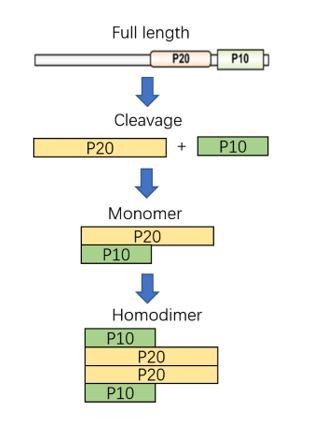
Apoptotic neurodegeneration in whitefly promotes the spread of TYLCV
Figures
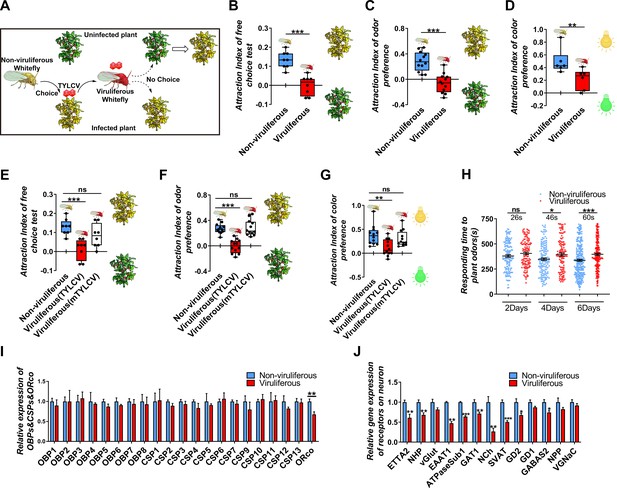
TYLCV impairs the host selectivity of whitefly between infected and uninfected host plants.
(A) Whitefly preferences change after TYLCV acquisition. (B–D) Whitefly attraction index (feed on infected or uninfected plants) of (B) free-choice assay with plants, n = 8, (C) dual-choice assay with plant odors, n = 12, and (D) dual-choice assay with colors (green and yellow), n = 6. (E–G) Whitefly attraction index (feed on artificial diet with/without purified virions) of (E) free-choice assay with plants, n = 8, (F) dual-choice assay with plant odors, n = 12, (G) dual-choice assay with plant colors, n = 12, (H) Responding time to the same plant odor in 12 min-monitoring was recorded. The difference of means ± SEM is labeled. Whiteflies on infected or uninfected plants were collected separately after 2 days, 4 days, or 6 days of feeding. (I) Relative gene expression of whitefly OBPs, CSPs, and ORco, n = 3–5. (J) Relative gene expression of whitefly neuron membrane receptors, n = 3–5. Box plots represent the median (bold black line), quartiles (boxes), as well as the minimum and maximum (whiskers). Values in bar plots represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
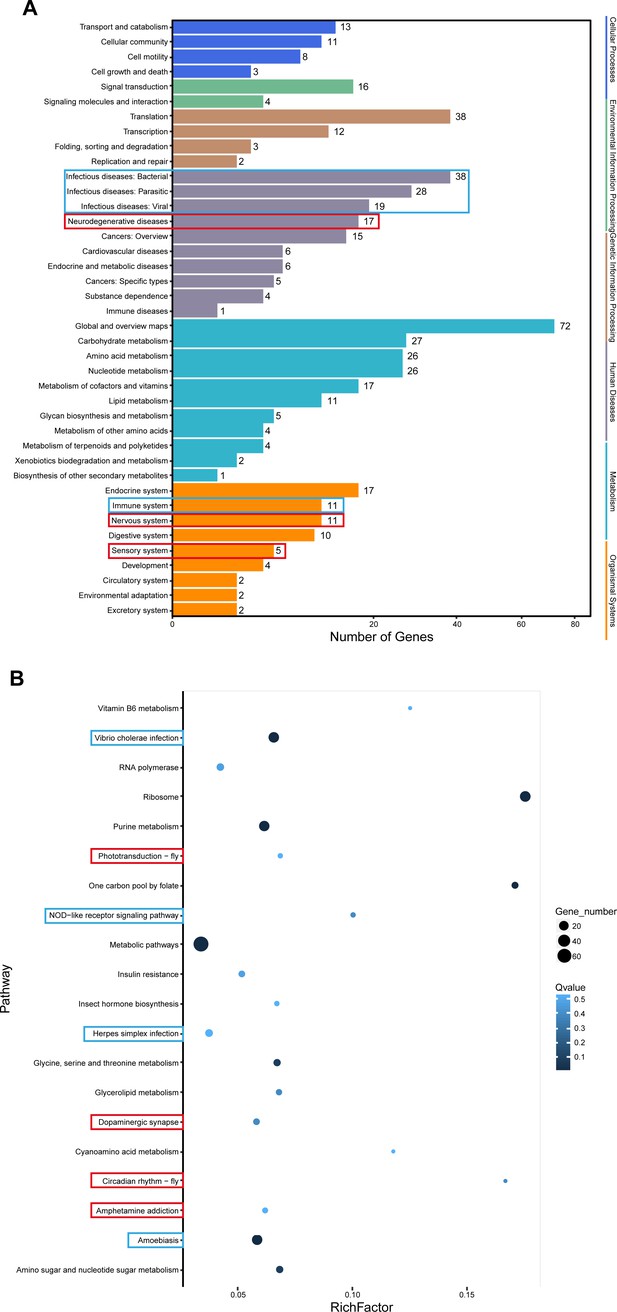
Whitefly immune system and nervous system response to TYLCV in head transcriptome analysis.
Blue frame represents immune-related changes and red frame represents nerve-related changes in (A) KEGG enrichment pathway and (B) differentially expressed gene enrichment analysis.
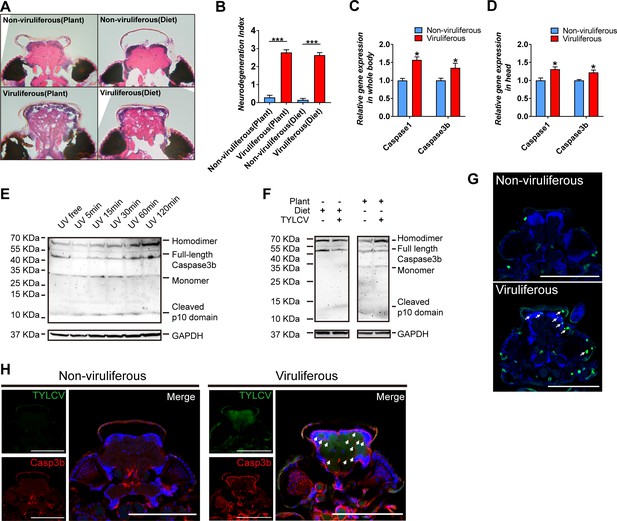
TYLCV induces apoptotic neurodegeneration in the brain of whitefly.
(A–B) Neurodegeneration of whitefly feeding on plant diet (infected or uninfected) and artificial diet (with or without TYLCV virions) were observed (A) and quantified (B) in head sections, nUninfected plant=21, nInfected plant=60, nArtificial diet=21, nDiet+TYLCV=60. (C–D) Both whitefly bodies with head and dissected heads were collected, and the relative gene expression of whitefly Caspase1 and Caspase3b were analyzed using qRT-PCR, n = 5. (E–F) A 2 hr time-course UV treatment was considered as a positive control to monitor Caspase3b cleavage and activation by non-reduced denaturing polyacrylamide gel electrophoresis (E). Both purified virions and infected plants would induce Caspase3b cleavage and activation (F). All bands of each sample were imaged from the same blot, n = 3. (G) Whitefly head sections were fixed and labeled with terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling. Green indicates TUNEL staining of the apoptotic cells. (H) Head sections were labeled with anti-TYLCV CP and anti-Caspase3b antibodies. All confocal images of the head section were dissected from whiteflies feeding on artificial diet. Scale bar = 100 μm, n > 15. Values in bar plots represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
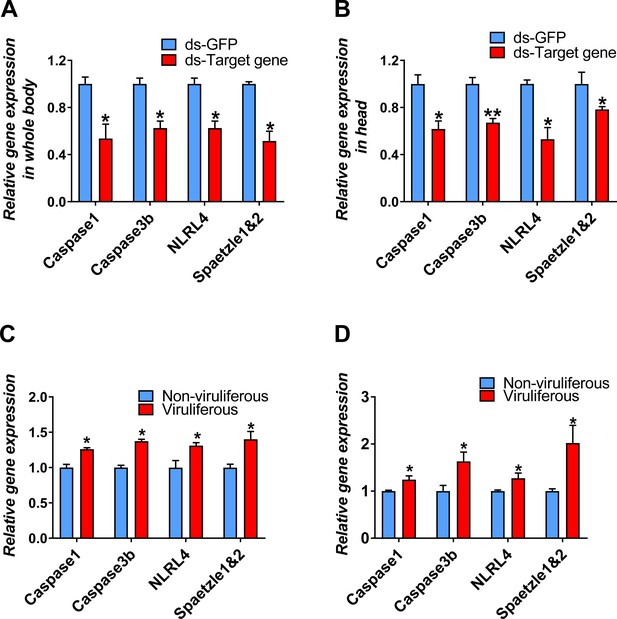
Relative gene expression in qRT-PCR analysis.
(A–B) RNA interference efficiency in (A) whole bodies and (B) heads. (C) TYLCV from infected benthamiana can also induce whitefly Caspase1, Caspase3b, NLRL4, and Spaetzle1 and 2 expression. (D) TYLCV can induce whitefly Caspase1, Caspase3b, NLRL4, and Spaetzle1 and 2 expression with only 12 hr virus acquisition. n = 5. Values represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
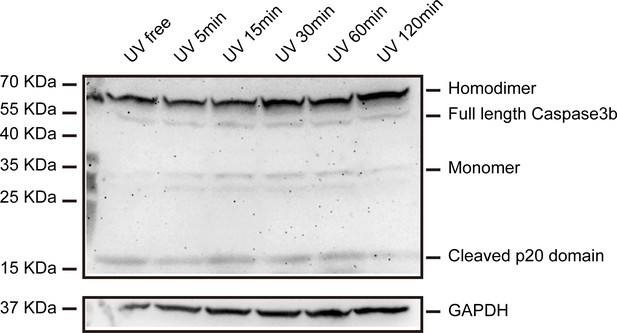
Dynamics of Caspase3b cleavage.
An anti-Caspase3b p20 antibody labeled p20 domain was also used in the time-course UV treatment experiment to monitor the dynamics of Caspase3b cleavage and the result was confirmed in Figure 2E using anti-Caspase3b p10 antibody.
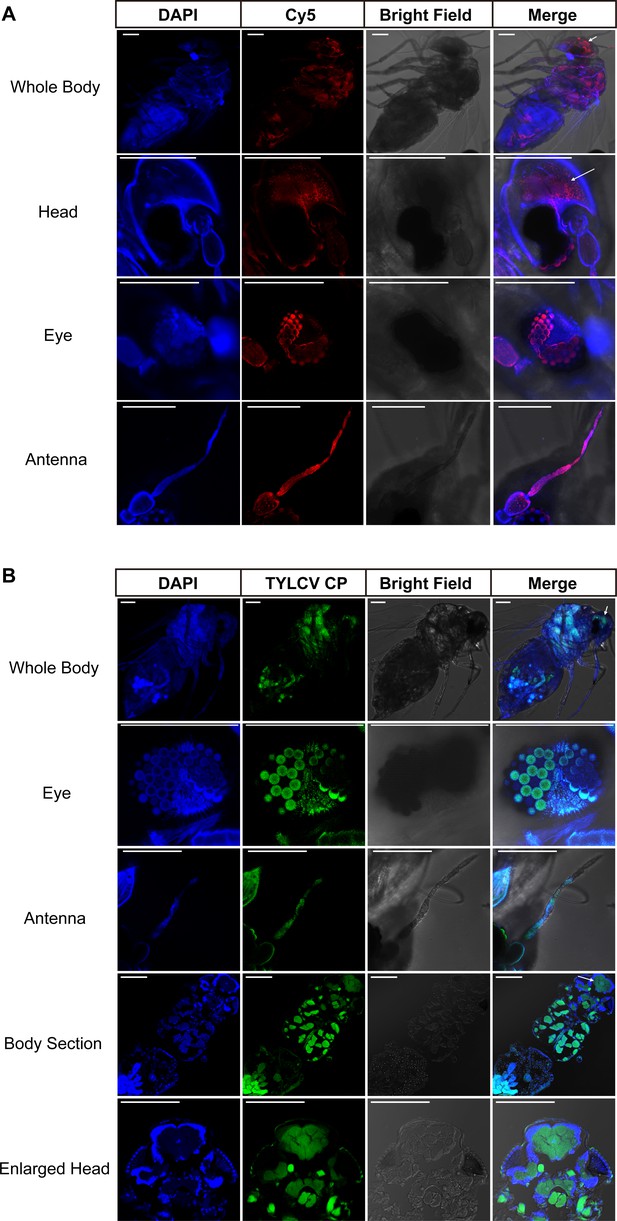
Localization of genome DNA and CP of TYLCV.
Localization of genome DNA and CP of TYLCV was observed in the whitefly brain using both (A) fluorescence in situ hybridization and (B) immunofluorescence. All samples were collected from artificial diet-containing TYLCV virions. Scale bar = 100 μm, n > 15.
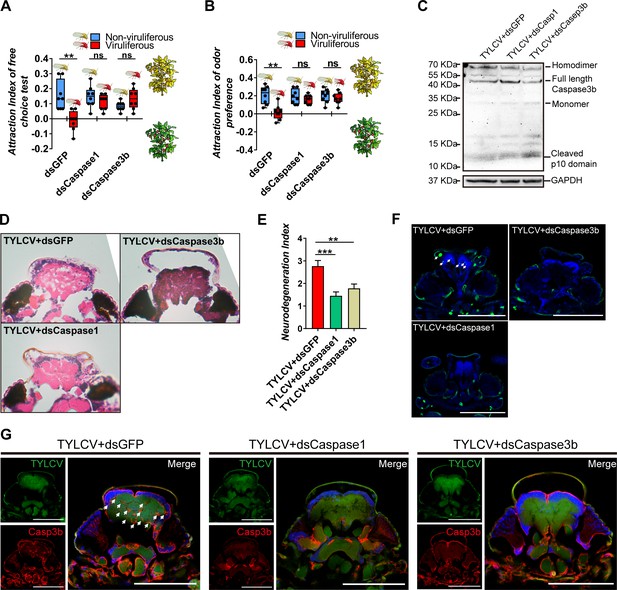
Silencing caspases alleviates virus-induced neurodegeneration.
(A–B) Whitefly attraction index (fed on artificial diet with dsRNA) of (A) free-choice assay with plants, n = 8, (B) dual-choice assay with plant odors, n = 8. (C) Caspase3b of whitefly treated with virions and dsRNA was detected using western blot. (D–E) Neurodegeneration of whitefly fed with virions and dsRNA was observed (D) and quantified (E) in head sections, nTYLCV+dsGFP=30, nTYLCV+dsCaspase1=31, nTYLCV+dsCaspase3b=41. (F–G) Head section images of whitefly feed with TYLCV and dsRNA. Interference with Caspase1 and Caspase3b alleviates brain apoptosis in TUNEL assay (F) and Caspase3b in immunofluorescence. Scale bar = 100 μm, n > 12. Box plots represent the median (bold black line), quartiles (boxes), as well as the minimum and maximum (whiskers). Values in bar plots represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).
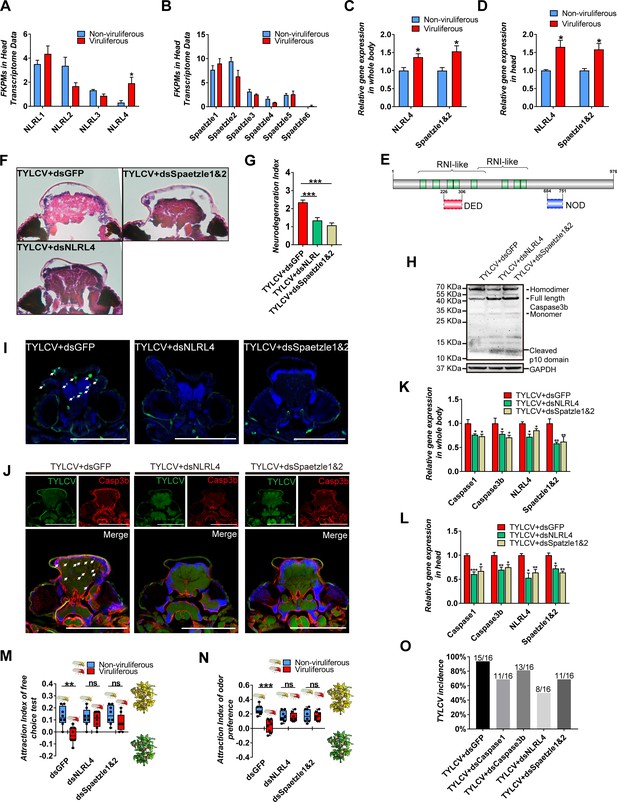
NLRL4 responses to TYLCV and induces neurodegeneration in whitefly.
(A–B) FKPMs of NLRLs (A) and Spaetzles (B) in whitefly head transcriptome data. (C–D) Relative gene expression of whitefly bodies (C) or heads (D) NLRL4 and Spaetzle1 and 2 were analyzed using qRT-PCR, n = 5. (E) Conserved domains of NLRL4 was predicted by InterPro and SMART. Green represents leucine-rich repeats, dashed boxes represent domain scores that are less significant than the required threshold. (F–G) Neurodegeneration of whitefly fed with virions and dsRNA was observed (F) and quantified (G) in head sections, nTYLCV+dsGFP=40, nTYLCV+dsNLRL4=49, nTYLCV+dsSpaetzle1&2 = 53. (H) Caspase3b of whitefly treated with virions and dsRNA was detected using western blot. (I–J) Head section images of whitefly fed with TYLCV and dsRNA. Interference with NLRL4 and Spaetzle1 and 2 alleviates brain apoptosis in TUNEL assay (F) and Caspase3b in immunofluorescence. Scale bar = 100 μm, n > 12. (K–L) Interference with NLRL4 or Spaetzle1 and 2 suppressed caspases expression in both bodies (K) and heads (L), n = 5. (M–N) TYLCV cannot alter whitefly preference in (M) free-choice assay, n = 8, and (N) odorant dual-choice assay, n = 8, after interference with NLRL4 or Spaetzle1 and 2. (O) Rescue whitefly preference impairs TYLCV transmission after interference with Caspase1, Caspase3b, NLRL4, or Spaetzle1 and 2. Box plots represent the median (bold black line), quartiles (boxes), as well as the minimum and maximum (whiskers). Values in bar plots represent mean ± SEM (*p<0.05, **p<0.01, ***p<0.001).

Virus transmission bioassay.
TYLCV infected and uninfected tomato plants were alternately placed in a circle for 4 hr transmission.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Tomato yellow leave curl virus) | Tomato yellow leave curl virus isolate SH2 infectious clone | Xueping Zhou, Institute of Plant Protection, CAAS | ||
| Strain, strain background (Tomato yellow leave curl virus) | Mutant Tomato yellow leave curl virus isolate SH2 infectious clone | Xiaowei Wang, Zhejiang University | ||
| Antibody | Mouse monoclonal anti-TYLCV CP | Jianxiang Wu, Zhejiang University | IF (1:500) | |
| Antibody | Mouse monoclonal anti-GAPDH | Proteintech | Cat# 60004–1-Ig; RRID:AB_2107436 | WB (1:5000) |
| Antibody | Rabbit polyclonal anti-Caspase3b p10 | This paper | Immunogen: YFRPKRPAIDL*C WB (1:3000) IF (1:500) | |
| Antibody | Rabbit polyclonal anti-Caspase3b p20 | This paper | Immunogen: LSQEDHSDADC WB (1:2000) | |
| Antibody | Alexa 488 goat anti-mouse IgG | Abcam | Cat#ab150113; RRID:AB_2576208 | IF (1:500) |
| Antibody | Alexa 555 goat anti-rabbit IgG | Abcam | Cat#ab150078; RRID:AB_2722519 | IF (1:500) |
| Commercial assay or kit | One Step TUNEL Apoptosis Assay Kit | Beyotime | Cat#C1088 | |
| Commercial assay or kit | Absolutely RNA Nanoprep Kit | Agilent | Cat#400753 | |
| Commercial assay or kit | RoomTemp Sample Lysis Kit | Vazyme | Cat#P073 | |
| Commercial assay or kit | TRIzol Reagent | Ambion | Cat#15596018 | |
| Commercial assay or kit | FastQuant RT Kit with gDNase | Tiangen | Cat#KR106 | |
| Commercial assay or kit | PowerUp SYBR Green Master Mix | Applied Biosystems | Cat#A25742 | |
| Commercial assay or kit | T7 RiboMAX Express RNAi System | Promega | Cat#P1700 | |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | GraphPad Prism software | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 |
Additional files
-
Supplementary file 1
KEGG and GO enrichment of head transcriptome, western blot labeled by a Caspase3b p20 antibody, immunofluorescence and FISH of TYLCV, RNA interference efficiency.
- https://cdn.elifesciences.org/articles/56168/elife-56168-supp1-v1.docx
-
Supplementary file 2
The list of differentially expressed genes (DEGs).
- https://cdn.elifesciences.org/articles/56168/elife-56168-supp2-v1.xls
-
Supplementary file 3
The list of all gene expression (FPKM).
- https://cdn.elifesciences.org/articles/56168/elife-56168-supp3-v1.xls
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56168/elife-56168-transrepform-v1.docx