Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction
Figures
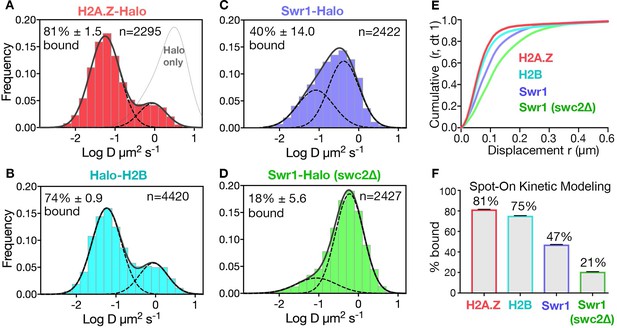
Diffusive behaviors of protein fusions to HaloTag (Halo) reveal chromatin-bound and free populations in live yeast.
(A, B) Normalized histograms and two-component Gaussian fits for H2A.Z-Halo (A) and Halo-H2B (B) show the log diffusion coefficient distributions. The Gaussian fit for HaloTag is shown for reference (‘Halo only’ in A). (C, D) Normalized histograms and two-component Gaussian fits for Swr1-Halo in WT cells (C) and the swc2Δ mutant (D). Solid line: sum of two-component fit; dashed line: individual component. Percent value of the slow component along with Bootstrap resampling errors and the number of trajectories (n) are indicated. (E) Cumulative distribution functions (CDF) of 10 ms displacements. (F) Spot-On results with fitting errors showing fractions of chromatin-bound molecules derived from modeling CDFs over 10–50 ms intervals. All molecules tracked with JF646 dye except Halo only, which was imaged with JF552.
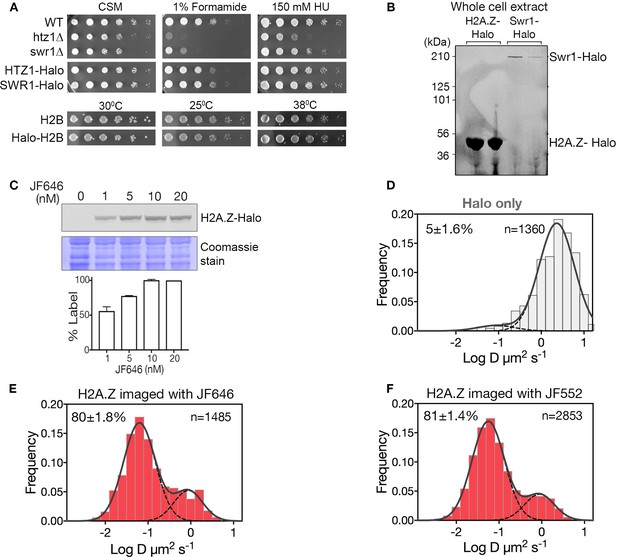
Cell growth, labeling and SPT analysis of Halo-tagged proteins.
(A) Growth of strains bearing SWR1, H2A.Z (HTZ1) and H2B (HTB1) fusions to HaloTag. Saturated cultures at optical density 1.0 were spotted (1:5 serial dilutions) on CSM plates with or without 1% formamide or 150 mM HU (hydroxyurea) and incubated for 2–3 days at the indicated temperatures. WT and mutant strains htz1Δ and swr1Δ are shown for comparison. (B) Cells expressing H2A.Z-Halo and Swr1-Halo were stained with JF646 and the cell lysate was resolved on SDS-PAGE. Fluorescent scan of duplicate lanes show specific labeling of Halo-tagged H2A.Z and Swr1 proteins. (C) SDS-PAGE shows 10 nM JF646 saturates H2A.Z-Halo in yeast cells in a routine 2 hr staining period. (D) Normalized histogram and two-component Gaussian fit for HaloTag (fused to NLS). The slow fraction is 5%. (E) Top: Profile of H2A.Z-Halo diffusivity in biological replicate, same condition as Figure 1A experiment. (F) Profile of H2A.Z-Halo diffusivity in cells stained with JF552. All molecules tracked with JF552 dye except (E), which was tracked with JF552.
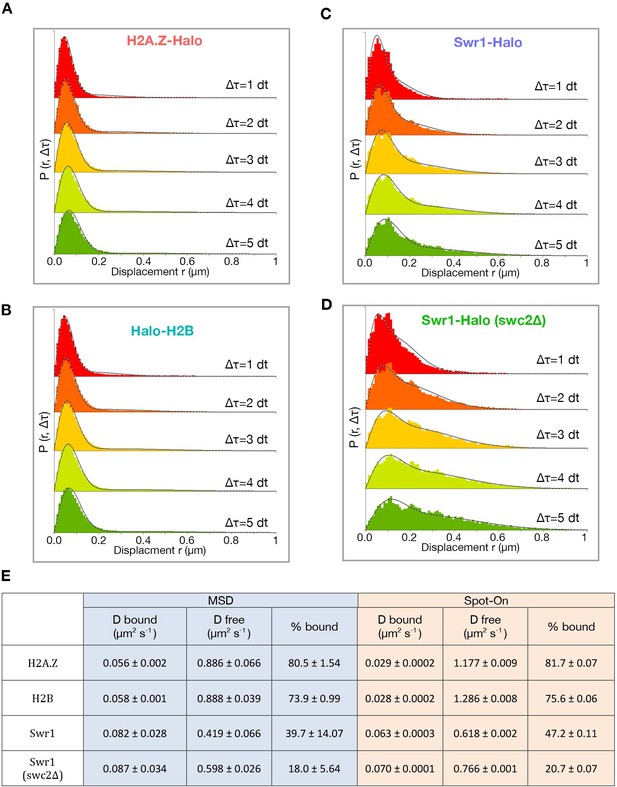
Spot-On kinetic modeling analysis.
(A) Histogram of displacements over time intervals of 10, 20, 30, 40 and 50 ms (dt 1–5) for H2A.Z. First four displacements were included for each track. Kinetic fitting shown as dashed line and measured displacements in color. Data were analyzed using the Spot-On web-interface (https://SpotOn.berkeley.edu). (B) Spot-On analysis of H2B. (C,D) Spot-On analysis of Swr1 in WT and swc2Δ cells. (E) Comparison of diffusive parameters for H2A.Z, H2B, Swr1 and Swr1 in the swc2Δ strain, extracted from MSD-based and Spot-On analytic platforms. Bootstrap resampling errors shown for MSD and fitting errors shown for Spot-On. All molecules tracked with JF646 dye.
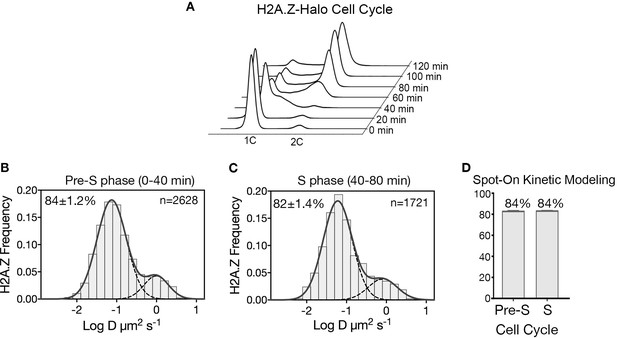
H2A.Z-Halo distribution in cell division cycle.
(A) FACS analysis shows DNA content of the synchronized cell population upon time of release from α-factor arrest, from 0’ – 120.’ (B, C) Normalized histogram and two-component Gaussian fit for H2A.Z-Halo in cells synchronized in pre-S phase, and in S phase. (D) Spot-On results shows both Pre-S and S phase cells have 84% chromatin bound H2A.Z. All molecules tracked with JF646 dye.
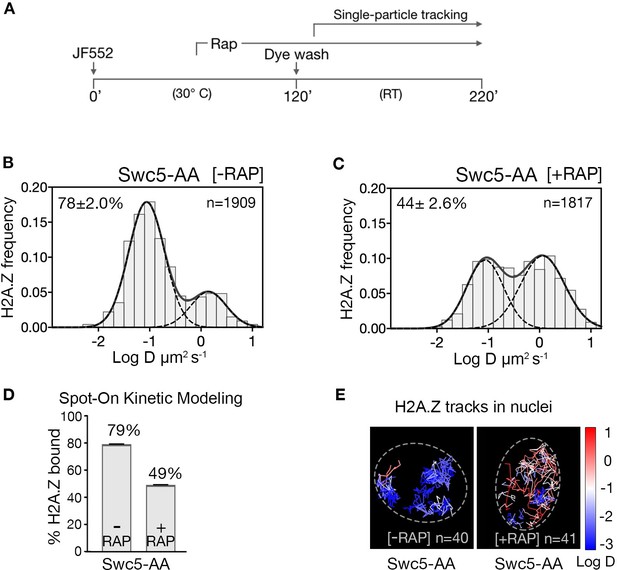
H2A.Z chromatin binding is substantially reduced upon abrogation of the deposition pathway by SWR1 inactivation.
(A) Time course of H2A.Z-Halo labeling, rapamycin treatment and image acquisition in Swc5-AA cells. Rapamycin treatment for an hour before SPT, and imaging performed in continued presence of rapamycin. (B, C) Normalized histograms and two-component Gaussian fits for H2A.Z-Halo imaged in the Swc5-AA cells. Imaging data were acquired in absence of rapamycin (B) or presence of rapamycin (C). Spot-On results show that Swc5 depletion causes a reduction in chromatin-bound H2A.Z. (E) Overlay of tracks, color-coded according to log diffusion coefficients, obtained from representative nuclei. Number of tracks (n) is indicated for each nucleus. All molecules tracked with JF552 dye.
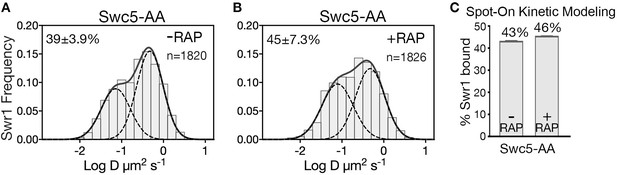
Swc5 is required post-recruitment for SWR1 activity.
(A,B) Normalized histograms and two-component Gaussian fits for Swr1-Halo imaged in Swc5-AA cells in the absence (A) or presence (B) of rapamycin. (C) Spot-On analysis of the same imaging data. Anchor away of Swc5 did not reduce the fraction of chromatin-bound Swr1, which is consistent with ChIP-PCR results showing Swr1 binds to gene promoters in absence of swc5 (Morillo-Huesca et al., 2010), and in vitro data showing efficient nucleosome binding by the purified SWR1(swc5∆) complex (Ranjan et al., 2013). All molecules tracked with JF646 dye.

Reduction in chromatin-bound H2A.Z in a time course of Swc5-AA.
(A) Schematic of cell staining and imaging conditions. (B,C) SPT data from different time points after rapamycin treatment were pooled for normalized diffusion histogram (B) and Spot-On analysis (C). (D–G) Data in Figure 2 re-analyzed for changes in level of chromatin-bound H2A.Z during the course of imaging. Data from first and second half of the imaging period were pooled, and analyzed using diffusion histograms (E,F) and Spot-On kinetic modeling (G).
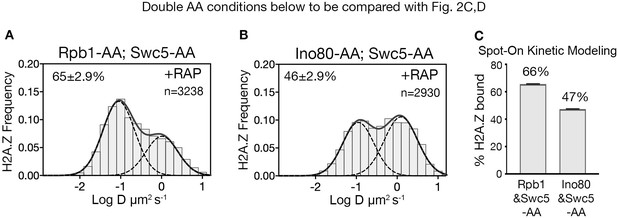
RNA polymerase II is critical for H2A.Z eviction.
(A) Normalized histograms and two-component Gaussian fits for H2A.Z-Halo imaged in cells co-depleted for Rpb1 and Swc5. (B) H2A.Z-Halo distributions in cells co-depleted for Ino80 and Swc5. (C) Spot-On results showing co-depletion of Rpb1 along with Swc5 inhibits H2A.Z eviction. All molecules tracked with JF552 dye.
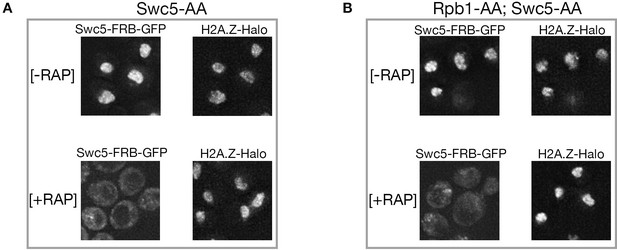
Efficient nuclear depletion of Swc5 in double anchor-away (SWC5-FRB; RPB1-FRB) strain.
(A, B) Deconvolution fluorescence microscopy shows nuclear depletion of Swc5-FRB-GFP in rapamycin-treated, single anchor-away cells, and in Swc5-FRB-GFP; Rpb1-FRBdouble anchor-away cells. Images of JF646-labeled H2A.Z-Halo locate nuclei. Z-axis steps of 200 µm were captured on a DeltaVision fluorescence microscope and 10 deconvoluted stacks were projected.
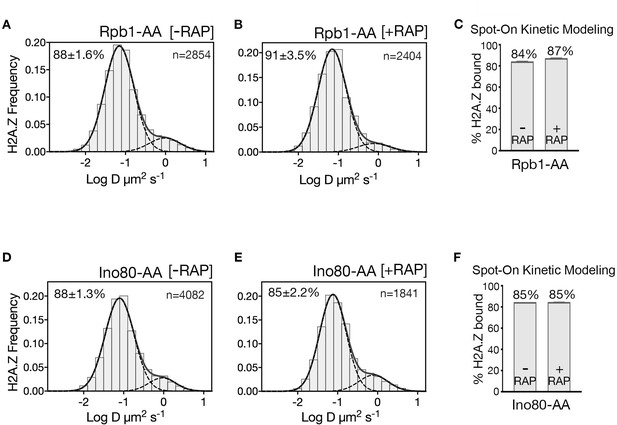
H2A.Z diffusion histograms in cells for single AA of Rpb1 and Ino80.
(A, B) H2A.Z-Halo distributions in Rpb1-AA strain without (A) and with (B) depletion of Rpb1. (C) Spot-On results for chromatin bound H2A.Z upon Rpb1 depletion. Single AA of Rpb1 causes only a slight increase in chromatin-bound H2A.Z, which is likely due to concomitant inhibition of SWR1’s biochemical activity at reduced concentration of free H2A.Z-H2B dimer (Wang et al., 2016). (D, E) H2A.Z distributions in single anchor-away of Ino80. (F) Spot-On results show Ino80 depletion has no effect on level of chromatin-bound H2A.Z. All molecules tracked with JF552 dye.
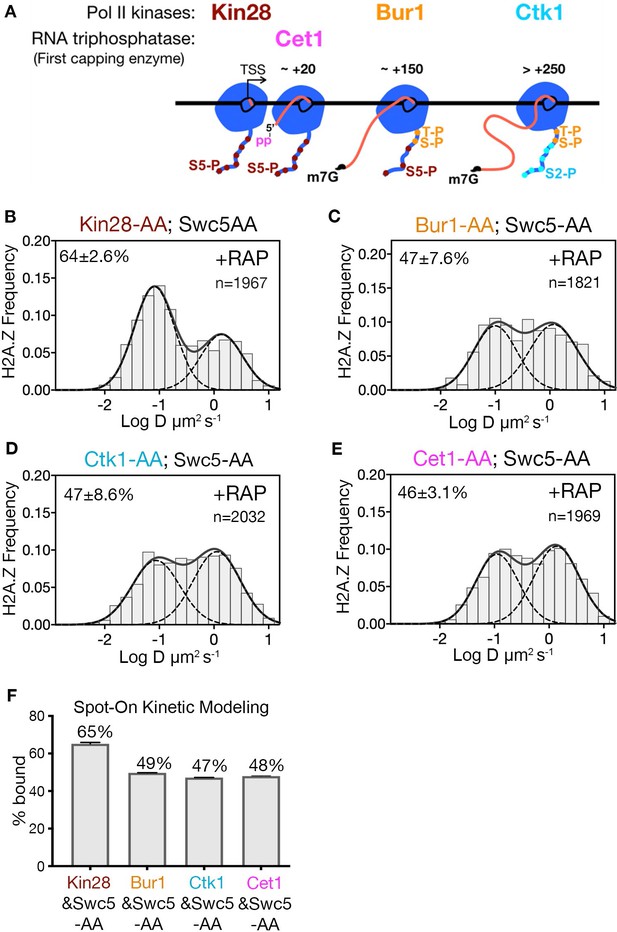
Kin28 phosphorylation of RNA polymerase II CTD is critical for H2A.Z eviction.
(A) Schematic representation shows the three Pol II kinases Kin28, Bur1 and Ctk1 recruited at initiation, early-elongation and elongation phases respectively of Pol II and corresponding phosphorylation of indicated Rpb1 CTD sites. Set1 is the first of the three RNA capping enzymes; it removes γ-phosphate from the RNA 5’end to generate 5’ diphosphate. (B, C, D, E) Normalized histograms and two-component Gaussian fits for H2A.Z-Halo imaged in cells co-depleted for Swc5 along with Kin28 (B), Bur1 (C), Ctk1 (D) and Cet1 (E). (F) Spot-On results show Kin28 is required to evict H2A.Z. All molecules tracked with JF552 dye.
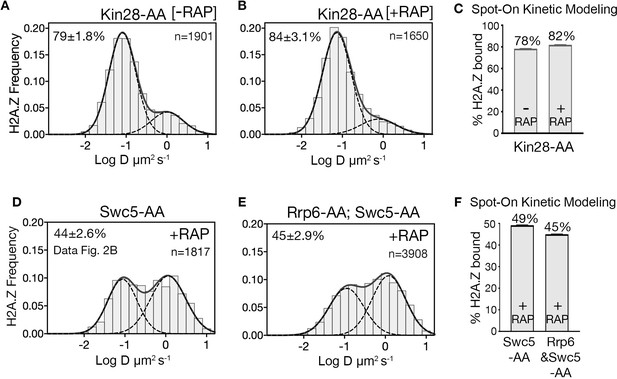
H2A.Z diffusion histograms in cells after single depletion of Kin28.
(A, B) H2A.Z-Halo distributions in Kin28-AA strain without (A) and with (B) depletion of Kin28. (C) Spot-On results for chromatin bound H2A.Z upon Kin28 depletion. (D) Reproduction of Gaussian fits for H2A.Z-Halo distributions in rapamycin-treated Swc5-AA cells (from Figure 2C). (E) H2A.Z-Halo distributions in cells co-depleted for Swc5 and Rrp6. (F) Spot-On results for chromatin-bound H2A.Z upon double depletion of Swc5 and Rrp6. All molecules tracked with JF552 dye.
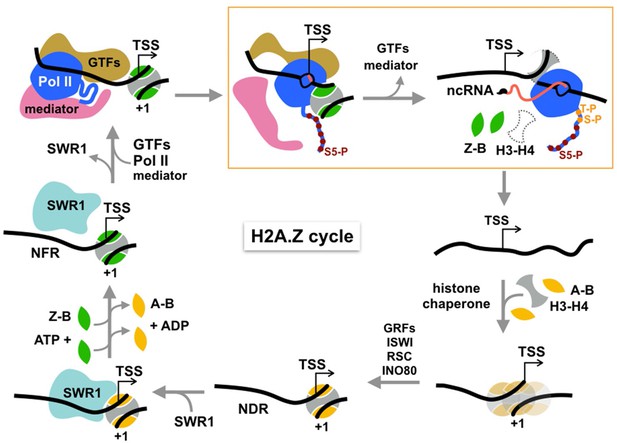
Cycle of H2A.Z eviction and deposition.
RNA polymerase II assembled genome-wide in the PIC and Rpb1 CTD Ser5 phosphorylated by Kin28 constitutively transcribes short noncoding RNAs (with m7G cap) and evicts H2A.Z-H2B dimers from the +1 nucleosome prior to termination. H2A.Z eviction should also occur in the course of mRNA transcription. Additional factors may be necessary for displacement of H3-H4 tetramer. The directional arrow indicates the annotated transcription start site. The gap is filled by histone chaperone-mediated deposition of canonical histones to reform an H2A-containing +1 nucleosome, which is positioned by chromatin remodelers and sequence-specific transcription factors, maintaining the NDR. This recruits SWR1 which is activated upon recognition of H2A-nucleosome and H2A.Z-H2B dimer substrates to activate one or two rounds of H2A.Z deposition. See text for discussion.
Videos
H2A.Z-Halo single molecules imaged in wild type cells.
Molecules were tracked with JF552 dye. Time is indicated on top in seconds and outline of nuclear regions are marked in yellow.
Swr1-Halo single molecules imaged in wild type cells.
Molecules were tracked with JF646 dye. Time is indicated on top in seconds and outline of nuclear regions are marked in yellow.
H2A.Z-Halo single molecules imaged after anchor-away of swc5.
Molecules were tracked with JF552 dye. Time is indicated on top in seconds and outline of nuclear regions are marked in yellow.
Additional files
-
Supplementary file 1
List of strains used in this study.
- https://cdn.elifesciences.org/articles/55667/elife-55667-supp1-v2.docx
-
Supplementary file 2
List of results from MSD and Spot-On analysis.
- https://cdn.elifesciences.org/articles/55667/elife-55667-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55667/elife-55667-transrepform-v2.docx





