HIV-1 Vpr induces cell cycle arrest and enhances viral gene expression by depleting CCDC137
Figures
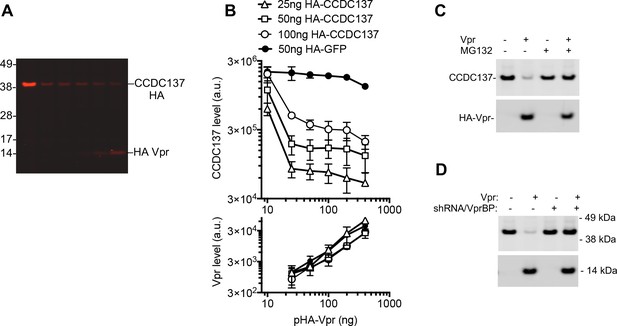
Depletion of CCDC137 by HIV-1 Vpr.
(A) Western blot analysis of 293 T cell lysates 28 hr after transfection (100 ng ng/well) with an HA-CCDC137 expression plasmid and (0 ng, 25 ng, 50 ng, 100 ng, or 200 ng/well) of an HA tagged Vpr expression plasmid. Representative of two experiments. (B) Western blot quantitation of HA-CCDC137 or HA-GFP, (upper panel) and HA-Vpr (lower panel) after transfection of 293 T cells with the indicated amounts (X-axis) of HA-Vpr and target protein (CCDC137-HA [open symbols] or HA-GFP [filled symbols]) expression plasmids. The mean and range of values from two independent experiments is plotted. (C) Western blot analysis of 293 T cell lysates at 28 hr after transfection with 150 ng of V5-tagged CCDC137 expression plasmid and 0 ng or 30 ng of HIV-1 Vpr expression plasmid. Cells were treated with 10 µM MG132 (+) or carrier (-) for 4 hr before harvest. Representative of four experiments. (D) Western blot analysis of cell lysates from empty pLKO vector (-) or VprBP shRNA (+) transduced 293 T cells after transfection of 150 ng of V5-tagged CCDC137 and 30 ng of HA-Vpr expression plasmid. Representative of three experiments.
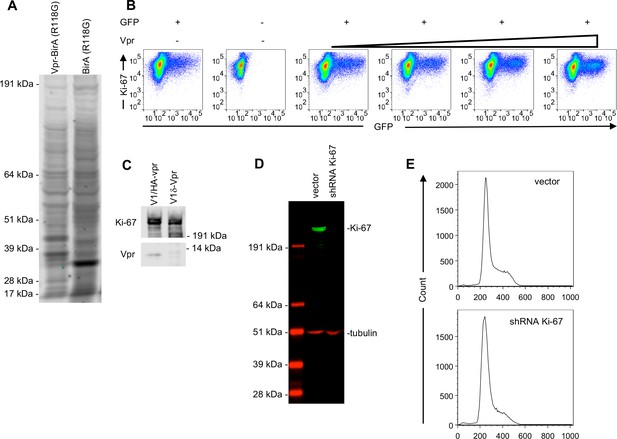
Discovery and analysis of proteins labelled with Vpr-BirA (R118G).
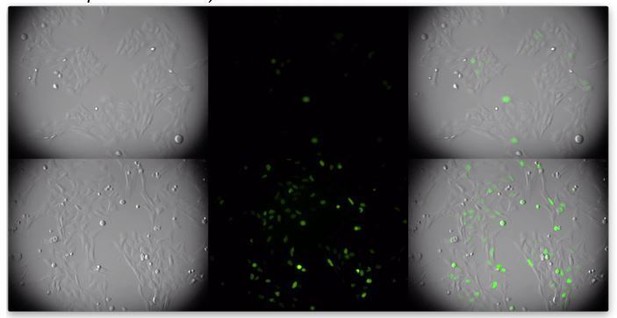
(A) Proteins recovered from lysates of MT-4 cells expressing Vpr-BirA (R118G) or BirA (R118G) using streptavidin beads. Biotinylated proteins were detected with IRDye 680RD-conjugated Streptavidin. Representative of two experiments. (B) Immunostaining/FACS analysis of endogenous Ki-67 levels in 293 T cells 28 hr after transfection (in 6-well plates) with 100 ng of a GFP expression plasmid and increasing amounts (0 ng, 50 ng, 100 ng, 200 ng, or 400 ng) of an HA-Vpr expression plasmid. Representative of two experiments. (C) Western blot analysis of endogenous Ki-67 and HA-Vpr expression in 293 T cells 48 hr after infection with V1/δ-Vpr or V1/HA-Vpr at an MOI of 2. Representative of two experiments. (D) Western blot analysis of Ki-67 and tubulin (loading control) levels in U2OS cells transduced with lentiviruses carrying no shRNA (vector) or shRNA against Ki-67 (shRNA Ki-67) after selection with puromycin. Representative of three experiments. (E) Analysis of DNA content by propidium iodide staining in U2OS cells transduced with lentiviruses carrying no shRNA (vector) or shRNA against Ki-67 (shRNA Ki-67) after selection with puromycin. Representative of three experiments.
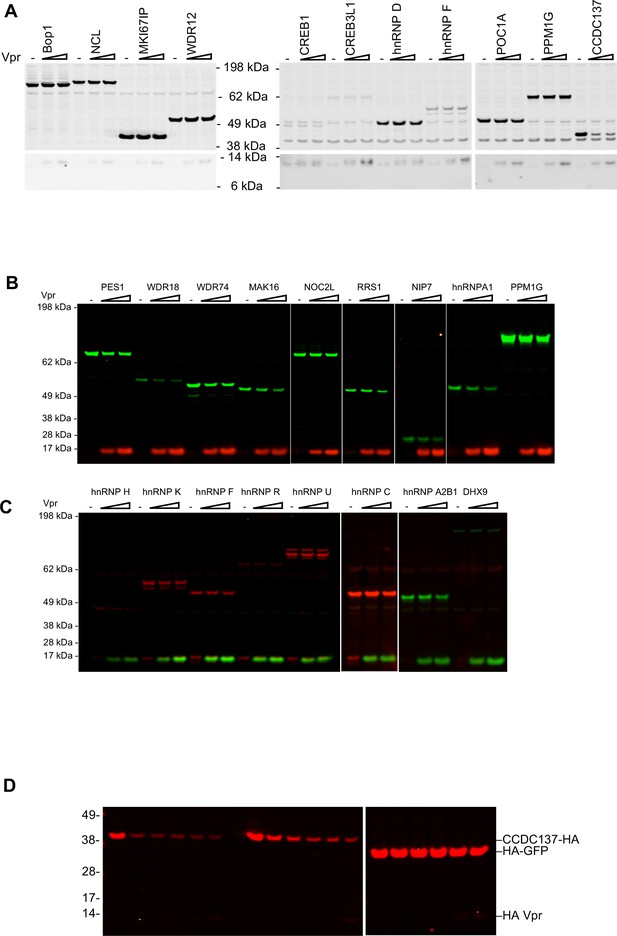
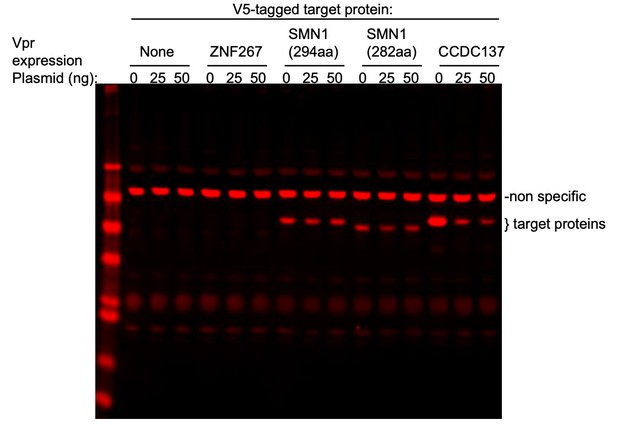
Focused screens for Vpr target proteins.
(A) Western blot analysis of 293 T cell lysates at 28 hr after transfection with 200 ng of plasmids expressing Flag-tagged Bop1, NCL, MKI67IP, or WDR12 (left panel), or V5-tagged CREB1, CREB3L1, hnRNPD, hnRNP F, POC1A, PPM1G, or CCDC137 (right panel) along with increasing amounts (0 ng, 25 ng, or 50 ng) of an HA-Vpr expression plasmid. (B) Western blot analysis of 293 T cell lysates at 28 hr after transfection of 200 ng of plasmids expressing V5-tagged PES1, WDR18, WDR74, MAK16, NOC2L, RRS1, NIP7, hnRNPA1, or PPM1G (shown in green) along with increasing amounts (0, 100 ng, or 300 ng) of HA-Vpr expression plasmid (shown in red). (C) Western blot analysis of 293 T cell lysates after transfection of plasmids expressing V5-tagged hnRNP F, hnRNP H, hnRNP K, hnRNP R, hnRNP U, or hnRNP C (shown in red) or HA-tagged hnRNP A2B1 or DHX9 (shown in green) with increasing amounts of HA-Vpr expression plasmid (shown in green). (D) Western blot analysis of 293 T cell lysates 28 hr after transfection of varying amounts (0 ng, 100 ng, 200 ng, or 400 ng/well) of an HA tagged CCDC137 expression plasmid or 50 ng of HA tagged GFP expression plasmid and increasing amounts (0 ng, 25 ng, 50 ng, 100 ng, or 200 ng/well) of an HA-Vpr expression plasmid. (A–D) Representative of two or three experiments.
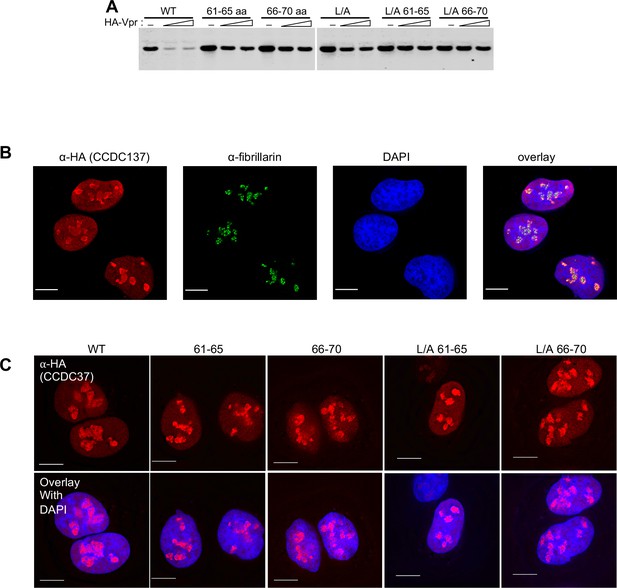
Derivation of Vpr-resistant CCDC137 mutants.
(A) Western blot analysis of 293 T cell lysates 28 hr after transfection with plasmids expressing HA-tagged CCDC137 encoding alanine substitutions at positions 61–65 or 66–70, in the LXXLL motif (L/A), or both, along with 0 ng, 50 ng or 100 ng of an HA-Vpr expression plasmid. Representative of three experiments. (B) Immunofluorescent staining of U2OS cells stably expressing V5-tagged wild-type CCDC137 (red). Endogenous fibrillarin was also immunostained (green) and DNA was stained with DAPI (blue). Scale bar: 10 μm. Representative of two experiments. (C) Immunostaining of U2OS cells stably expressing HA-tagged wild-type (WT) or mutant CCDC137 bearing Ala substitutions at positions 61 to 65 (61-65), 66 to 70 (66-70) alone or in combination with LXXLL motif mutations (L/A). Scale bar: 10 μm. Representative of two experiments.
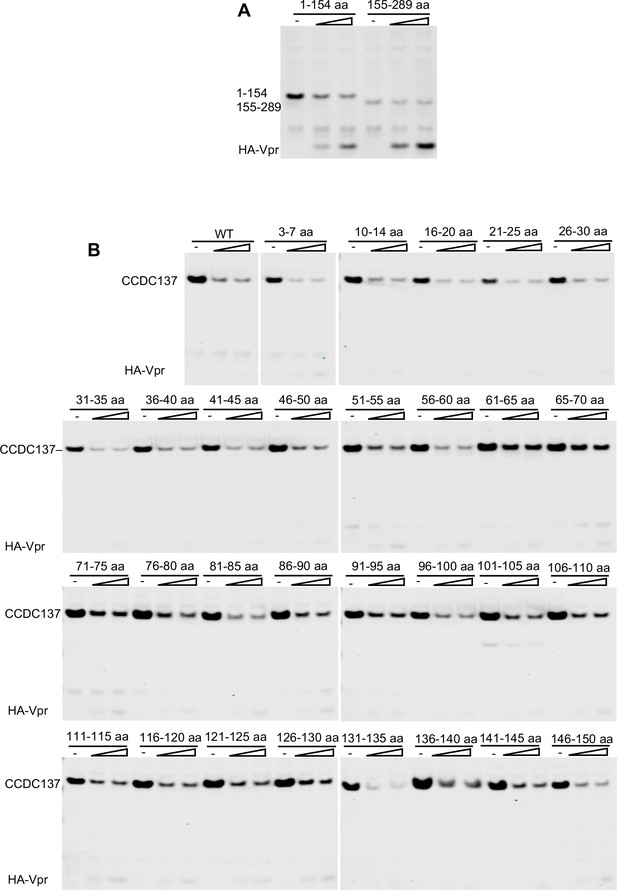
Mapping of degrons in CCDC137 for Vpr-induced depletion.
(A) Western blot analysis (at 28 hr post transfection) following co-transfection of plasmids expressing truncated CCDC137-HA (1–154 aa and 155–289 aa) with increasing amounts (0 ng, 50 ng, or 100 ng) of an HA-Vpr expression plasmid. Representative of two experiments. (B) Western blot analysis (at 28 hr post transfection) following co-transfection of CCDC137-HA expression plasmids with increasing amounts (0 ng, 25 ng, or 50 ng) of HA-Vpr expression plasmids. The CCDC137 proteins encoded alanine scanning mutations at the indicated positions. Each indicated 5-aa stretch was replaced with Ala except where Ala was present in the original sequence. Representative of two experiments.
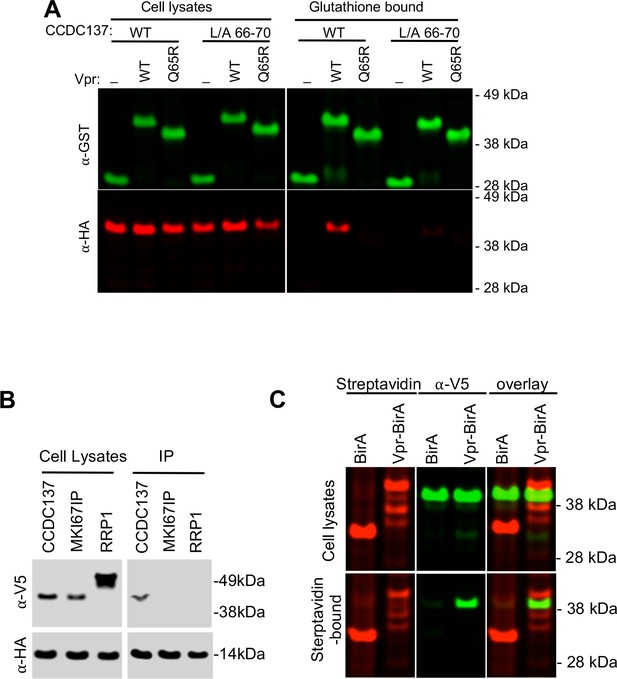
Physical association of Vpr with CCDC137.
(A) Western analysis of cell lysates and glutathione-agarose-bound fractions following transfection of 293 T cells with plasmids expressing GST (-), GST-Vpr WT, or GST-Vpr mutant Q65R along with plasmids expressing HA-tagged wild-type (WT) or Vpr-resistant CCDC137 (L/A 66–70). Representative of three experiments. (B) Western blot analysis of cell lysates and immunoprecipitates (IP) following cotransfection of 293 T cells with plasmids expressing V5-tagged CCDC137, MKI67IP, or RRP1 along with a HA-Vpr expression plasmid. Immunoprecipitation was performed with an anti-HA monoclonal antibody and protein G beads. Representative of three experiments. (C) Western analysis of cell lysates and streptavidin bead-bound proteins following transfection of 293 T cells stably expressing V5-tagged CCDC137 with plasmids expressing BirA (R118G) or Vpr-BirA (R118G). Cells were treated with 50 µM biotin at 24 hr after transfection, and 10 µM MG132 at 40 hr post transfection, and harvested at 44 hr post transfection. Biotinylated proteins were detected with IRDye 680RD Streptavidin (red, left panel) and CCDC137 was detected with anti-V5 antibody (green, middle panel). Representative of three experiments.
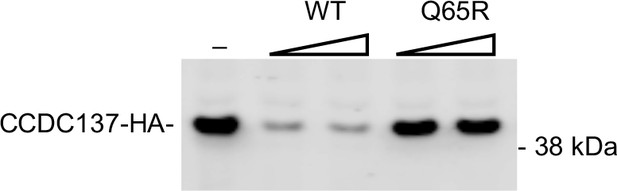
Vpr (Q65R) does not deplete CCDC137 293 T cells were transfected with HA-tagged CCDC137 expression plasmid along with varying amounts (0 ng, 25 ng, or 50 ng) of HA-tagged wild-type NL4-3 Vpr (WT), mutant Q65R (Q65R), expression plasmids.
Twenty-eight hours post transfection, cells were harvested for Western blot analysis. Representative of four experiments.
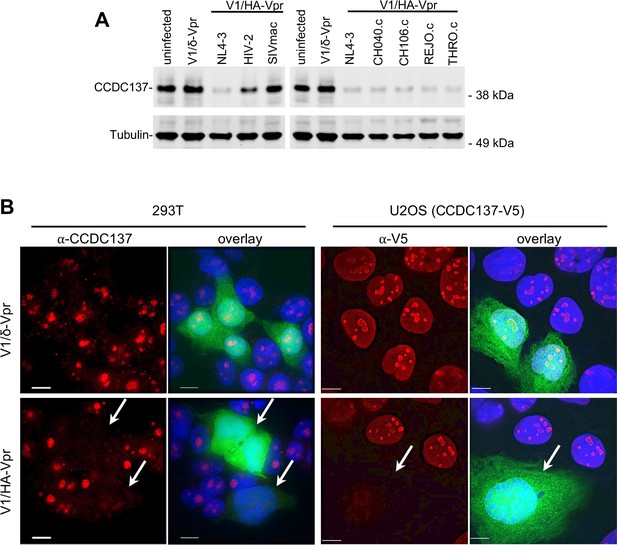
Depletion of CCD137 in HIV-1 infected cells.
(A) Western analysis of 293 T cells 48 hr after infection (MOI = 2) with minimal HIV-1 viruses (V1) carrying no Vpr (V1/δVpr), or Vpr proteins from HIV-1NL4-3, HIV-2, SIVmac, or HIV-1 strains (CH040.c, CH106.c, REJO.c, or THRO.c). Representative of three experiments. (B) Immunofluorescent detection of endogenous (293 T cells, left) or V5-tagged (U2OS cells, right) CCDC137 at 48 hr after infection with V1/δ-Vpr (upper) or V1/HA-Vpr (lower). Infected, GFP-positive are indicated by arrows. Scale bar = 10 μm. Additional examples in Figure 4—figure supplement 2. Representative of three experiments.
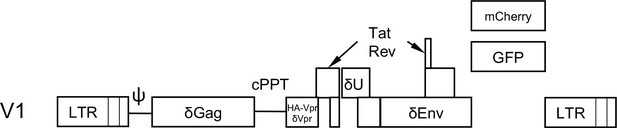
V1, a minimal HIV-1 genome.
Schematic representation of the minimal HIV-1 genomes used herein that contain GFP or mCherry reporter genes to mark infected cells, and assess HIV-1 gene expression using live cell imaging or FACS. Large deletions are introduced into the Gag, Pol, Vif and Env ORFs, while the Vpu ORF contains a premature termination codon. In V1/δVpr, the Vpr ORF contains a deletion while in V1/HA-Vpr an intact Vpr ORF from HIV-1NL4-3, is present.
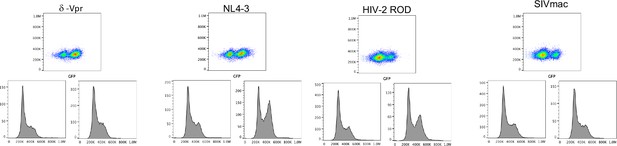
Cell cycle effects of HIV-1, HIV-2, and SIVmac Vpr.
Analysis of DNA content by propidium iodide staining in U2OS cells 48 hr after infection with the minimal HIV-1 viruses (V1) carrying no Vpr (V1/δ-Vpr) or Vpr from HIV-1NL4-3, HIV-2ROD, or SIVMAC. The upper plot in each group of three FACS plots depicts forward scatter (Y axis) vs GFP fluorescence (X-axis). Cells were gated based on GFP expression. The DNA content as assessed by propidium iodide fluorescence (X-axis) versus cell count (Y-axis) is displayed for the uninfected (GFP-) population (lower left histograms) and infected (GFP+) population (lower right histograms).
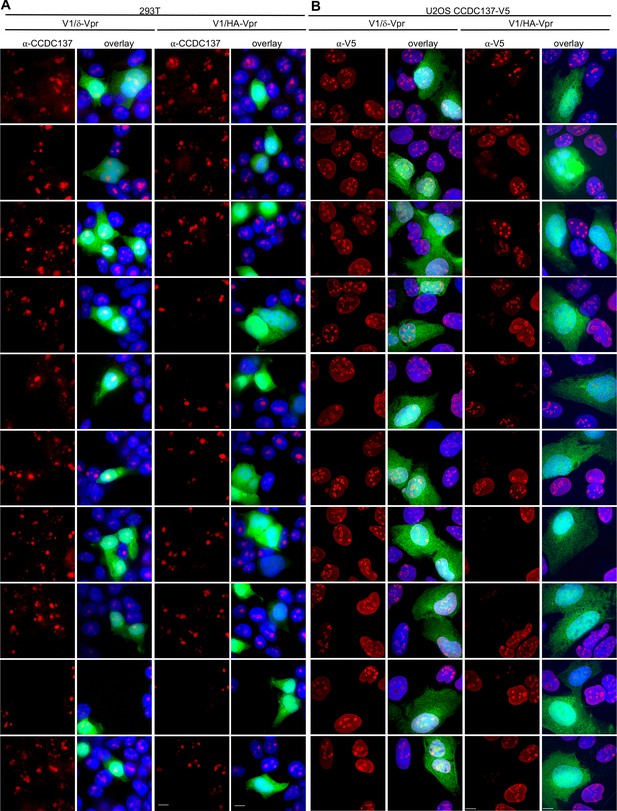
Immunofluorescent detection of CCDC137 depletion by Vpr.
(A, B) Gallery of images acquired following immunofluorescent staining to detect endogenous CCDC137 in 293 T cells (A) or ectopic V5-tagged CCDC137 in U2OS cells (B) at 48 hr after infection with V1/δ-Vpr (left) or V1/HA-Vpr (right) at low MOI. Scale bar: 10 μm. Representative of three experiments.
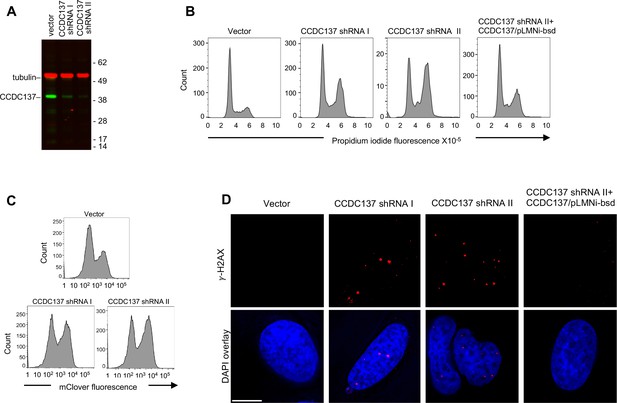
Induction of cell cycle arrest and DNA damage response by depletion of CCDC137.
(A) Western blot analysis of 293 T cells after transduction with pLKO lentivirus vectors carrying no shRNA (vector) or an shRNAs targeting one of two sequences on CCDC137 mRNA (I, II) and selection with puromycin for 40 hr. Representative of three experiments. (B) Propidium iodide (PI) DNA content staining of 293 T cells transduced with shRNA-carrying lentiviruses as in (A). Rightmost panel, 293 T cells were also transduced with a retroviral vector (pLMNiBsd) expressing exogenous CCDC137. Representative of four experiments. (C) FACS analysis of a U2OS-derived cell clone stably expressing mClover-hGeminin(1–110 aa), following transduction and selection as in (A). Representative of two experiments. (D) Immunofluorescent staining of γ-H2AX foci (red) in U2OS cells following transduction as in (B). Scale bar: 10 μm. Representative of four experiments.
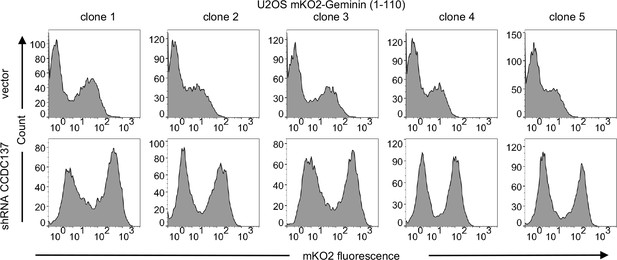
Cell cycle arrest induced by CCDC137 depletion.
FACS analysis of U2OS-derived cell clones stably expressing an mKusabira-Orange2 (mKO2)-hGeminin (1–110 aa) fusion protein, following transduction with lentiviruses carrying no shRNA (vector, upper panels) or a CCDC137-targeting shRNA I (lower panels) and selection with puromycin for 40 hr. Representative of two or three experiments for each clone.
G2/M arrest induced by CCDC137 depletion.
U2OS expressing mClover-hGeminin (1–110 aa) were transduced with a pLKO lentivirus vector containing no shRNA (upper row), or shRNA targeting CCDC137 (lower row). Images are phase contrast (left), mClover-hGeminin (green, center) and an overlay (right), and were acquired commencing at 36 hr post transduction, and every 30 min for the subsequent 72 hr.
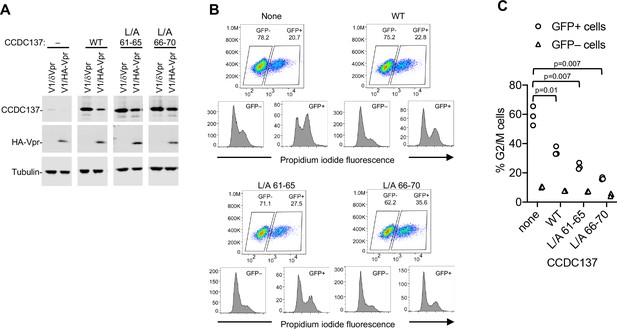
Vpr-resistant CCDC137 attenuates Vpr-induced G2/M arrest.
(A) Western blot analysis of U2OS cells stably expressing doxycycline-inducible wild-type or Vpr depletion-resistant CCDC137 after doxycycline treatment for 24 hr and infection (MOI = 2) with V1/HA-Vpr or V1/δ-Vpr for 48 hr. Representative of three experiments. (B) DNA content assay of WT or mutant CCDC137-expressing cells infected (MOI = 0.3) with V1/HA-Vpr. Upper plots depict forward scatter (Y axis) vs GFP fluorescence (X-axis) and percentages of GFP+ and GFP- cells. Lower plots depict PI staining in the uninfected (GFP-, lower left) and infected (GFP+, lower right) populations. Representative of three experiments. (C) Quantitation and statistical analysis of the effects of WT or Vpr-resistant mutant CCDC137 overexpression on Vpr-induced cell cycle arrest. The percentage of cells in G2/M, as determined by DNA content analysis is plotted for three independent experiments. The indicted p-values were calculated for infected (GFP+) cells expressing no CCDC137 versus WT or mutant CCDC137 using a t-test with Welch’s correction.
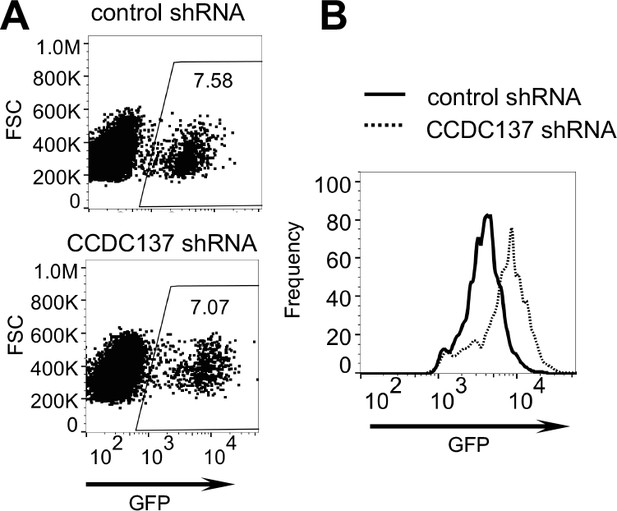
CCDC137 depletion increases HIV-1 gene expression in U2O2 cells.
(A, B) HIV-1(GFP) expression in U2OS cells transduced with an empty lentiviral vector (solid line) or CCDC137-targeting shRNA (dotted line) and selected with puromycin prior to infection with V1/δ-Vpr/GFP for two days. (B) represents histogram of GFP fluorescence in infected cells, gated as shown in (A). Numbers in (A) represent % of cells within the gate. Representative of three experiments.
G2/M arrest and HIV-1 gene expression induced by Vpr in U2OS cells.
U2OS cells expressing mClover-hGeminin (1–110 aa) were infected with V1/δ-Vpr (upper row) or V1/HA-Vpr (lower row) carrying mCherry. Images (from left to right) are phase contrast, mCherry (red), mClover-hGeminin (green) and an overlay of all channels, and were acquired commencing 18 hr after viral infection, and every 30 min for the subsequent 48 hr.
HIV-1 (V1/mCherry) expression in CCDC137 depleted U2OS cells.
U2OS expressing mClover-hGeminin (1–110 aa) were transduced with lentiviruses containing no shRNA (upper row), or shRNAs targeting CCDC137 (lower row). At 28 hr post transduction, cells were infected with V1/δ-Vpr carrying mCherry. Images (from left to right) are phase contrast, mCherry (red), mClover-hGeminin (green) and an overlay of all channels, and were acquired commencing 12 hr after V1/δ-Vpr/mCherry infection and every 30 min for the subsequent 60 hr.
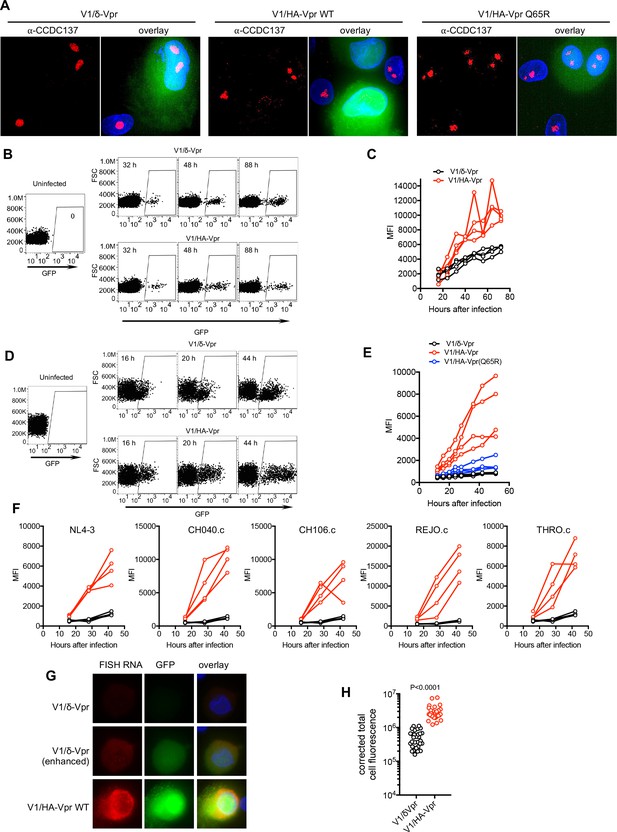
Vpr increases HIV-1 gene expression in primary cells.
(A) Example of immunofluorescent staining to detect endogenous CCDC137 and GFP expression in primary macrophages at 48 hr after infection with V1/δ-Vpr (left), V1/HA-Vpr (center) or V1/HA-Vpr (Q65R) (right) at low MOI. Scale bar: 10 μm. Additional examples are shown in Figure 8—figure supplement 1. Representative of two experiments, each with three macrophage donors. (B) FACS analysis of GFP expression in activated primary CD4+ cells after infection with V1/δ-Vpr or V1/HA-Vpr. A representative donor is shown. Representative of three experiments, each with three or four donors. (C) GFP levels (mean fluorescence intensity (MFI), gated on infected cells) in primary CD4+ cells from four donors after infection with V1/δ-Vpr or V1/HA-Vpr. Representative of three experiments, each with three or four donors. (D) FACS analysis of GFP expression in macrophages after infection with V1/δ-Vpr or V1/HA-Vpr. A representative donor is shown. Representative of five experiments, each with three or four donors. (E) GFP levels (MFI, gated on infected cells) in macrophages from four donors after infection with V1/δ-Vpr, V1/HA-Vpr or V1/HA-Vpr(Q65R). Representative of five experiments, each with three or four donors. (F) FACS analysis of GFP expression macrophages after infection with V1/δ-Vpr (Black) or V1/HA-Vpr (Red) derivatives encoding Vpr proteins from several different transmitted founder virus strains. The mean fluorescent intensity (MFI) of infected cells, gated as in (D) for four macrophage donors is plotted. Representative of two experiments, each with three or four donors. (G) Representative images of primary macrophages infected with V1/δ-Vpr or V1/HA-Vpr and subjected to fluorescent in situ hybridization (FISH) based detection of HIV-1 RNA using probes directed at the GFP sequence. The FISH signal is displayed in red and GFP protein signal is displayed in green. The upper (V1/δ-Vpr) and lower (V1/HA-Vpr) rows are displayed with the same gain and brightness/contrast settings. The center (V1/δ-Vpr) row is a duplicate of the upper row, displayed with enhanced brightness to enable visualization of the FISH and GFP signals. Representative of two experiments, each with three donors. (H) GFP RNA levels (total cell fluorescence) determined by FISH analysis of macrophages infected with V1/δ-Vpr or V1/HA-Vpr. Each symbol represents a single cell from a representative donor, from one of two experiments, each with three donors. P-value is calculated using a Welch’s t-test.
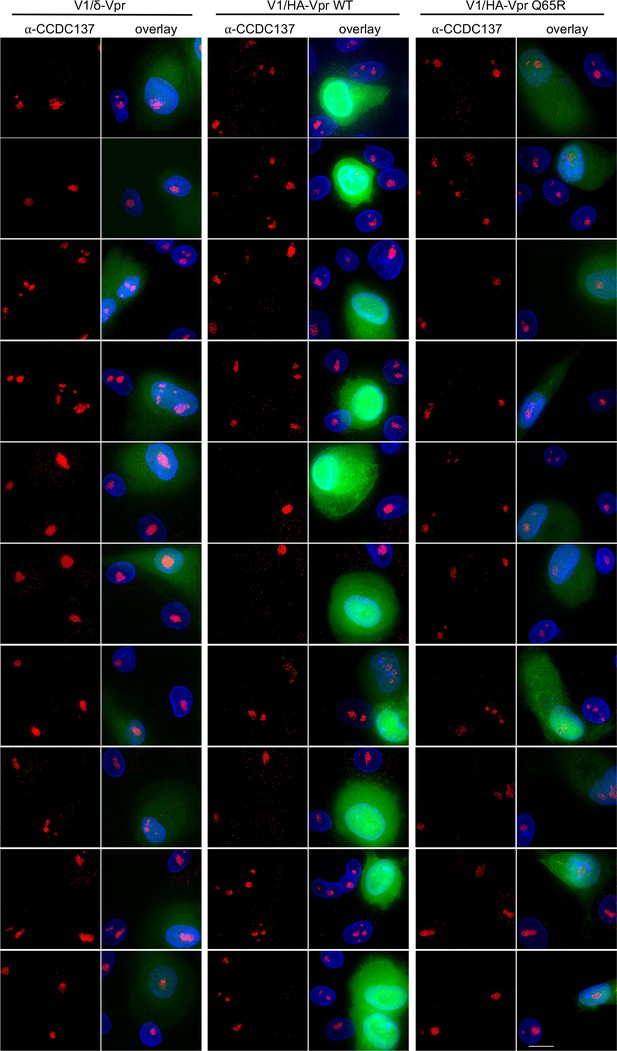
Immunofluorescent detection of CCDC137 depletion by Vpr and increased V1/GFP expression in macrophages.
Gallery of images following immunofluorescent staining to detect endogenous CCDC137 and GFP expression in primary macrophages at 48 hr after infection with V1/δ-Vpr (left), V1/HA-Vpr (center) or V1/HA-Vpr (Q65R) (right) at low MOI. Scale bar: 10 μm. Representative of two experiments, each with three macrophage donors.
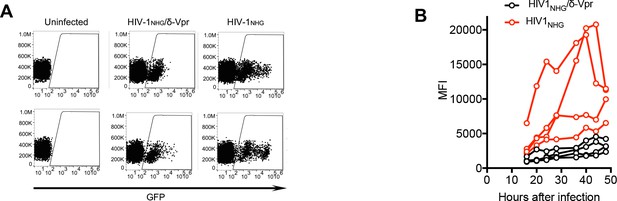
Enhancement of HIV-1 gene expression in macrophages by Vpr.
(Α) FACS analysis of GFP expression in primary macrophages after infection with HIV-1NHG/δ-Vpr or HIV-1NHG. Representative of two experiments, each with three or four donors. (Β) FACS analysis of GFP expression in primary macrophages after infection with HIV-1NHG/δ-Vpr (Black) or HIV-1NHG (Red). The mean fluorescent intensity (MFI) of infected cells, gated as in (A) for four macrophage donors is plotted. Representative of two experiments, each with three or four donors.
HIV-1 (V1/δ-Vpr vs V1/HA-Vpr) expression in macrophages (donor #1).
Freshly prepared human macrophages were infected with V1/δ-Vpr (upper row) or V1/HA-Vpr (lower row) carrying GFP. Images are phase contrast (left), GFP (green, center) and an overlay (right), and were acquired commencing at 24 hr after viral infection, and every 30 min for the subsequent 60 hr.
HIV-1 (V1/δ-Vpr vs V1/HA-Vpr) expression in macrophages (donor #2).
Freshly prepared human macrophages were infected with V1/δ-Vpr (upper row) or V1/HA-Vpr (lower row) carrying GFP. Images are phase contrast (left), GFP (green, center) and an overlay (right), and were acquired commencing at 24 hr after viral infection, and every 30 min for the subsequent 60 hr.
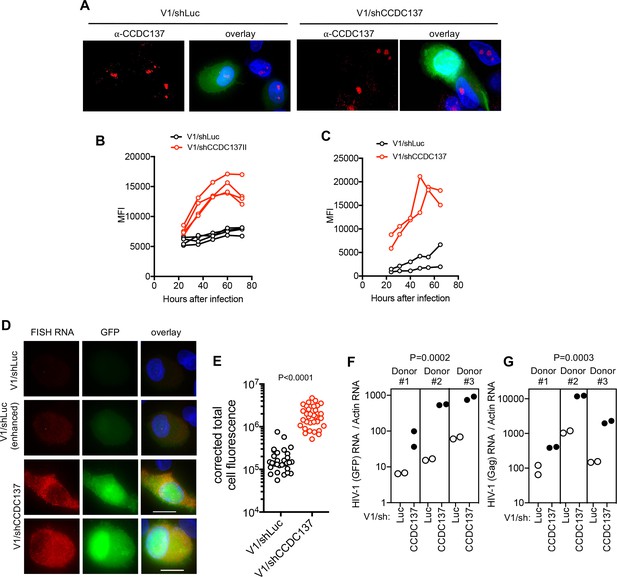
CCDC137 depletion increases HIV-1 gene expression in primary cells.
(A) GFP expression and Immunofluorescent staining to detect endogenous CCDC137 in primary macrophages at 48 hr after infection with V1/sh (left) or V1/shCCDC137 II (right) at low MOI. Scale bar: 10 μm. Additional examples are shown in Figure 9—figure supplement 2. Representative of two experiments, each with three donors. (B) GFP expression in primary CD4+ T-cells after infection with V1/shLuc or V1/shCCDC137. MFI (gated on infected cells) for four donors is shown. Representative of three experiments, each with three or four donors. (C) GFP expression in macrophages after infection with V1/shLuc or V1/shCCDC137. MFI of infected cells for two donors is shown. Representative of three experiments, each with two to four donors. (D) Representative images of primary macrophages infected with V1/shLuc or V1/shCCDC137II and subjected to fluorescent in situ hybridization (FISH) based detection of HIV-1 RNA using probes directed at the GFP sequence. The FISH signal is displayed in red and GFP protein signal is displayed in green. The upper (V1/shLuc) and lowest two (V1/shCCDC137II) rows are displayed with the same gain and brightness/contrast settings. The second (V1/shLuc) row is a duplicate of the upper row, displayed with enhanced brightness to enable visualization of the FISH and GFP signals. Representative of two experiments, each with three donors. (E) GFP RNA levels (total cell fluorescence) determined by FISH analysis of macrophages infected with V1/shLuc or V1/shCCDC137. Each symbol represents a single cell from a representative donor from one of two experiments, each with three donors. P-value is calculated using a Welch’s t-test. (F) qRT-PCR measurement of HIV-1 RNA (GFP) levels in three macrophage donors after infection with V1/shLuc or V1/shCCDC137II. Each symbol represents a technical replicate. Representative of two experiments, each with three donors. P-value is calculated using a ratio paired t-test for the three displayed donors. (G) qRT-PCR measurement of HIV-1 RNA (Gag) levels in three macrophage donors after infection with V1/shLuc or V1/shCCDC137II. Each symbol represents a technical replicate. Representative of two experiments, each with three donors. P-value is calculated using a ratio paired t-test for the three displayed donors.
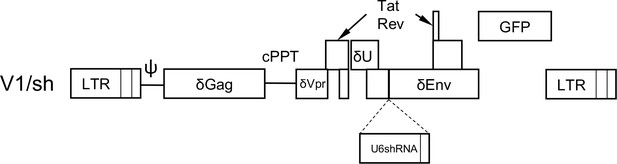
V1/sh, a minimal HIV-1 genome carrying a reporter gene and an shRNA expression cassette.
A variant of V1, termed V1/sh contains an expression cassette for an shRNA driven by a U6 promoter embedded in the truncated env gene, enabling measurement of HIV-1 reporter gene expression in shRNA expressing cells.
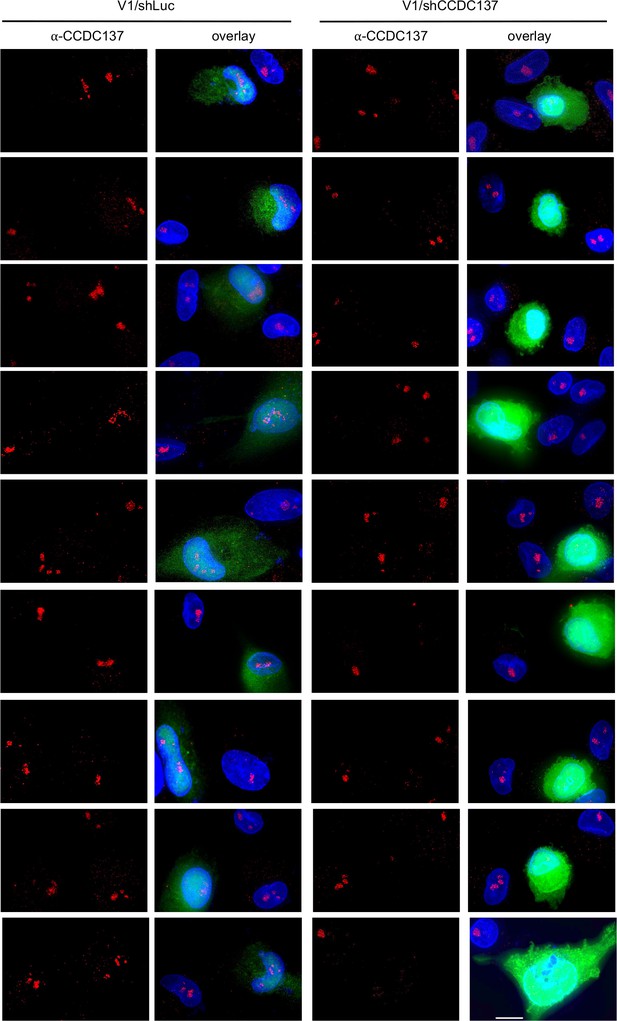
Immunofluorescent detection of CCDC137 depletion by shRNA and increased.
GFP expression in macrophages Gallery of images of immunofluorescent staining to detect endogenous CCDC137 as well as GFP expression in primary macrophages at 48 hr after infection with V1/sh (left) or V1/shCCDC137 II (right) at low MOI. Scale bar: 10 μm. Representative of two experiments, each with three donors.
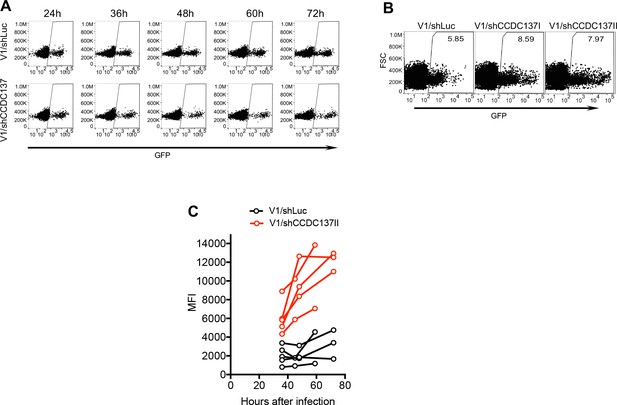
Enhancement of HIV-1 gene expression in macrophages and CD4+ T-cells by shRNA-mediated CCDC137 depletion.
(A) FACS analysis of GFP levels in primary CD4+ cells at the indicated time points after infection with V1/shLuc or V1/shCCDC137. A representative donor is shown from one of two experiments, each with three donors. (B) FACS analysis of GFP levels in macrophages after infection with V1/shLuc or V1/shCCDC137. A representative donor is shown from one of three experiments, each with three or four donors. (C) FACS analysis of GFP expression in macrophages from four additional donors after infection with V1/shLuc or V1/shCCDC137. MFI of infected cells is plotted. Representative of two experiments, each with two to four donors.
HIV-1 (V1/shLuc vs V1/shCCDC137) expression in macrophages (donor #3).
Freshly prepared human macrophages were infected with V1/shLuc (upper row) or V1/shCCDC137 (lower row) carrying GFP. Images are phase contrast (left), GFP (green, center) and an overlay (right), and were acquired commencing at 20 hr after viral infection, and every 30 min for the subsequent 38 hr.
HIV-1 (V1/shLuc vs V1/shCCDC137) expression in macrophages (donor #4).
Freshly prepared human macrophages were infected with V1/shLuc (upper row) or V1/shCCDC137 (lower row) carrying GFP. Images are phase contrast (left), GFP (green, center) and an overlay (right), and were acquired commencing at 20 hr after viral infection, and every 30 min for the subsequent 38 hr.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CCDC137 | This paper | NCBI Gene ID: 339230 | |
| Cell line (Homo-sapiens) | 293T | ATCC | CRL-3216 | |
| Cell line (Homo-sapiens) | U2OS | ATCC | HTB-96 | |
| Antibody | anti-FLAG | Sigma | Cat# F3165 | WB (1:1000) Monoclonal ANTI-FLAG M2 antibody produced in mouse |
| Antibody | anti-HA | BioLegend | Cat# 901515 | WB (1:1000) Anti-HA.11 Epitope Tag Antibody |
| Antibody | anti-Ki67 | Abcam | Cat# ab16667 | IF (1:500); WB (1:1000) Rabbit Anti-Ki67 antibody [SP6] |
| Antibody | anti-V5 | Invitrogen | Cat# R960-25 | WB (1:1000); IF (1:200) Mouse Monoclonal |
| Antibody | anti-V5 | Thermo Fisher Scientific | Cat# PA1-993 | WB (1:1000) Rabbit Polyclonal |
| Antibody | anti-CC137 | Abcam | Cat# ab185368 | WB (1:500); IF (1:200) Rabbit polyclonal to CC137 |
| Antibody | anti-CC137 | Abcam | Cat# ab183864 | WB (1:500) Rabbit polyclonal to CC137 |
| Antibody | anti-gamma H2AX phosphor S139 | Abcam | Cat# ab11174 | IF (1:500) Rabbit polyclonal to gamma H2A.X (phospho S139) |
| Recombinant DNA reagent | V1/δ-Vpr | This paper | a minimal proviral plasmid | |
| Recombinant DNA reagent | V1/HA-Vpr | This paper | a minimal proviral plasmid expressing HA-tagged Vpr | |
| Recombinant DNA reagent | V1/Vpr | This paper | a minimal proviral plasmid expressing Vpr | |
| Recombinant DNA reagent | V1/mCherry | This paper | mCherry replaces GFP in V1 | |
| Recombinant DNA reagent | pLKO.1 TRC | Addgene | Cat# 10878 | Lentiviral construct to express the shRNA. |
| Recombinant DNA reagent | pLKOΔ-puro | Busnadiego et al., 2014 | LKO-derived lentiviral expression vector | |
| Recombinant DNA reagent | pLKOΔ-puro CCDC137 | This paper | tetracycline-inducible CCDC137 expression pLKO vector | |
| Peptide, recombinant protein | GM-CSF | Thermo Fisher | Cat# PHC2011 | Macrophage differentiation |
| Commercial assay or kit | FxCycle PI/RNase Staining Solution | Invitrogen | Cat#: F10797 | |
| Commercial assay or kit | Power SYBR Green RNA-to-CT 1-Step Kit | Thermo Fisher | Cat# 4389986 | |
| Chemical compound, drug | MG 132 | Sigma Aldrich | Cat#:M7449 | |
| Software, algorithm | MetaMorph | Molecular Devices | ||
| Software, algorithm | Prism | Graphpad |
Additional files
-
Supplementary file 1
List of proximal/interacting proteins identified using Mass Spectrometry analysis of biotinylated proteins in BirA (R118G)-fused Vpr-expressing cells.
The table includes UniProtKB accession number, description, mean peak area, scores (the sum of the highest ions score for each distinct peptide), percent coverage (calculated by dividing the number of amino acids in all found peptides by the total number of amino acids in the entire protein sequence), the number of distinct peptides used for identification in a protein, and peptide spectrum matches (PSM, the total number of identified peptide sequences for the protein). The table contains results from duplicate samples.
- https://cdn.elifesciences.org/articles/55806/elife-55806-supp1-v1.xlsx
-
Supplementary file 2
List of oligonucleotides and their sequences and cDNA and accession numbers employed in the molecular construction of expression plasmids used in this study.
- https://cdn.elifesciences.org/articles/55806/elife-55806-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55806/elife-55806-transrepform-v1.docx