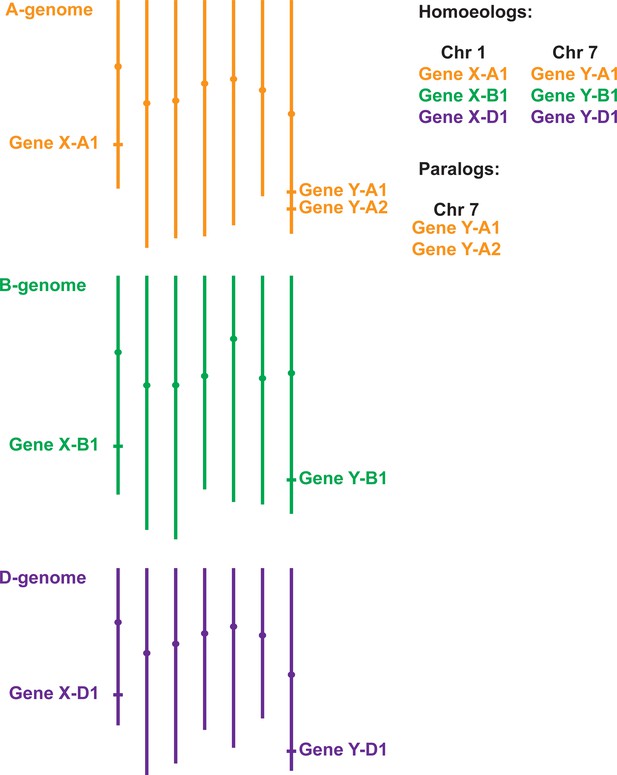
Gene homology within polyploid wheat.
Due to two separate hybridisation events, genes in polyploid wheat will be present in multiple copies called homoeologs, which usually have similar chromosome locations. In the example of hexaploid bread wheat illustrated here, Gene X has homoeologs on chromosomes 1A, 1B and 1D. Duplicated genes, called paralogs (e.g. two copies of Gene Y on chromosome 7A), have evolved either within wheat or in one of its ancestral species. Most paralogs arise from intra-chromosomal duplications, although inter-chromosomal duplications can also occur.
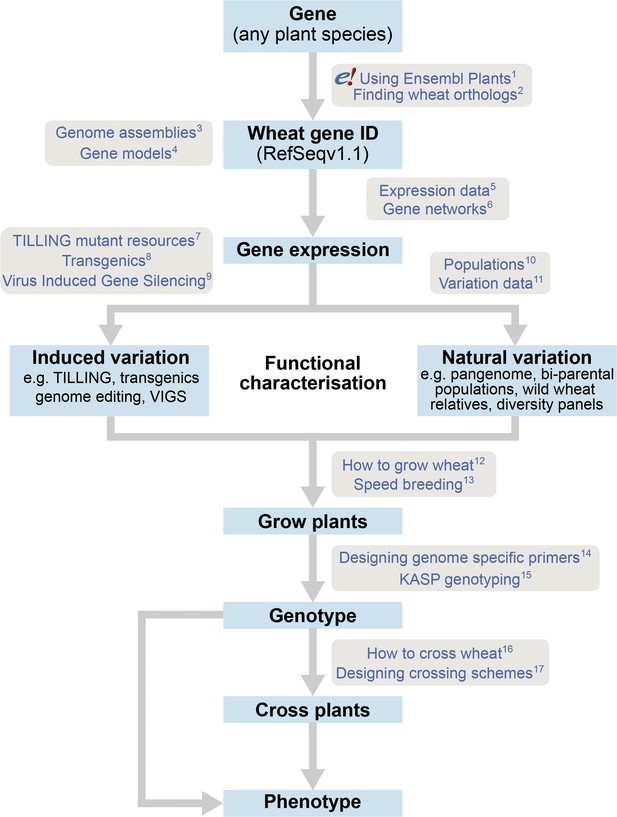
The roadmap for gene characterisation in wheat.
Overview of a proposed strategy to take a gene from any plant species, identify the correct wheat ortholog(s) using Ensembl Plants (https://plants.ensembl.org) and determine gene expression using expression browsers and gene networks. Suggestions for functional characterisation are provided including induced variation such as mutants, transgenics or Virus-Induced Gene Silencing (VIGs). In addition, publicly available populations incorporating natural variation are available. Finally steps for growing, genotyping and crossing plants are outlined. Links to detailed tutorials and further information are provided and can be found on www.wheat-training.com. (1) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/EnsemblPlants-primer.pdf (2) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/Finding-wheat-orthologs.pdf (3) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/Genome_assemblies.pdf (4) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/Gene-models.pdf (5) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/Expression-browsers.pdf (6) www.wheat-training.com/wp-content/uploads/Genomic_resources/pdfs/Gene-networks.pdf (7) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Selecting-TILLING-mutants.pdf (8) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Transgenics.pdf (9) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Virus_Induced_Gene_Silencing.pdf (10) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Populations.pdf (11) www.wheat-training.com/wp-content/uploads/Genomic_resources/Variation-data.pdf (2) www.wheat-training.com/wp-content/uploads/Wheat_growth/pdfs/Growing_Wheat_final.pdf (13) www.wheat-training.com/wp-content/uploads/Wheat_growth/pdfs/Speed_breeding.pdf (14) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Designing-genome-specific-primers.pdf (15) https://www.biosearchtech.com/support/education/kasp-genotyping-reagents/running-kasp-genotyping-reactions (16) http://www.wheat-training.com/wp-content/uploads/Wheat_growth/pdfs/How-to-cross-wheat-pdf.pdf (17) www.wheat-training.com/wp-content/uploads/Functional_studies/PDFs/Designing-crossing-schemes.pdf.
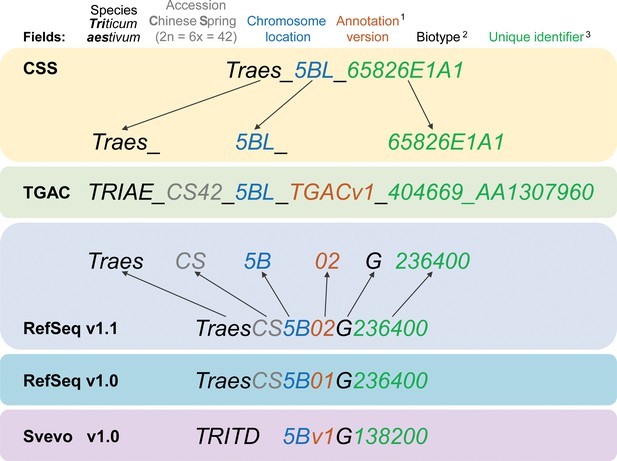
Gene model ID nomenclature description from the five available gene annotations for domesticated polyploid wheat.
Here, one gene is used as an example to highlight the differences in gene ID nomenclature. Fields represented in the nomenclature are shown at the top with matching colours for the corresponding features in the gene names. Yellow background shows the CSS gene names with dark grey arrows pointing towards the corresponding field in the TGAC gene annotation (TGACv1, green background). Blue backgrounds show the gene nomenclatures for RefSeqv1.0 and v1.1 annotations (as used in Ensembl Plants), while the lilac background shows the nomenclature for Svevo v1.0 (modern durum wheat). (1) Two annotation versions are available for the RefSeqv1.0 genome assembly: RefSeqv1.0 (release annotation) and RefSeqv1.1 (improved annotation). These are differentiated by the annotation version number; ‘01’ for RefSeqv1.0 and ‘02’ for RefSeqv1.1. Otherwise, the annotations follow the same rules. (2) In the RefSeq and Svevo annotations, the biotype is represented by an additional identifier, where G = gene. (3) In the RefSeqv1.0 and v1.1 annotation, identifiers are progressive numbers in steps of 100 reflecting the relative position between gene models. For example, gene TraesCS5B02G236400 would be adjacent to gene TraesCS5B02G236500. However, it is important to note that the relative positions of genes may change in future genome releases as the assembly is improved, for example, if scaffolds are rearranged. In these cases, the gene order would no longer be retained. In the gene annotation for the tetraploid durum wheat cv. Svevo, the species name is TRITD (TRITicum Durum) and gene identifiers increase in steps of 10, rather than by steps of 100 as in the RefSeq hexaploid wheat annotation. Note that RefSeqv1.0 and v1.1 comprises High Confidence (HC) and Low Confidence (LC) gene models. Low Confidence gene models are flagged by the ‘LC’ at the end (not shown). HC and LC genes which otherwise display the same unique identifier are not the same locus and are not in sequential order. Hence, TraesCS5B02G236400 and TraesCS5B02G236400LC are both located on chromosome 5B, but are not the same gene nor are they physically adjacent. Similarly, genes from homoeologous chromosomes with the same subsequent numeric identifier are not necessarily homoeologous genes. For example, TraesCS5A02G236400, TraesCS5B02G236400 and TraesCS5D02G236400 are not homoeologous genes.
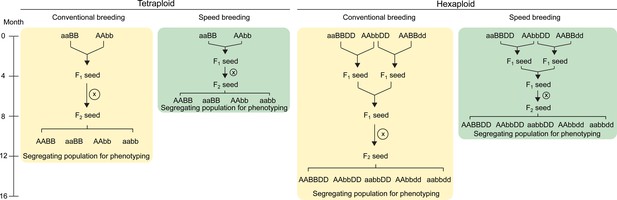
Crossing scheme to combine TILLING or CRISPR/Cas9 single mutants in wheat.
In tetraploid wheat, mutations in the A and B genome homoeologs can be combined through a single cross. The F1 plants are self-pollinated to produce a segregating F2 population which contains homozygous double and single mutants, as well as wild type plants (screening using molecular markers required; only four genotypes shown). These F2 progeny can be characterised for the phenotype of interest. The use of ‘speed breeding’ (Watson et al., 2018), reduces the time taken to reach this phenotyping stage from 12 (yellow) to 7.5 months (green). In hexaploid wheat, a second round of crossing is required to combine the mutant alleles from all three homoeologs. The F2 progeny segregating for the three mutant alleles can be genotyped using molecular markers to select the required combination of mutant alleles (only five genotypes shown; all factorial combinations are possible). Speed breeding reduces the time taken to generate triple homozygous mutants for phenotyping to 10 months (green), compared to 16 months in conventional conditions (yellow). Self-pollination is represented by an X inside a circle. Combinations of wild type alleles from the A (AA), B (BB) and D (DD) genomes, as well as the mutant alleles from each genome (aa, bb and dd, respectively) are indicated.
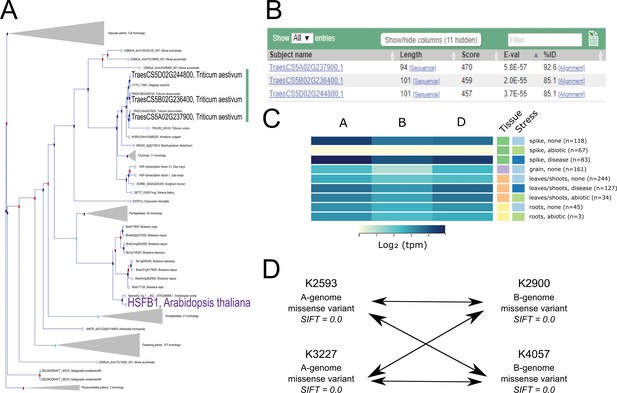
Case study exemplifying use of available resources for gene functional characterisation in wheat.
(A). The Ensembl Plants Gene Tree illustrates the identification of the wheat triad (green bar) most closely related to AtHSFB1 (shown in purple). (B) Using Os09g0456800 (the rice ortholog of AtHSFB1) as a BLASTp query against wheat predicted proteins independently identifies the same wheat triad. (C) Examination of RNA expression data from www.wheat-expression.com shows that the wheat triad is most highly expressed in the spike, with differential expression in abiotic and disease stress conditions. The samples are identified by tissue of origin (spike, green; grain, purple; leaves/shoots, orange; roots, yellow) and stress (none, light blue; abiotic, green; disease, dark blue) as they are on the website. (D) After identification of suitable wheat TILLING mutants, A and B genome homoeologs are combined via this example crossing scheme, demonstrating the four crosses required between the two selected mutations in each homoeolog. Note that the functional validation proposed in (D) is carried out using the tetraploid mutant population.
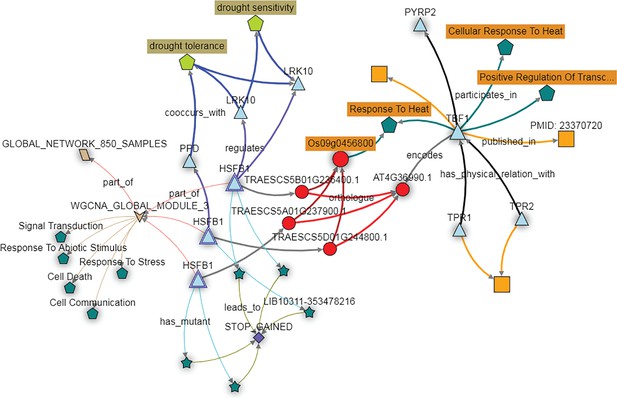
The KnetMiner network illustrates the putative role of the wheat TBF1 orthologs in responding to abiotic stress.
The wheat orthologs of the Arabidopsis gene TBF1, here depicted as three copies of the gene HSFB1 (light blue triangles) fall in expression module three (brown arrow; WGCNA module 3). The genes in this module are enriched for GO terms such as ‘Response to Stress’ and ‘Response to Abiotic Stimulus’ (dark green pentagons). The HFSB1 homoeologs are predicted to regulate other genes (blue triangles) in the GENIE3 network (purple connecting arrows) which are associated with the drought tolerance trait ontology terms (light green pentagon). PTC mutations are available for all three HFSB1 homoeologs (dark green stars connecting with STOP GAINED SNP effect) in the Cadenza population.
Tables
Comparison of annotated genome assemblies in hexaploid and tetraploid wheat.
| CSS | TGACv1 | RefSeqv1.0 | Durum wheat | Wild emmer wheat | |
|---|---|---|---|---|---|
| Publication | Mayer et al., 2014 | Clavijo et al., 2017 | The International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018 | Maccaferri et al., 2019 | Avni et al., 2017 |
| Contigs/Chromosomes | >1 million | 735,943 | 21 chromosomes + ChrU | 14 chromosomes + ChrU | 14 chromosomes + ChrU |
| Mean scaffold size | 7.7 kbp | 88.7 kbp | Chromosomes | Chromosomes | Chromosomes |
| Assembly Size | 10.2 Gbp | 13.4 Gbp | 14.6 Gbp | 10.5 Gbp | 10.5 Gbp |
| Order | Synteny/genetic order† | Large Bins | Physical order | Physical order | Physical order |
| Coding genes* | 133,090 HC 88,998 LC | 104,091 HC 103,660 LC | 107,891 HC 161,537 LC | 66,559 HC 303,404 LC | 67,182 HC 271,179 LC |
| Assembly-related resources | Archive Ensembl Plants | Archive Ensembl Plants | Ensembl Plants GrainGenes, URGI | Ensembl Plants GrainGenes | Ensembl Plants GrainGenes |
| TILLING mutants | TILLING mutants | ||||
| expVIP, wheatExp | expVIP | expVIP, eFP | |||
| Accession | Chinese Spring | Chinese Spring | Chinese Spring | Svevo | Zavitan |
-
*Number of high confidence (HC) and low confidence (LC) genes which are defined based on multiple criteria outlined in the published papers. Care must be taken when interpreting their nomenclature (see Figure 3).
†Chromosome arm assignment was derived from chromosome flow-sorting, while approximate intra-chromosomal ordering was established using synteny derived from grasses (GenomeZipper) and genetic mapping (POPSEQ) (Mascher et al., 2013; Mayer et al., 2014).
Natural variation resources available in wheat.
| Collection | Short description | Number of accessions | Genotyping | Data/seed availability | More information/Reference |
|---|---|---|---|---|---|
| Wild wheat relatives and progenitor species | |||||
| Seeds of Discovery | Wheat and wild relative accessions held by ICARDA and CIMMYT | 80,000 accessions: 56,342 domesticated hexaploid (eight taxa); 18,946 domesticated tetraploid (eight taxa); 3,903 crop wild relatives included all known 27 wild species from Aegilops-Triticum species complex and 11 genomic constitutions. | DArT-seq | CIMMYT Dataverse http://hdl.handle.net/11529/10548030 Germinate data warehouse http://germinate.cimmyt.org/wheat. Records for all germplasm accessions can also be accessed at https://ssl.fao.org/glis/ | https://seedsofdiscovery.org/ |
| Open Wild Wheat | Accessions of Aegilops tauschii (D genome progenitor) | 265 accessions | Whole genome shotgun sequenced (10-30x) | Sequencing: https://opendata.earlham.ac.uk/wheat/under_license/toronto/; Seed: https://www.seedstor.ac.uk/search-browseaccessions.php?idCollection=38 | www.openwildwheat.org; Arora et al., 2019 |
| Wild wheat introgression lines | Introgression lines from Aegilops caudata, Aegilops speltoides, Amblyopyrum muticum, Thinopyrum bessarabicum, Thinopyrum elongatum, Thinopyrum intermedium, Thinopyrum ponticum, Triticum timopheevii, Triticum urartu, rye and wheat cultivars (Chinese Spring, Higbury, Paragon, Pavon 76) | 153 stable homozygous introgression lines available | 35K Axiom Wheat Relative Genotyping array + 710 KASP markers (Grewal et al., 2020) | Genotype: https://www.nottingham.ac.uk/wrc/germplasm-resources/genotyping.aspx; Seed: https://www.seedstor.ac.uk/ (accessions WR0001-WR0155) | www.nottingham.ac.uk/WISP; Grewal et al., 2018a; Grewal et al., 2018b, King et al., 2018, King et al., 2017 |
| Synthetic hexaploid wheat | |||||
| Synthetic hexaploid wheat | Sythetic hexaploid wheats generated using Aegilops tauschii (DD) + European tetraploid (AABB) wheat | 50 synthetic hexaploid wheats + pre-breeding accessions; backcross populations with Robigus and Paragon also available | 35K Axiom breeders array | Genotype: https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/axiom_download.php Seed: https://www.seedstor.ac.uk/ (store codes WS0001-WS0232) | https://www.niab.com/research/research-projects/designing-future-wheat |
| Wheat diversity panels | |||||
| Watkins historic collection of landrace wheats | World collection of wheat landraces grown as farmer saved seed before the 1930s. Genetically stable collection developed by two generations of single seed descent | 829 accessions (core set of 119 represent majority of assayed genotypic variation). F4:5 mapping populations against Paragon, mainly for the core set. | 35K Axiom breeders array (Allen et al., 2017); subset exome sequenced (Gardiner et al., 2018) | Genotype: https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/axiom_download.php Seed: https://www.seedstor.ac.uk/ (store codes WATDE0001-WATDE1063) | http://wisplandracepillar.jic .ac.uk/results_resources.htm ; Wingen et al., 2014; Wingen et al., 2017 |
| GEDIFLUX (Genetic Diversity Flux) collection | Western European winter wheat varieties that individually occupied over 5% of national acreage from 1945 to 2000. Bi-parental populations with Paragon (ongoing) | 479 accessions | 35K Axiom breeders array | Genotype: https://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/axiom_download.php; Seed: https://www.seedstor.ac.uk/ (store codes WGED0001- WGED0729) | http://wisplandracepillar.jic.ac.uk/results_resources.htm; Wingen et al., 2014 |
| NIAB wheat association mapping panel | Bread wheat varieties released between 1916–2007. Predominantly UK varieties (68%), also other North Western European countries e.g. France (10%) and Germany (8%) | 480 accessions | 90K SNP array | Seed, Genotype and Pedigree: https://www.niab.com/research/research-projects/resources | Fradgley et al., 2019 |
| OzWheat diversity panel | Genetic diversity in Australian wheat breeding (colonial landraces 1860s, first Australian-bred cultivars 1890s, CIMMYT-derived semi dwarfs 1960s, post 2000 wheat) | 285 accessions | 90K SNP array + additional 26K SNPs from transcriptome data | Seed and Genotype: contact Shannon Dillon from CSIRO (Shannon.Dillon@csiro.au) | |
| Vavilov wheat collection | Hexaploid wheat accessions including landraces, historic breeding lines and cultivars. Pure lines generated by single seed descent | 295 accessions | DArtT-seq (34,311 polymorphic markers) | Genotype: Lee Hickey at The University of Queensland (l.hickey@uq.edu.au) Seed: Australian Grains Genebank (sally.norton@ecodev.vic.gov.au) | Riaz et al., 2017 |
| WHEALBI wheat panel | Worldwide wheat accessions including diploid and tetraploid wild relatives, old hexaploid landraces and modern elite cultivars | 487 accessions | Exome capture (~600,000 genetic variants in ~40,000 genes; 12,000 genes identified as putative presence/absence variation compared to RefSeqv1.0) | Genotype:https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/iwgsc_refseqv1.0_Whealbi_GWAS.zip; Seed: https://www.gbif.org/dataset/a52ca10a-136a-4072-a6de-3ec6e7852365 | Pont et al., 2019 |
| Global Durum Wheat (GDP) panel | Diversity used in durum wheat breeding programs globally, including landraces and modern varieties | 1,056 accessions | 90K SNP array | Genotype: ms in preparation; Seed: ICARDA genebank http://indms.icarda.org Filippo Bassi, F.Bassi@cgiar.org | |
| Tetraploid wheat Global Collection (TGC) | Wild emmer wheat, domesticated emmer, durum wheat landraces and other tetraploid wheat sub-species (Triticum aethiopicum, Triticum carthlicum, Triticum polonicum, Triticum turanicum, Triticum turgidum, Triticum karamyschevii and Triticum petropavlovsky) | 1,856 accessions | 90K SNP array | Genotype: GrainGenes; Seed: on request for non-commercial use from University of Bologna (marco.maccaferri@unibo.it and roberto.tuberosa@unibo.it) | Maccaferri et al., 2019 |
| MAGIC populations | |||||
| CSIRO, Aus | 4-way (parents Baxter, Chara, Westonia, Yitpi); 8-way (parents Baxter, Westonia, Yitpi, AC Barrie (Canada), Xiaoya54 (China), Volcani (Israel), Pastor (Mexico), Alsen (USA)) | 1,500 (4-way) and 3,000 (8-way) RILs | 90K SNP array, microsatellite and DArT markers > 20,000 SNPs mapped in each population | Seed and Genotype: on request from CSIRO (Bill.Bovill@csiro.au) | Huang et al., 2012; Shah et al., 2019 |
| NIAB, UK | 8-way (parents Alchemy, Brompton, Claire, Hereward, Rialto, Robigus, Xi19, Soissions); 16-way (Banco, Bersee, Brigadier, Copain, Cordiale, Flamingo, Gladiator, Holdfast, Kloka, Maris Fundin, Robigus, Slejpner, Soissons, Spark, Steadfast, Stetson) | NIAB 8-way MAGIC:>1,000 RILs; NIAB 16-way MAGIC: ~600 RILs | 35K Axiom breeders array. Genome sequence (Claire, Robigus, others underway). Exome capture sequence of 16-way parents. Skim-seq of all RILs underway. | Claire and Robigus genomes: https://opendata.earlham.ac.uk/opendata/data/Triticum_aestivum/EI/v1.1/ Genotyping and Seed: https://www.niab.com/research/research-projects/resources | Mackay et al., 2014; Gardner et al., 2016 |
| Germany | 8-way (Event, Format, BAYP4535, Potenzial, Ambition, Bussard, Firl3565, Julius) | 394 F6:8 RILs | 5,435 SNPs from SNP array | Genotype and pedigree: http://doi.org/10.14459/2018mp1435172 (click the ‘open attachment browser’ link); Seed: Bavarian State Research Centre for Agriculture (Freising, Germany) | Stadlmeier et al., 2018 |
| Germany | WM-800, 8-way (Patras, Meister, Linus, JB Asano, Tobak, Bernstein, Safari, Julius) | 910 F4:6 RILs | 15K Infinium iSelect SNP array | Genotype and pedigree: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6069784; Seed: on request from Martin Luther University, Germany (klaus.pillen@landw.uni-halle.de) | Sannemann et al., 2018 |
| Durum | 4-way (Claudio (Italy), Colosseo (Italy), Neodur (France), Rascon/2*Tarro (advanced CIMMYT line)) | 334 F7:8 RILs | 90K SNP array | Genotype and pedigree: https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.12424; Seed: on request for non-commercial use from University of Bologna (marco.maccaferri@unibo.it and roberto.tuberosa@unibo.it) | Milner et al., 2016 |
Tetraploid and hexaploid wheat genome assemblies that are currently available, in addition to the Chinese Spring reference hexaploid genome.
| Variety | Habit | Origin | Availability † |
|---|---|---|---|
| Hexaploid wheat | |||
| CDC Landmark | spring | Canada | 10+ Genome Project |
| CDC Stanley | spring | Canada | 10+ Genome Project |
| Paragon | spring | UK | 10+ Genome Project |
| Cadenza | spring | UK | 10+ Genome Project |
| LongReach Lancer | spring | Australia | 10+ Genome Project |
| Mace | spring | Australia | 10+ Genome Project |
| Synthetic W7984 | spring | Mexico | Chapman et al. (2015) |
| Weebill | spring | Mexico | 10+ Genome Project |
| ArinaLrFor | winter | Switzerland | 10+ Genome Project |
| Julius | winter | Germany | 10+ Genome Project |
| Jagger | winter | US | 10+ Genome Project |
| Robigus | winter | UK | 10+ Genome Project |
| Claire | winter | UK | 10+ Genome Project |
| Norin61 | winter | Japan | 10+ Genome Project |
| SY Mattis | winter | France | 10+ Genome Project |
| Spelt (PI190962) | winter | Europe | 10+ Genome Project |
| Tetraploid wheat | |||
| Zavitan* | - | Israel | Avni et al. (2017) |
| Svevo | spring | Italy | Maccaferri et al., 2019 |
| Kronos | spring | US | 10+ Genome Project |
-
*‘Zavitan’ is a tetraploid wild emmer (T. dicoccoides) accession.
†Varieties included within the 10+ Wheat Genomes Project can be accessed through the Earlham Grassroot Genomics portal (https://wheatis.tgac.ac.uk/grassroots-portal/blast) and the 10+ Wheat Genomes project portal (http://webblast.ipk-gatersleben.de/wheat_ten_genomes) (subset of varieties in each). The ‘Svevo’ genome can be accessed through https://www.interomics.eu/durum-wheat-genome and Ensembl Plants. ‘Synthetic W7984’ and ‘Zavitan’ can be accessed through the Grassroot Genomics, and Ensembl Plants, respectively.





