Convergent changes in muscle metabolism depend on duration of high-altitude ancestry across Andean waterfowl
Figures
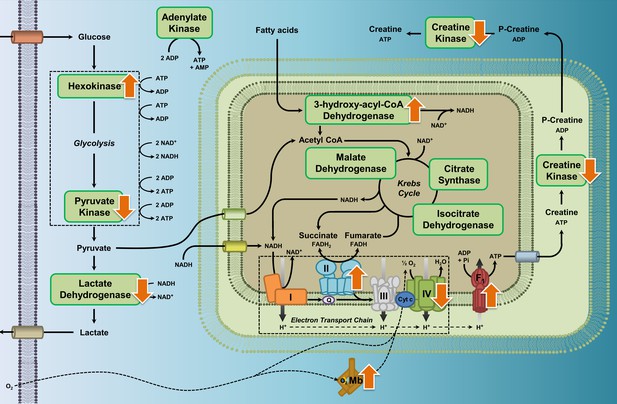
Enzyme pathway diagram illustrating where we observed differences in metabolic enzyme activity and myoglobin content in high-altitude waterfowl compared to their close low-altitude relatives.
In addition to the observed increases in myoglobin content, increases in the activities of hexokinase, ATP synthase, HOAD, and complex II (succinate dehydrogenase), and decreases in activities of pyruvate kinase, lactate dehydrogenase, creatine kinase and complex IV (cytochrome c oxidase), we observed no changes in activity for the enzymes citrate synthase, isocitrate dehydrogenase, malate dehydrogenase, complex I (NADH-ubiquinone oxidoreductase), and adenylate kinase.

Simplified phylogenetic tree, generated using maximum parsimony and constrained to the same topology as the global waterfowl phylogeny published by Gonzalez et al., 2009.
(see Figure 2—figure supplement 1). Branch lengths are measured as the total number of nucleotide substitutions in the 5’ end of the mtDNA control region.

Phylogeny of the waterfowl based on Gonzalez et al.
Gonzalez et al., 2009 showing the placement of seven high-altitude waterfowl lineages in this study.

Metabolic enzyme activities for (A) lactate dehydrogenase (LDH), (B) pyruvate kinase (PK), (C) creatine kinase (CK), (D) hexokinase (HK), and (E) 3-hydroxyacyl-CoA dehydrogenase (HOAD), measured in the pectoralis of high- and low-altitude waterfowl.
The diagonal represents the line of equality (x = y). Values are shown as mean ± SEM U/g tissue (n = 8–12). High-altitude values are significantly different overall from the corresponding low-altitude values when p<0.05 in Wilcoxon’s Signed-Rank Tests, which were carried out including (*) and excluding (†) ruddy ducks.
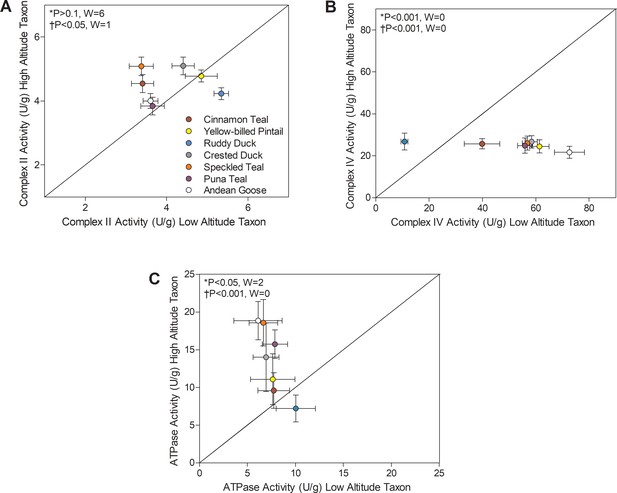
Mitochondrial enzyme activities for (A) Complex II, (B) Complex IV, and (C) ATP synthase measured in the pectoralis of high- and low-altitude waterfowl.
The diagonal represents the line of equality (x = y). Values are shown as mean ± SEM U/g tissue (n = 8–12). High-altitude values are significantly different overall from the corresponding low-altitude values when p<0.05 in Wilcoxon’s Signed-Rank Tests, which were carried out including (*) and excluding (†) ruddy ducks.
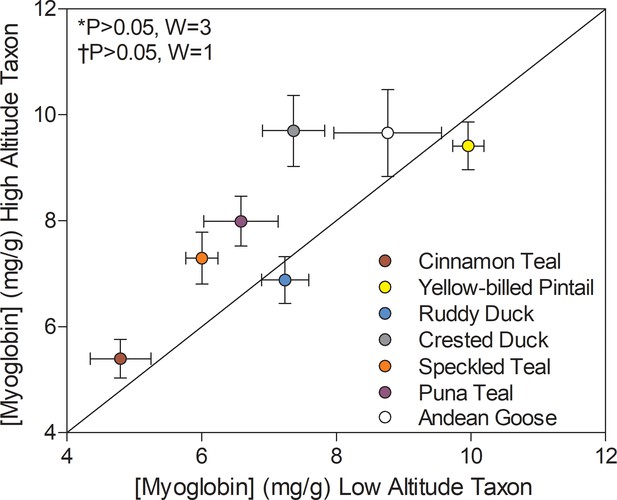
Myoglobin content measured in the pectoralis of high- and low-altitude waterfowl.
The diagonal represents the line of equality (x = y). Values are shown as mean ± SEM mg/g tissue (n = 8–12). High-altitude values are significantly different overall from the corresponding low-altitude values when p<0.05 in Wilcoxon’s Signed-Rank Tests, which were carried out including (*) and excluding (†) ruddy ducks.
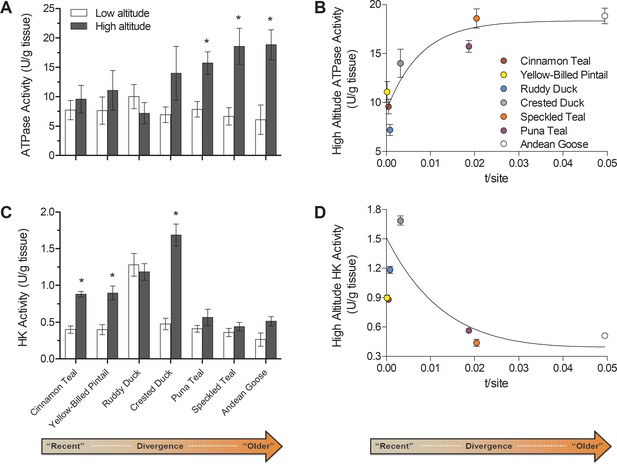
Changes over evolutionary time at altitude of (A) ATP synthase activity and (C) hexokinase activity measured in thepectoralis of seven high- and low-altitude waterfowl pairs.
Values are given as the mean ± SEM U/g tissue (n = 8–12). * - Significantly different activity in high-altitude ducks compared to low-altitude ducks (two-factor ANOVA followed by the Bonferroni post-tests; p<0.05). (B) ATP synthase and (D) Hexokinase activities in each high-altitude taxon plotted against the t/site value between each high-low pair.
Tables
Seven species of Andean ducks showing classification level, ΦST, time since divergence (t/site), and the approximate time (T) ago in years they became established at high altitude based on coalescent analysis.
ΦST and t/site were calculated using previously published mtDNA sequences. T in years was calculated using the substitution rate published by Peters et al., 2005 of 4.8 × 10−8 substitutions/site/year.
| Cinnamon teal | Yellow-billed pintail | Ruddy duck | Crested duck | Puna teal (H) Silver teal (L) | Speckled teal | Andean goose (H) Magellan goose (L) |
|---|---|---|---|---|---|---|
| New | New | New | Intermediate | Established | Established | Established |
| Subspecies | Populations | Subspecies | Subspecies | Species | Subspecies | Species |
| ΦST = 0.07 | ΦST = 0.05 | ΦST = 0.38 | ΦST = 0.85 | ΦST = 0.93 | ΦST = 0.77 | ΦST = 1.0 |
| t/site = 0.000143116 | t/site = 0.00052227 | t/site = 0.000806087 | t/site = 0.003174242 | t/site = 0.017886364 | t/site = 0.019886364 | t/site = 0.04547956 |
| T (years) = 2982 | T (years) = 10,898 | T (years) = 16,793 | T (years) = 66,130 | T (years) = 372,633 | T (years) = 414,219 | T (years) = 947,491 |
| Capture range HA = 3812 m LA = 0–13 m | Capture range HA = 3812 m LA = 3 m | Capture range HA = 3812 m LA = 480–507 m | Capture range HA = 4281–4655 m LA = 760–1050 m | Capture range HA = 3812 m LA = 410–485 m | Capture range HA = 4209–4657 m LA = 760–1050 m | Capture range HA = 4368–4806 m LA = 0–27 m |
| HA (n = 8) LA (n = 8) | HA (n = 8) LA (n = 10) | HA (n = 6) LA (n = 10) | HA (n = 12) LA (n = 10) | HA (n = 11) LA (n = 10) | HA (n = 11) LA (n = 10) | HA (n = 12) LA (n = 8) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Chemical compound, drug | Glucose | Sigma G8270 | D-(+)-Glucose ≥99.5% (GC) | Enzyme assay reagent |
| Chemical compound, drug | ATP | Sigma A2383 | Adenosine 5’-triphosphate disodium salt hydrate Grade I,≥99%, from microbial | Enzyme assay reagent |
| Chemical compound, drug | MgCl2 | Sigma M8266 | Magnesium Chloride anhydrous,≥98% | Enzyme assay reagent |
| Chemical compound, drug | NADP+ | BioShop Canada NAD007 | B-NADP, Disodium trihydrate,>95% | Enzyme assay reagent |
| Chemical compound, drug | G6PDH | Roche 10127655001 | Glucose-6-Phosphate Dehydrogenase (G6P-DH) grade I, from yeast | Enzyme assay reagent |
| Chemical compound, drug | LDH | Roche 10127876001 | L-Lactate Dehydrogenase (L-LDH) from rabbit muscle | Enzyme assay reagent |
| Chemical compound, drug | PEP | Sigma P7002 | Phosphoenolpyruvic acid trisodium salt hydrate ≥97% (enzymatic) | Enzyme assay reagent |
| Chemical compound, drug | ADP | Sigma A5285 | Adenosine 5’-diphosphate monopotassium salt dehydrate bacterial,≥95%, powder | Enzyme assay reagent |
| Chemical compound, drug | Pyruvate | Sigma P2256 | Sodium pyruvate ReagentPlus,≥99% | Enzyme assay reagent |
| Chemical compound, drug | NADH | BioShop Canada NAD002 | NADH ß-NICOTINAMIDE ADENINE REDUCED | Enzyme assay reagent |
| Chemical compound, drug | Oxaloacetate | Sigma O4126 | Oxaloacetic acid ≥97% (HPLC) | Enzyme assay reagent |
| Chemical compound, drug | Acetyl CoA | BioShop Canada ACO201 | ACETYL COENZYME A, Trilithium Salt | Enzyme assay reagent |
| Chemical compound, drug | DTNB | Sigma D218200 | 5,5’-Dithiobis(2-nitrobenzoic acid) ReagentPlus, 99% | Enzyme assay reagent |
| Chemical compound, drug | Isocitrate | Sigma I1252 | DL-Isocitric acid trisodium salt hydrate ≥93% | Enzyme assay reagent |
| Chemical compound, drug | CoQ10 | Sigma C9538 | Coenzyme Q10, ≥98% (HPLC) | Enzyme assay reagent |
| Chemical compound, drug | Rotenone | Sigma R8875 | Rotenone, ≥95% | Enzyme assay reagent |
| Chemical compound, drug | BSA | Sigma A6003 | Bovine Serum Albumin lyophilized powder, essentially fatty acid free,≥96% (agarose gel electrophoresis) | Enzyme assay reagent |
| Chemical compound, drug | KCN | Sigma 60178 | Potassium cyanide BioUltra,≥98.0% (AT) | Enzyme assay reagent |
| Chemical compound, drug | Succinate | Sigma S2378 | Sodium succinate dibasic hexahydrate ReagentPlus,≥99% | Enzyme assay reagent |
| Chemical compound, drug | DCPIP | Sigma D1878 | 2,6-Dichloroindophenol sodium salt hydrate, BioReagent | Enzyme assay reagent |
| Chemical compound, drug | DUB | Sigma D7911 | Decylubiquinone, ≥97% (HPLC) | Enzyme assay reagent |
| Chemical compound, drug | CytcCH2 | Sigma C7752 | Cytochrome c from equine heart ≥95% based on Mol. Wt. 12,384 basis | Enzyme assay reagent |
| Chemical compound, drug | Oligomycin | Sigma O4876 | Oligomycin from Streptomyces diastatochromogenes ≥90% total oligomycins basis (HPLC) | Enzyme assay reagent |
| Chemical compound, drug | HK | Roche 11426362001 | Hexokinase (HK) | Enzyme assay reagent |
| Chemical compound, drug | Acetoacetyl CoA | Sigma A1625 | Acetoacetyl coenzyme A sodium salt hydrate Cofactor, for acyl transfer | Enzyme assay reagent |
| Chemical compound, drug | Creatine | Sigma C3630 | Creatine monohydrate, ≥98% | Enzyme assay reagent |
| Chemical compound, drug | PK | Roche PK-RO | Pyruvate Kinase (PK) from rabbit muscle | Enzyme assay reagent |
| Chemical compound, drug | KH2PO4 | P5378 | Potassium phosphate monobasic, ReagentPlus | Assay buffer reagent |
| Chemical compound, drug | EGTA | Sigma E4378 | Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid, ≥97.0% | Assay buffer reagent |
| Chemical compound, drug | EDTA | Sigma EDS | Ethylenediaminetetraacetic acid BioUltra, anhydrous,≥99% (titration) | Assay buffer reagent |
| Chemical compound, drug | Triton-X 100 | Sigma X100 | Triton X-100 laboratory grade | Assay buffer reagent |
| Software, algorithm | Geneious | Biometters Ltd., Auckland, NZ | Used for sequence alignment | |
| Software, algorithm | PAUP | Version 4, Sinauer Associates, Sunderland, Massachusetts, USA | Used to generate branch lengths | |
| Software, algorithm | MESQUITE | https://www.mesquiteproject.org/ | Used to analyze phylogenetic contrasts | |
| Software, algorithm | PDAP module | http://mesquiteproject.org/pdap_mesquite/ | Used to analyze phylogenetic contrasts | |
| Software, algorithm | IM | https://bio.cst.temple.edu/~hey/software | Used to calculate divergence |
Additional files
-
Supplementary file 1
Supporting data tables, statistical analyses, and methodology.
(a) Maximal activities (µmol/g tissue/min), body mass (g) and myoglobin (Mb; mg/g tissue) concentration in pectoralis muscle. (b) Two-factor ANOVA results of maximal activities (µmol/g tissue/min), mass (g) and myoglobin (Mb; mg/g tissue) concentration in pectoralis muscle. (c) Two-factor ANOVA results of maximal activities (µmol/g tissue/min), mass (g) and myoglobin (Mb; mg/g tissue) concentration in pectoralis muscle excluding data for ruddy ducks from the subfamily Oxyurinae. (d) Test of covariance for enzyme activity (µmol/g tissue/min) or myoglobin content (Mb; mg/g tissue) and body mass (g). (e) Test of covariance for enzyme activity (µmol/g tissue/min) or myoglobin content (Mb; mg/g tissue) and body mass (g) excluding data for ruddy ducks from the subfamily Oxyurinae. (f) Correlation analyses of phylogenetic independent contrasts of bird mass (g), myoglobin (Mb) content (mg/g tissue), or enzyme activity (µmol/g tissue/min) versus altitude (m). (g) Correlation analyses of phylogenetic independent contrasts of bird mass (g), myoglobin (Mb) content (mg/g tissue), or enzyme activity (µmol/g tissue/min) versus altitude (m) excluding data for ruddy ducks from the subfamily Oxyurinae. (h) Assay conditions for enzymatic measurements. (i) List of GenBank gene accession numbers for mtDNA control region used in the construction of the phylogenetic tree. (j) Maximal activities (µmol/g tissue/min) in pectoralis muscle from surface, intermediate and deep tissue sampling locations.
- https://cdn.elifesciences.org/articles/56259/elife-56259-supp1-v3.docx
-
Supplementary file 2
Complete list of accession numbers, databases and source references used to create the phylogenetic tree.
- https://cdn.elifesciences.org/articles/56259/elife-56259-supp2-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56259/elife-56259-transrepform-v3.pdf





