SynGAP isoforms differentially regulate synaptic plasticity and dendritic development
Figures
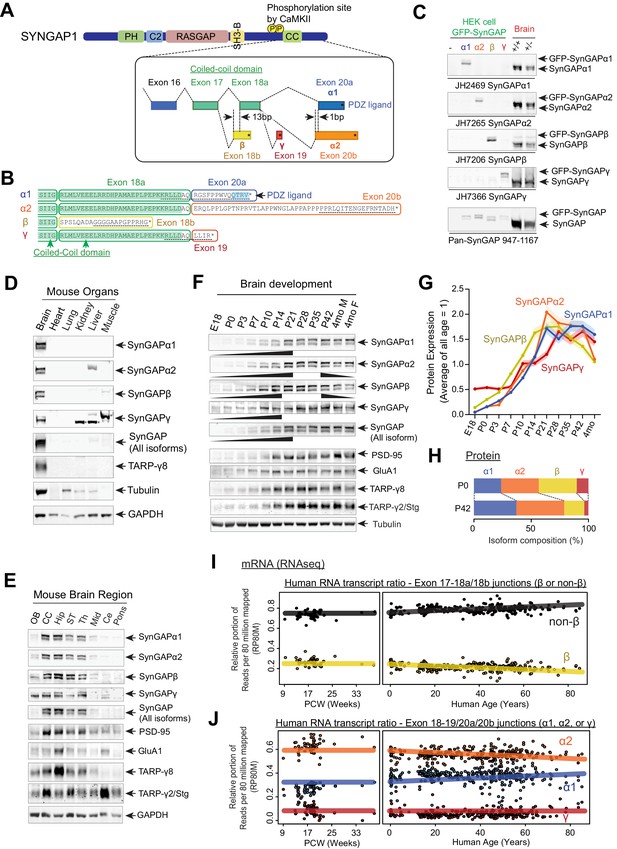
SynGAP isoforms are differentially expressed during brain development.
(A) Schematic of SYNGAP1 splicing at the C-terminus. SYNGAP1 is alternatively spliced within exons 18–20 to generate four unique C-terminal isoforms designated as α1, α2, β, and γ. (B) C-terminal amino-acid sequences of SynGAP isoforms encoding select protein domains. Coil-Coil domain (yellow) and PDZ ligand-binding domain (blue). Targeted epitopes of isoform-specific SynGAP antibodies (JH2469, JH7265, JH7206, and JH7366) are indicated as dotted lines. (C) Specificity of SynGAP isoform-specific antibodies. Immunoblots of SynGAP isoform expression in lysates prepared from HEK 293 T cells expressing individual GFP-tagged SynGAP isoforms and lysates prepared from brain tissue obtained from WT and Syngap1 +/- mice were shown. Quantification of relative SynGAP isoform levels with respect to total SynGAP expression measured from immunoblot were shown in Figure 1—figure supplement 1A. Two-way ANOVA followed by Tukey's post hoc test (Isoform F(4,30) = 1.900; p=0.13, Genotype F(1,30) = 451.2; p<0.001, Interaction F(4,30)=1.900; p=0.13, n = 4 each condition) was performed. Error bar indicates ± SEM. (D) Endogenous expression and distribution of SynGAP isoforms in various organs. Immunoblots of qualitative distribution of SynGAP isoforms in lysates prepared from various organ tissues of WT mice were shown. Asterisks indicate non-specific bands that are also detected in tissue from knockout mice. Two-way ANOVA followed by Tukey's post hoc test (Tissue F(5,144) = 1433; p<0.0001, Isoform F(7,144) = 229.3; p<0.0001, Interaction F(35,144) = 25.45; p<0.0001, n = 4 each condition) was performed. Heat map of immunoblots was displayed in Figure 1—figure supplement 1B. The amount of protein in the brain is standardized as 1.0. (E) Western blot of endogenous levels of individual SynGAP isoforms and other synaptic proteins in lysates prepared from several brain regions obtained from WT and Syngap1 +/- mice. (OB: Olfactory bulb, CC: Cerebral cortex, Hip: Hippocampus, ST: Striatum, Th: Thalamus, Mid: Midbrain, Ce: Cerebellum). Two-way ANOVA followed by Tukey's post hoc test (Brain regions F(7, 264)=1048; p<0.0001, Molecules F(10,264) = 8.0 x 10 −12; p>0.9999, Interaction F(70.264) = 59.06; p<0.0001, n = 4 each condition) was performed. Graph showing the mean values of each signal was displayed in Figure 1—figure supplement 1C. (F–H) Developmental expression profiles of individual SynGAP isoforms and related synaptic proteins. (F) Immunoblots of SynGAP isoform expression measured in forebrain tissue lysates prepared from WT and Syngap1 +/- mice at different developmental ages. (G) Quantification of immunoblots representing relative enrichments along developmental stage. The mean values of each signal were plotted in the graph. (H) Quantification of absolute SynGAP isoform abundance at P0 and P42 from C and G. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Developmental stage F(10,330) = 397.4; p<0.0001, Molecule F(9,330) = 2.116; p=0.027, Interaction F(90,330) = 26.18; p<0.0001, n = 4 each condition) was performed. (I) mRNA expression of the β and non-β SYNGAP1 isoforms across age in human dorsolateral prefrontal cortex. The relative portion of RNAseq reads spanning the exon 17–18 junction supporting either isoform was plotted against human age (post-conception weeks and years) with a linear regression. (J) mRNA expression of the α1, α2, and γ SYNGAP1 isoforms across age. The relative portion of RNAseq reads spanning the exon 18–19 junction (γ) or 18–20 (α1, α2) junction supporting each isoform was plotted against human age. Reads per 80 million mapped (RP80M) of RNAseq data are shown in Figure 1—figure supplement 1E,F.
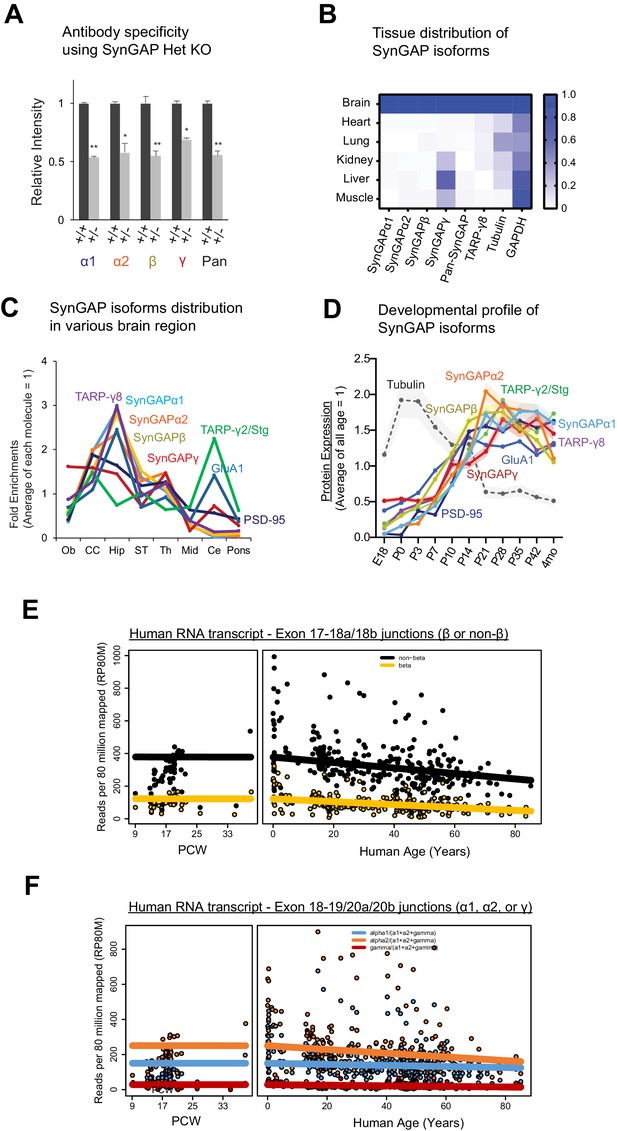
SynGAP isoforms are differentially expressed during brain development.
(A) Quantification of Figure 1C immunoblot showing specificity of SynGAP isoform-specific antibodies. Graph of relative SynGAP isoform levels with respect to total SynGAP expression measured from immunoblot was displayed. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Isoform F(4,30) = 1.900; p=0.13, Genotype F(1,30) = 451.2; p<0.001, Interaction F(4,30)=1.900; p=0.13, n = 4 each condition) was performed. Error bar indicates ± SEM. (B) Quantification of Figure 1D immunoblot showing endogenous expression and distribution of SynGAP isoforms in various organs. Heat map of immunoblot was displayed. The amount of protein in the brain is standardized as 1.0. (C) Quantification of Figure 1E immunoblot showing western blot of endogenous levels of individual SynGAP isoforms and other synaptic proteins in lysates prepared from several brain regions obtained from WT. (OB: Olfactory bulb, CC: Cerebral cortex, Hip: Hippocampus, ST: Striatum, Th: Thalamus, Mid: Midbrain, Ce: Cerebellum). Two-way ANOVA followed by Tukey's post hoc test (Brain regions F(7, 264)=1048; p<0.0001, Molecules F(10,264) = 8.0 x 10 −12; p>0.9999, Interaction F(70.264) = 59.06; p<0.0001, n = 4 each condition) was performed. The mean values of each signal were plotted in the graph. (D) Complete set of quantification for immunoblots representing relative enrichments along developmental stage corresponds to Figure 1G. The mean values of each signal were plotted in the graph. Error bars indicate ± SEM. (E) mRNA expression of the β and non-β SYNGAP1 isoforms across age in human dorsolateral prefrontal cortex. RNAseq reads spanning the exon 17–18 junction supporting either isoform were normalized to sequencing depth and plotted against human age (post-conception weeks and years) with a linear regression. (F) mRNA expression of the α1, α2, and γ SYNGAP1 isoforms across age. RNAseq reads spanning the exon 18–19 junction (γ) or exon 18–20 junction (α1, α2) supporting each isoform were normalized to sequencing depth and plotted against human age.
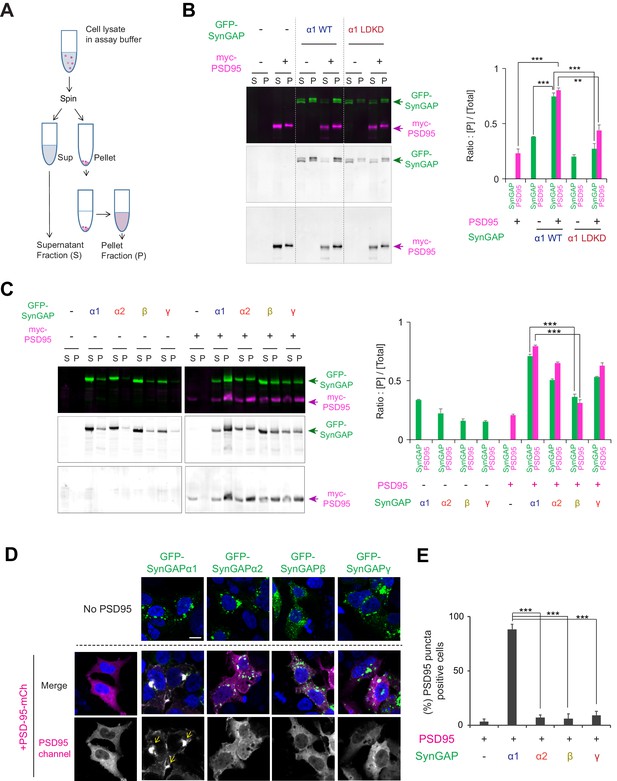
Condensation properties and subcellular localization of various SynGAP isoforms in live cells.
(A) Schematic diagram of LLPS sedimentation assay. HEK 293 T cell lysates were centrifuged and fractionated into insoluble pellet (P) and soluble supernatant (S) fractions. (B) Representative immunoblot probing levels of GFP-SynGAP (WT or phase separation mutant) and myc-PSD-95 in phase-separated supernatant and pellet lysate fractions obtained from HEK cells expressing myc-PSD-95 and either GFP-SynGAP-WT or GFP-SynGAP-LDKD constructs. (Right panel) Quantification of pellet fraction ratios obtained from averaged immunoblots as the representative example shown in (A). Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Molecules F (1,30)=3.026; p=0.09, Transfections F(4,30) = 280.7; p<0.0001, Interaction F(4,30)=59.69; p<0.0001, n = 4, ***p<0.001, **p<0.01, *<0.05) was performed. (C) Representative western blot probing levels of individual SynGAP isoforms in phase-separated supernatant and pellet lysate fractions obtained from HEK cells expressing myc-PSD-95 and individual GFP-tagged SynGAP isoforms. (Right panel) Quantification of pellet fraction ratios obtained from averaged western blots as the representative example shown in (D). Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Transfections F(8,54) = 812,2; p<0.0001, Molecules F (1,54)=50.88; p<0.0001, Interaction F(8,54) = 101.5; p<0.0001, n = 4, ***p<0.001, **p<0.01, *<0.05) was performed. (D, E) Representative confocal images of living HEK cells expressing myc-PSD-95 alone or myc-PSD-95 and individual SynGAP isoforms. Scale Bar, 10 μm (D). (E) Quantification of the averaged percentage of PSD-95-positive puncta identified in images of living HEK cells as shown in (D). Error bars indicate ± SEM. One-way ANOVA ANOVA followed by Tukey's post hoc test (Transfections F (4, 15)=96.77; p<0.0001, n = 4 independent coverslip, ***p<0.001, **p<0.01, *<0.05) was performed.
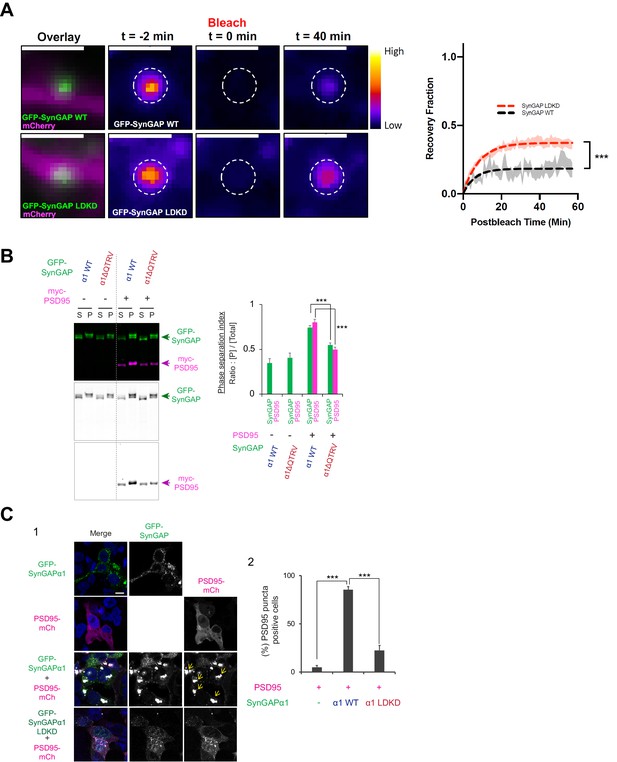
Condensation properties and subcellular localization of various SynGAP isoforms in live cells.
(A) (Left) Representative confocal time course images of dendritic spines of cultured neurons before and after photobleaching of shRNA-resistant GFP-SynGAP (WT or LDKD) under conditions of endogenous SynGAP knockdown. The left-most images display and overlay of the GFP signal from GFP-SynGAP and mCherry reporting the transfection of the SynGAP shRNA, two minutes before photobleaching. Time course images show relative fluorescence intensity before (t = −2 min), immediately following (t = 0 min), and 40 min (t = 40 min) following photobleaching with a 488 nm laser. Scale bar = 2 μm. (Right) Quantification of fluorescence recovery following photobleaching normalized to pre-bleach mean fluorescence intensity. Dashed-line curves (black = GFP SynGAP WT; red = GFP SynGAP LDKD) represent the results of single-order exponential fitting following nonlinear regression analysis. Shadows around each curve represent the SEM for the raw values. Curve plateaus were used to estimate the mobile fraction. The plateau of GFP-SynGAP WT recovery was 0.183 (95% CI = 0.169–0.199), and the plateau of GFP-SynGAP LDKD recovery was 0.372 (95% CI = 0.362–0.384). Plateaus were compared using the extra sum-of-squares F test. GFP-SynGAP WT n = 16 bleached spines from 4 neurons; GFP-SynGAP LDKD n = 19 bleached spines from 3 neurons. ***p<0.0001, F = 82.99 (1, 2026). (B) Representative immunoblot probing levels of GFP-SynGAP (WT or PDZ deletion mutant) and myc-PSD95 in phase-separated supernatant and pellet lysate fractions obtained from HEK cells expressing myc-PSD95 and either GFP-SynGAP-WT or GFP-SynGAP-ΔQTRV (PDZ ligand deletion) constructs. (Right panel) Quantification of pellet fraction ratios obtained from averaged immunoblots as the representative example shown in left panel. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Molecules F(1,24) = 195.00; p<0.0001, Transfections F (3,24)=456.7; p<0.0001, Interaction F(3,24)=71.27; p<0.0001, n = 4, ***p<0.001, **p<0.01, *<0.05) was performed. (C) Representative confocal images of fixed HEK cells expressing myc-PSD95 alone or myc-PSD95 and either GFP-SynGAP-WT or GFP-SynGAP-LDKD constructs (F1). Scale Bar, 10 μm. Arrowheads indicate PSD95- and SynGAP-α1-containing puncta (>1 μm). (F2) Quantification of the averaged percentage of PSD95-positive puncta identified in images of fixed HEK cells as shown in (F1). Error bars indicate ± SEM. One-way ANOVA followed by Tukey's post hoc test (Transfections F(2, 9)=126.8; p<0.0001, n = 4 independent coverslip, ***p<0.001, **p<0.01, *<0.05) was performed.
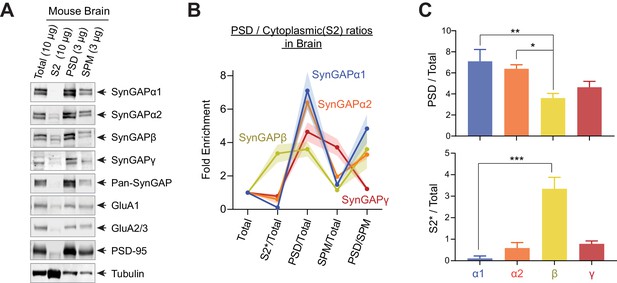
Subcellular localization of various SynGAP isoforms in the brain.
(A) Immunoblot probing endogenous levels of individual SynGAP isoforms and other synaptic proteins in forebrain tissue lysates obtained from adult mice subjected to postsynaptic density fractionation. (B, C) Averaged enrichment of SynGAP isoforms in subcellular fractions in comparison to their levels within the total homogenate fraction, S2 fractions, and PSD fractions. Error bars indicate ± SEM. Kruskal-Wallis test followed by Dunn’s multiple comparison (PSD: H(4) = 15.98; p=0.0011, S2: H(4) = 18.23, p=0.0004, n = 4–7 independent samples for each molecules, Dunn’s multiple comparison ***p<0.001, **p<0.01, *<0.05) was performed.
Subcellular localization of various SynGAP isoforms in the brain.
Complete set of quantification for immunoblot probing endogenous levels of individual SynGAP isoforms and other synaptic proteins in forebrain tissue lysates obtained from adult mice subjected to postsynaptic density fractionation that corresponds to Figure 3. Error bars indicate ± SEM.
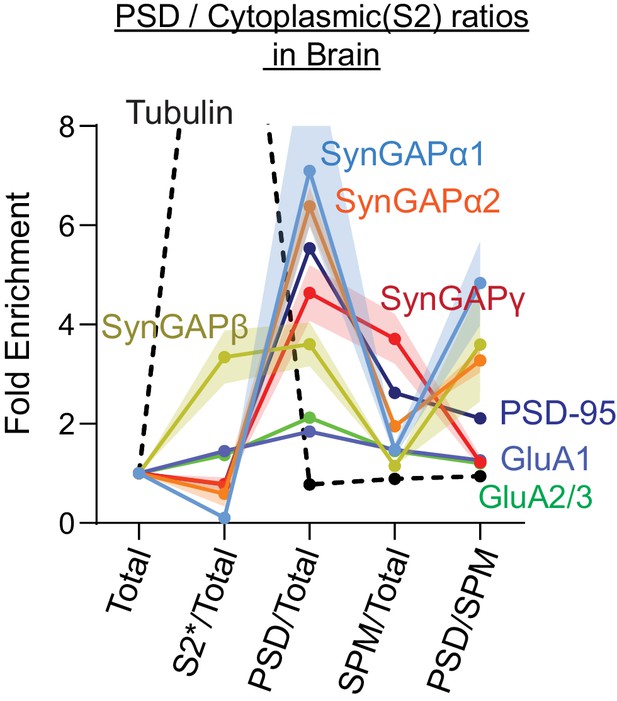
SynGAP isoforms differentially regulate the activity of small G proteins.
(A) Representative immunoblot detecting levels of active GTP-bound Ras following co-immunoprecipitation of active Ras by pulldown of Raf1 in response to expression of individual SynGAP isoforms in HEK cell lysates. (B) Representative immunoblot detecting levels of active GTP-bound Rap1 following co-immunoprecipitation of active Rap1 by pulldown of Ral1 in response to expression of individual SynGAP isoforms in HEK cell lysates. (C) Representative immunoblot detecting levels of active GTP-bound Rac1 following co-immunoprecipitation of active Rac1 by pulldown of PAK1 in response to expression of individual SynGAP isoforms in HEK cell lysates. (D, E) Quantification of averaged percent reduction of active GTP-bound forms of Ras, Rap1, and Rac1 normalized to total (active + inactive) levels in response to expression of individual SynGAP isoforms expressed in HEK cell lysates. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (SynGAP isoforms F(6,105) = 62.76; p<0.0001, small G proteins F (2,105)=7.414; p=0.0010, Interaction F(12,105) = 2.207; p=0.016, n = 6, ***p<0.001, **p<0.01, *<0.05) was performed.
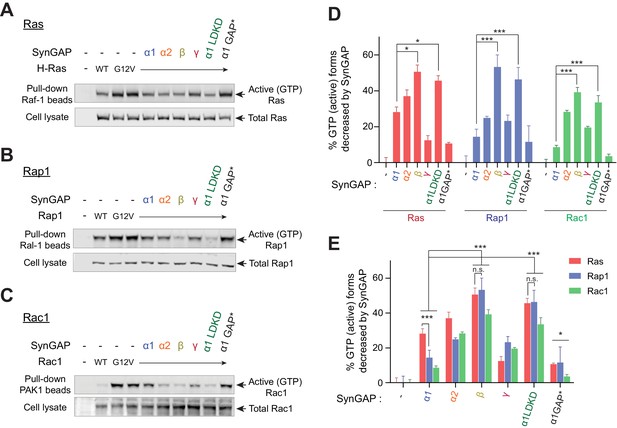
Dispersion dynamics of various SynGAP isoforms from synaptic spines during LTP.
(A) Live confocal images of hippocampal neurons expressing individual GFP-tagged SynGAP isoforms and mCherry during basal conditions and chemLTP conditions. Yellow arrows indicate dendritic spines that enlarged following chemical LTP treatment. Blue arrowheads mark dendritic spines that did not enlarge after chemical LTP treatment. Scale Bar, 5 μm. (B) Quantification of averaged relative change in the GFP-SynGAP isoform at synaptic spines following chemLTP treatment. Synaptic localization of SynGAP isoforms was determined by calculating the ratio of GFP intensity within dendritic spine heads and dividing by GFP intensity localized to the dendritic shaft at the base of the dendritic spine. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (SynGAP isoforms F(3,296) = 21.43; p<0.0001, chemLTP F (1,296)=119.9; p<0.0001, Interaction F(3,296) = 6.607; p<0.0001, n = 37–39 spines from 4 independent experiments, ***p<0.001, **p<0.01, *<0.05) was performed. Error bar indicates ± SEM.
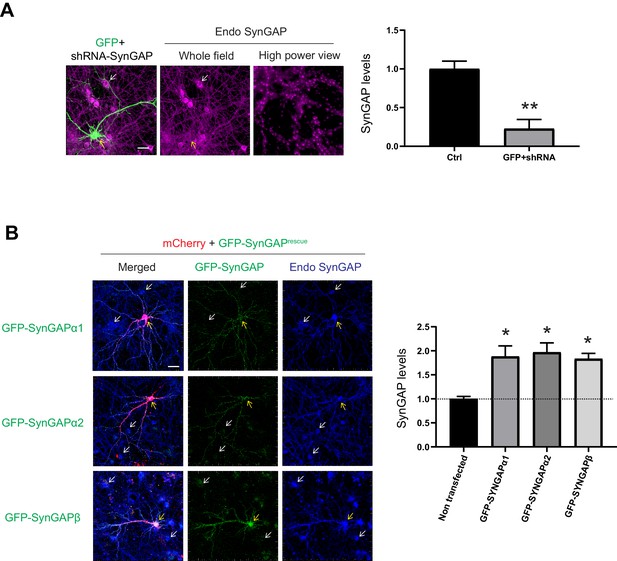
shRNA efficiency and rescue SynGAP construct titration assay.
(A) Efficiency of shRNA-SynGAP construct. Hippocampal neurons were transfected with shRNA-SynGAP#5 construct together with GFP (marker for transfected cell) and stained for pan-SynGAP antibody (**p<0.01, 77.3 ± 0.1% reduction of SynGAP expression upon our shRNA-SynGAP#5 construct, n = 4 cells, unpaired T-test (two-tailed), Error bars indicate ± SEM). Scale Bar, 10 μm. (B) Expression levels of our shRNA-resistant SynGAP isoform construct. Hippocampal neurons were transfected with mCherry and GFP-SynGAP isoform construct and stained for SynGAP isoform specific antibodies described in Figure 1 (α1 88.3 ± 0.2%, α2 97.0 ± 0.2%, β 83.5 ± 0.1% for GFP-SynGAP portion, n = 4 cells). Dotted line showed endogenous SynGAP expression levels. Error bars indicate ± SEM. One-way ANOVA followed by Tukey's post hoc test (Transfections F(4,10) = 126.8; p<0.01, n = 3 cells, *p<0.05) was performed. Scale Bar, 10 μm.
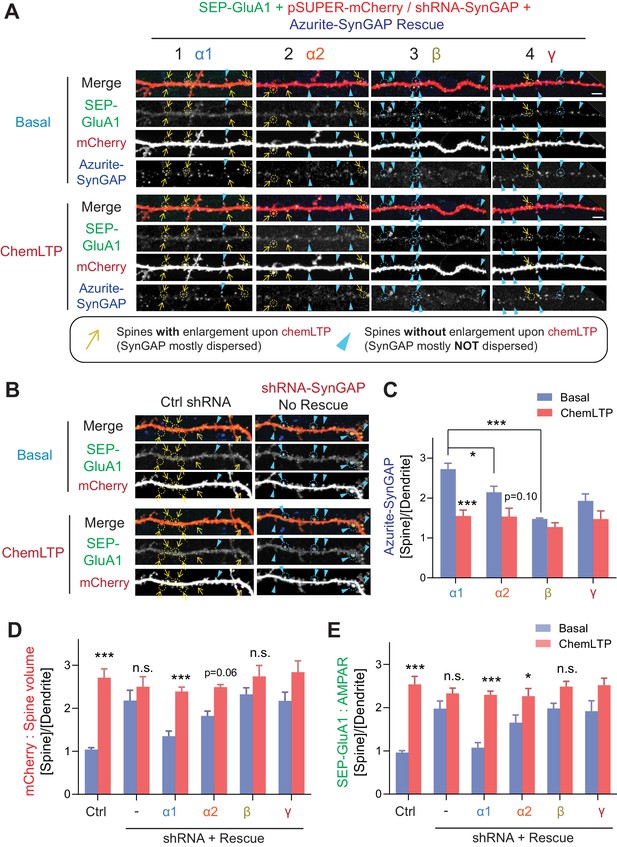
SynGAP-α1 rescues AMPA receptor trafficking and structural plasticity deficits in SynGAP-depleted hippocampal neurons.
(A, B) Representative live confocal images of cultured hippocampal neurons expressing SEP-GluA1, mCherry, and individual Azurite-tagged SynGAP isoforms in basal and chemLTP conditions. Endogenous SynGAP was knocked-down and replaced with individual Azurite-tagged SynGAP isoforms. Yellow arrows indicate dendritic spines that exhibited LTP-induced enlargement. Scale Bar, 5 μm. (C–E) Quantification of averaged SEP-GluA1, mCherry, Azurite-SynGAP intensity in dendritic spines on hippocampal neurons expressing individual SynGAP isoforms before and after chemLTP treatment. Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (ChemLTP F(1,242) = 501.1 [GluA1], 426.4 [mCherry], 219.4 [SynGAP], p<0.001; Genotype F(5,242) = 30.68 [GluA1], 35.71 [mCherry], 553.7 [SynGAP], p<0.001; Interaction (5,240)=15.02 [GluA1], 18.57 [mCherry], 553.7 [SynGAP], p<0.001; n = 47–49 spines from 4 independent coverslips each condition, ***p<0.001, **p<0.01, *<0.05) was performed. Error bar indicates ± SEM.
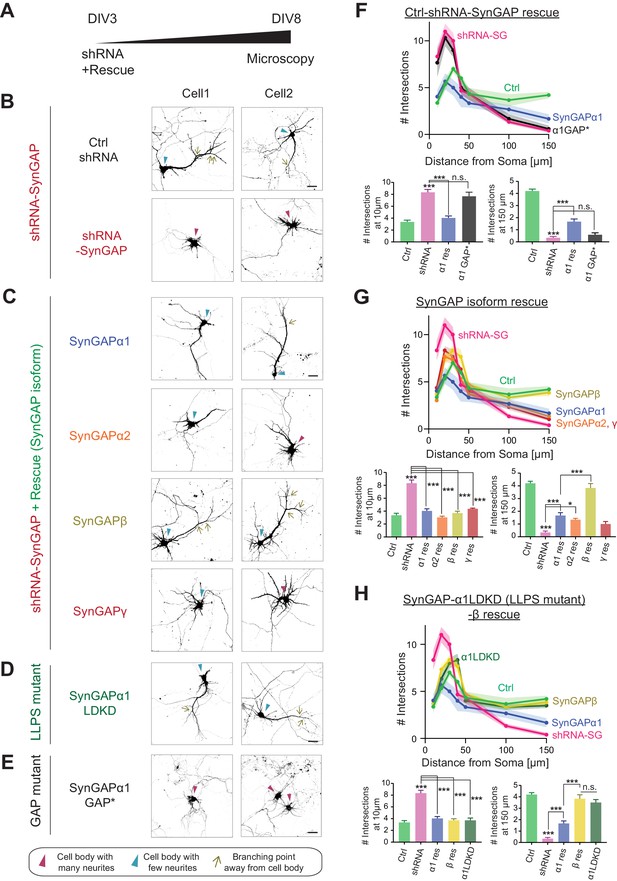
SynGAP-β rescues aberrant dendritic arbor development in SynGAP-depleted neurons.
(Α) Schematic of experimental timeline for assessing the effects of individual SynGAP isoform expression on dendritic development. Neuronal morphology was evaluated by observing co-transfected DsRed at DIV 8. (Β-E) Representative images of dendritic arbors of young cultured hippocampal neurons expressing DsRed upon SynGAP knockdown (B) and upon SYNGAP1 knockdown plus expressing individual SynGAP isoforms (C), SynGAPα1 LLPS mutant (D), or SynGAPα1 GAP mutant (E). Scale Bar, 20 μm. (F–H) Sholl analysis of dendritic branches presented as the mean number of intersections plotted as a function of distance from the center of the cell body (center = 0). Error bars indicate ± SEM. Two-way ANOVA followed by Tukey's post hoc test (Distance F(6,952) = 288.6, p<0.001; Genotype F(7,952) = 21.96, p<0.001; Interaction (42,952)=14.83, p<0.001, n = 18) was performed. Error bar indicates ± SEM.
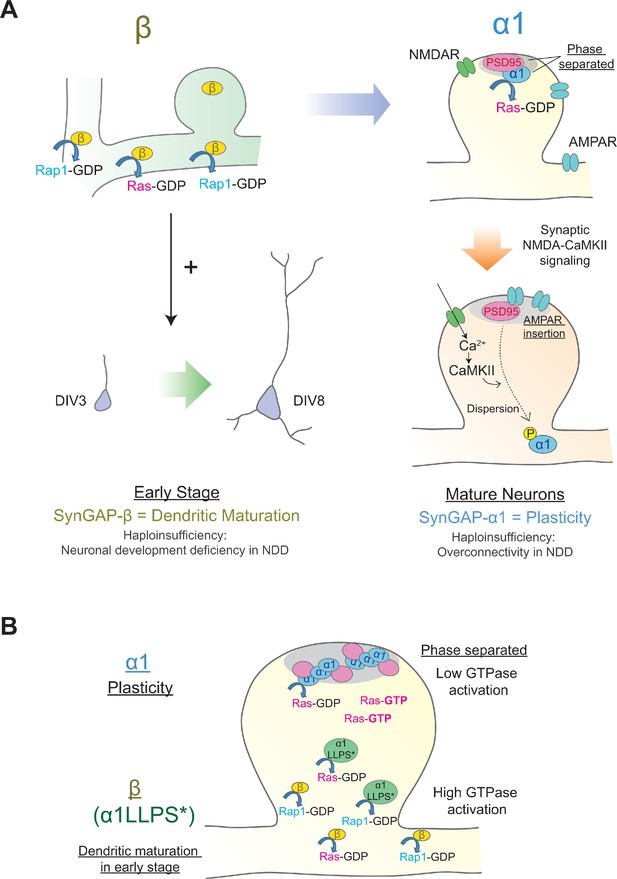
Distinct roles of individual SynGAP isoforms in neuronal development and synaptic plasticity.
(Α) Schematics illustrating isoform-specific roles for SynGAP in neuronal maturation and synaptic plasticity. SynGAP-β is expressed early in development, has the lowest LLPS propensity resulting in cytosolic localization, possesses the highest GAP activity in cells, and promotes normal dendritic development. SynGAP-β deficiency may be relevant to the neuronal development deficits in neurodevelopmental disorders (NDD). SynGAP-α1 is expressed later in development, undergoes strongest LLPS in spines resulting in dense expression in the PSD at the basal state, and is rapidly dispersed upon synaptic NMDAR-CaMKII activation. SynGAP-α1 deficiency may be relevant to the synaptic plasticity deficits and overconnectivity in NDD. (B) Schematics illustrating the phase-separation and the localization/functions of SynGAP isoforms. SynGAP-α1 is tightly packed at PSD by phase separation and has the ability of low GTPase activations. In contrast, SynGAP-β is less phase-separated and localized more in cytoplasmic region in synapses and dendritic shafts. It has a strong ability of GTPase activation. The phase-separation mutant of SynGAP-α1 LLPS*) behaves similarly to SynGAP-β.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Rattus norvegicus) | SynGAP-α1 | NM_001113409.3 | ||
| Gene (Rattus norvegicus) | SynGAP-α2 | AF050183.2 | ||
| Gene (Rattus norvegicus) | SynGAP-β | AB01692.1 | ||
| Gene (Rattus norvegicus) | SynGAP-γ | AF058789.2 | ||
| Strain, strain background Mus musculus | SynGAP KO mice | Kim et al., 2003 | Backcrossed with C57BL6 | |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216 | |
| Biological sample Mus musculus | Mouse whole brain or brain region | C57BL6, male and female | ||
| Biological sample (Rattus norvegicus) | Rat Hippocampal Primary Neuron | Days in vitro 3–21 | ||
| Antibody | Rabbit polyclononal antibody | Kim et al., 1998 | JH2469 | Anti-SynGAPα1 1:1000 |
| Antibody | Rabbit polyclononal antibody | JH7265 | Anti-SynGAPα2 1:1000 | |
| Antibody | Rabbit polyclononal antibody | JH7266 | Anti-SynGAPβ 1:1000 | |
| Antibody | Rabbit polyclononal antibody | JH7366 | Anti-SynGAPγ 1:1000 | |
| Peptide, recombinant protein | SynGAPα2 C-tail | Johns Hopkins Sequencing Facility | RH376 | CPPRLQITENGEFRNTADH |
| Peptide, recombinant protein | SynGAPβ C-tail | Johns Hopkins Sequencing Facility | RH371 | CGGGGAAPGPPRHG |
| Peptide, recombinant protein | SynGAPγ C-tail | Johns Hopkins Sequencing Facility | RH377 | CRLLDAQLLIR |
| Sequence-based reagent | Primer: SG upstream sense for ScaI cloning | IDT | SJ22 | ACTGTAGCCTGGGTGTCCAATATG |
| Sequence-based reagent | Primer: alpha 2 SG reverse | IDT | SJ24 | ggattgcggccgcCTAGTGGTCTGCGGTGTTCCG |
| Sequence-based reagent | Primer: beta SG reverse | IDT | SJ25 | ggattgcggccgcTCAGCCATGGCGGGGTGGTCC |
| Sequence-based reagent | Primer: gamma SG reverse | IDT | SJ23 | ggattGCGGCCGCttacctgatgaggagCTGAGCGTCGAGCAGCCT |
| Genetic reagent (Rattus norvegicus) | GFP-SynGAP-α1 | Araki et al., 2015 | EGFP: N terminal tag | |
| Genetic reagent (Rattus norvegicus) | GFP-SynGAP-α2 | EGFP: N terminal tag | ||
| Genetic reagent (Rattus norvegicus) | GFP-SynGAP-β | EGFP: N terminal tag | ||
| Genetic reagent (Rattus norvegicus) | GFP-SynGAP-γ | EGFP: N terminal tag | ||
| Genetic reagent (Rattus norvegicus) | GFP-SynGAP-α1 LDKD | Zeng et al., 2016 | EGFP: N terminal tag; 2-point mutations, L1202D and K1252D | |
| Genetic reagent (Rattus norvegicus) | shRNA-SynGAP#5 | Araki et al., 2015 | shRNA sequence: CCT GGA TGA AGA CTC CAT TAT | |
| Commercial assay or kit | Imject Maleimide-Activated mcKLH | Pierce | 77605 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Pierce | 23225 | |
| Chemical compound, drug | DL-AP5 | TOCRIS | 0105 | |
| Chemical compound, drug | Glycine | TOCRIS | 0219 | |
| Chemical compound, drug | Strychnine | SIGMA | P1675 | |
| Chemical compound, drug | Picrotoxin | TOCRIS | 1128 | |
| Chemical compound, drug | Tetrodotoxin citrate | TOCRIS | 1069 | |
| Software, algorithm | Prism 8 | GraphPad | ||
| Software, algorithm | ImageJ 1.8.0_112 | NIH | Particle Analysis |





