Transparent soil microcosms for live-cell imaging and non-destructive stable isotope probing of soil microorganisms
Figures
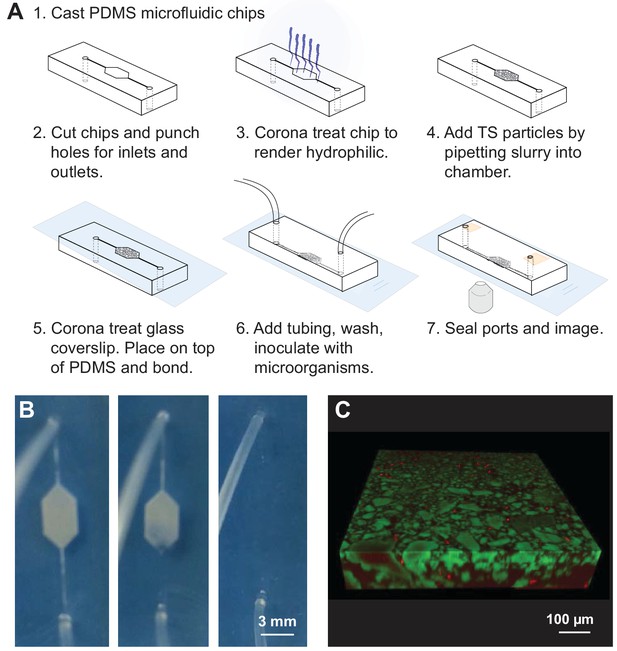
Transparent soil (TS) microcosms.
(A) Manufacture process of microcosm fluidics chambers. (B) 20% ethanol added after chip manufacture hydrates dry, hydrophobic Nafion and renders it transparent. Microfluidics chamber (3 × 5 mm hexagon, with 200 µm wide channels) filled with Nafion and attached by tubing to syringe with 20% ethanol, held in syringe pump. As ethanol is slowly flowed into the microcosm by the syringe pump, the Nafion hydrates and becomes transparent. Rehydrated Nafion can then be washed with media, washing away ethanol and rendering microcosms suitable for cell culture. (C) Three-dimensional confocal rendering of fluorescently labeled E. coli cells visualized to 100 µm depth in Nafion-based TS microcosm by confocal microscopy. Sulforhodamine-stained Nafion particles (false-colored green), and E. coli cells constitutively expressing cyan fluorescent protein (PspacC-cfp, false-colored red). Scale bar = 100 µm.
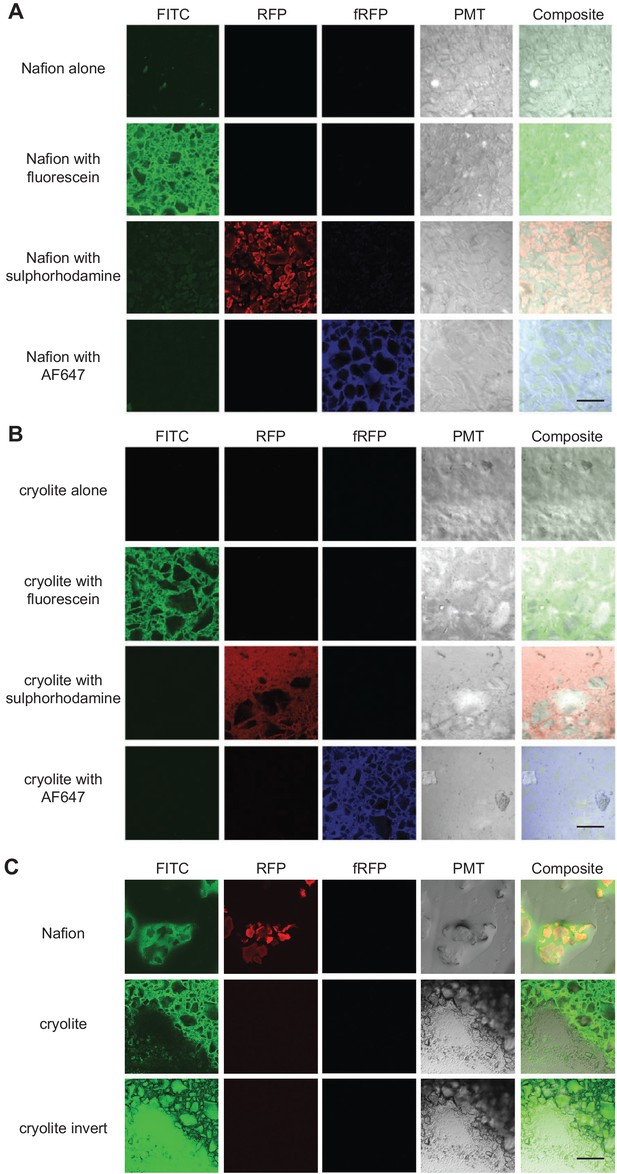
Visualization of TS matrices.
Nafion (A) and cryolite (B) were packed into a microwell microscope slide, saturated with water containing the fluorophore indicated, and imaged on a confocal laser scanning microscope with the noted filter sets. A single Z-slice about 10 µm deep into the TS matrix is shown here. ‘Cryolite with fluorescein inverted’ is the inverted image of ‘cryolite with fluorescein’, highlighting particles rather than the pore space. Partially hydrated microcosms (C) have air-filled pockets (black). Image size of each square is 850 × 850 µm. Scale bars are 250 µm.
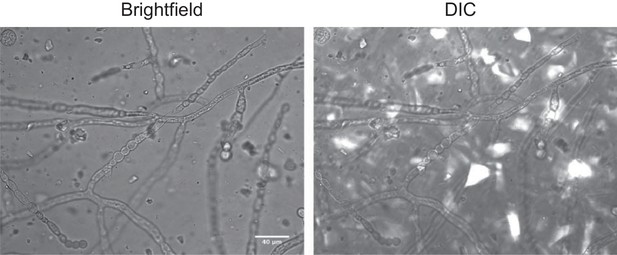
Cryolite crystals are invisible under brightfield illumination, but visible under DIC.
M. fragilis fungus was grown from spores for 24 hr at 30°C in a microfluidic chamber packed with crystalline cryolite and saturated with aqueous minimal salts growth medium (MSN, minimal salts with free ammonium and 2% glucose). Images of the fungus were taken through the cryolite matrix, about 25 µm up from the cover slip. Images taken on Zeiss Axio Observer widefield light microscope, 40x objective, water immersion lens, under brightfield or DIC illumination. Scale bar = 40 µm.
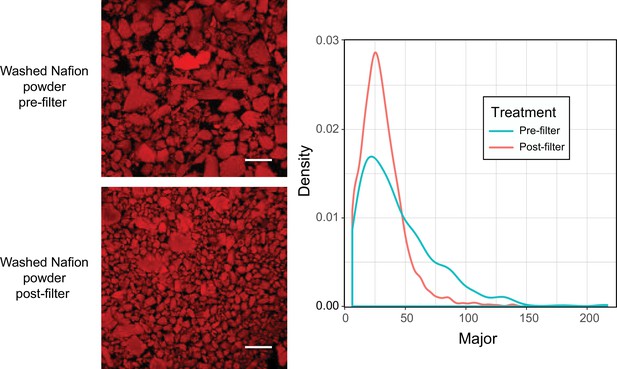
Particle size distribution of PowdION Nafion powder before and after filtering through 40 µm cell strainer.
Commercially available Nafion powder was washed (see Materials and methods) and filtered through 40 µm cell strainer. Particles were stained with sulforhodamine to render particles fluorescent in the RFP channel (false-colored red). Particles were imaged by confocal microscopy and 15 µm z-stacks flattened into maximum intensity projections. Images were analyzed in Fiji image analysis software by binarizing and applying watershed process, then calculating particle area size, represented in density plot. Scale bar = 100 µm.
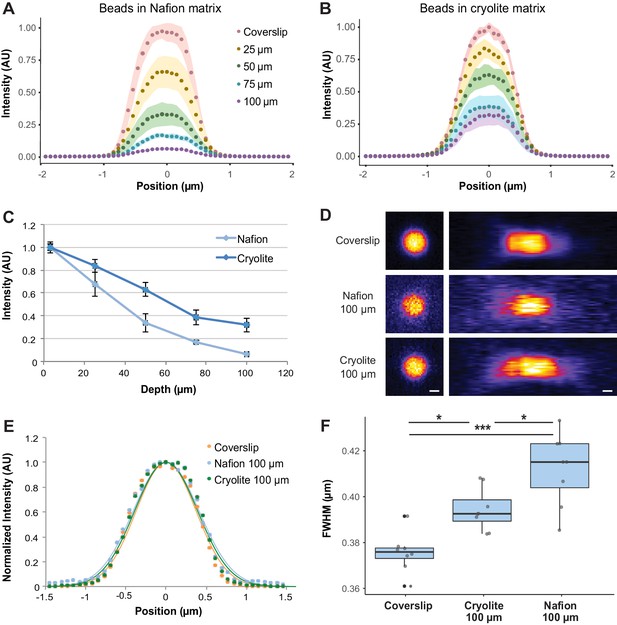
Optical properties of TS microcosms.
1 µm FITC-fluorescent beads were mixed into TS matrices saturated with water and imaged by confocal microscopy. Average lateral intensity profiles of beads at different depths within a Nafion (A) or cryolite (B) matrix (n = 8 beads per depth) are shown. (C) Maximum intensities of beads at different depths (n = 8 beads per depth) indicate a greater decay of bead brightness with depth in Nafion than in cryolite. (D) Images of 1 µm beads acquired at the coverslip and 100 µm into Nafion and cryolite microcosms, lateral (xy; left panels) and axial (xz; right panels) views. Image intensity normalized for each image; intensities are not comparable between categories. Scale bar is 0.5 µm. (E) Normalized average lateral intensity profiles of beads at coverslip and at 100 µm depth and fitted Gaussian models. (F) Full-width half-maximum (FWHM) values derived from fitted Gaussian curves of lateral intensity profiles of individual beads are a measure of spatial resolution. FWHM of beads at the coverslip are significantly lower than FWHM of beads 100 µm deep in cryolite (Tukey-Kramer HSD p-value=0.01014) and Nafion microcosms (Tukey-Kramer HSD p-value = 8.6 × 10−6), and FWHM for beads in cryolite is significantly lower than beads in Nafion at the same height (Tukey-Kramer HSD p-value=0.0160).
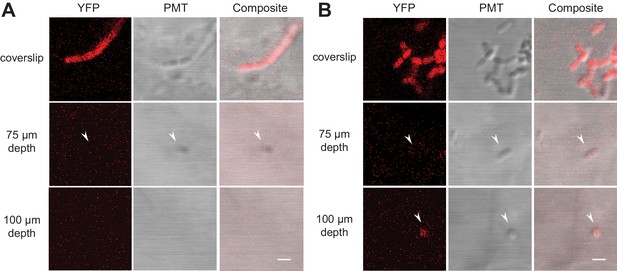
Fluorescently labeled bacteria in TS microcosms.
B. subtilis 3610 cells expressing constitutive YPet were inoculated into TS microcosms in MSgg, incubated at room temperature (22°C) for 48 hr, and Z-stacks acquired by confocal microscopy. Cells were not fixed, but imaged live. Single slices (0.75 µm thick) acquired at the coverslip, 75 µm, and 100 µm deep into (A) Nafion and (B) cryolite TS microcosms are shown (frames from Videos 1, 2, 3 and 4, YFP and DIC channels only). White arrows indicate examples of single visible bacteria. Scale bars = 1 µm.
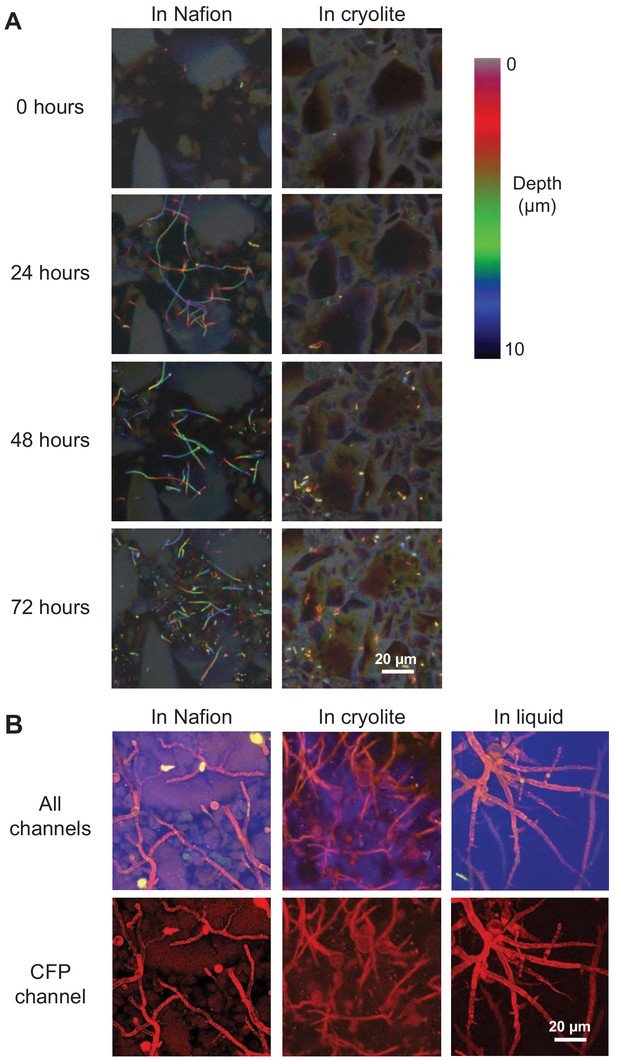
Non-destructive imaging of B. subtilis and M. fragilis in TS over time.
(A) Cells of a B. subtilis 3610 eps-tasA biofilm mutant strain expressing constitutive YPet were inoculated into TS microcosms in MSgg, incubated at room temperature (22°C), and imaged over time. Images are ten-micron thick Z-stacks flattened into color-coded Z-projections. Filaments in Nafion (left) are chains of single cells, some of which have segmented, and some of which have not yet expressed autolysins for segmentation and thus appear as filaments (Chen et al., 2009). Scale bar = 20 µm. (B) M. fragilis spores were inoculated into microcosms with or without TS and incubated for 48 hr at 30°C in MSN minimal salts with 2% glucose. Confocal micrographs of 25 µm Z-stacks flattened into maximum intensity projections; sulforhodamine-stained Nafion (false-colored green), M. fragilis autofluorescence in YFP channel (false-colored red), PMT channel (false-colored blue). Scale bar = 20 µm.
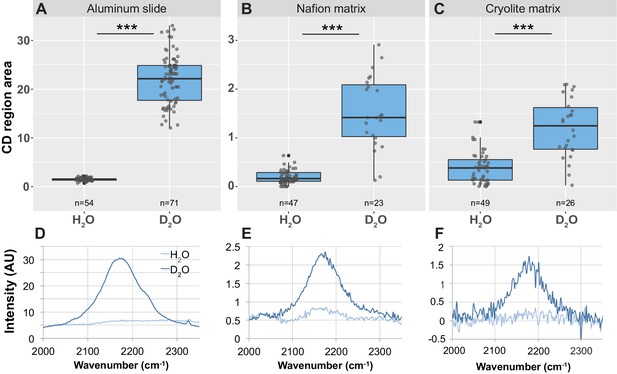
Detection of CD peak shifts and D2O labeling in B. subtilis cells in TS microcosms.
B. subtilis 3610 cells were grown in minimal salts growth medium made with either regular water or 50% heavy water (deuterium oxide, D2O). Cells were then either spotted onto an aluminum slide (A, D) or inoculated into separate Nafion (B, E) or cryolite (C, F) microcosms. Raman spectra of single cells were obtained by microspectroscopy on the aluminum slide, or within the TS matrix from cells embedded anywhere from 15 to 85 µm deep within the matrix. Average background subtracted spectra cells grown in H2O or D2O show a broad peak in the CD region of the spectrum between 2050 and 2250 in D2O-labeled cells (D, E, F). CD region for individual cell spectra was calculated as the area under the curve between 2150 and 2200 cm−1. Each dot represents CD region for an individual background-subtracted cell spectrum (A, B, C). Each boxplot represents a single separate biological replicate (e.g. all H2O-grown cells inoculated into Nafion came from a single culture, and all H2O-grown cells inoculated into cryolite came from a separate single culture). D2O-labeled cells have larger CD area than H2O-labeled cells on all three substrates (Welch’s t-test p-value<1.3 × 10−5 for all three substrates).
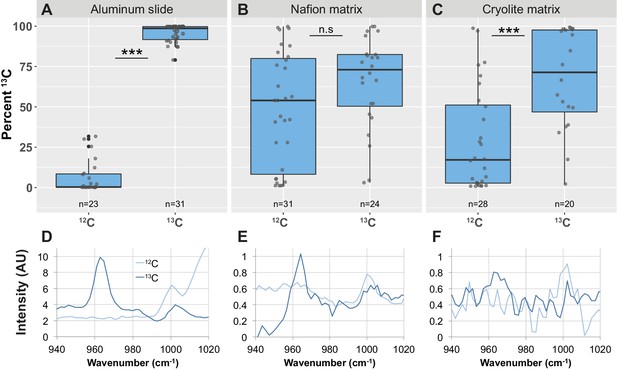
Detection of phenylalanine peak shifts and 13C enrichment in B. subtilis cells in TS microcosms.
B. subtilis 3610 cells were grown in minimal salts growth medium made with either regular (12C) glucose or 13C glucose. Cells were then either spotted onto an aluminum slide (A, D) or inoculated into separate Nafion (B, E) or cryolite (C, F) microcosms. Raman spectra of single cells were obtained by microspectroscopy on the aluminum slide, or within the TS matrix from cells embedded anywhere from 15 to 85 µm deep within the matrix. Percent 13C is calculated individually for each background subtracted cell spectrum (see Materials and methods). Each dot represents Percent 13C for an individual background-subtracted cell spectrum (A, B, C). Each boxplot represents a single separate biological replicate (e.g. all 12C-grown cells inoculated into Nafion came from a single culture, and all 12C-grown cells inoculated into cryolite came from a separate single culture). Average background subtracted spectra cells grown in 12C or 13C glucose are shown (D, E, F). On aluminum slides, 13C-labeled cells show a significantly higher Percent 13C than 12C-labeled cells (A; Welch’s t-test p-value<2.2×10−16). In Nafion TS, the Percent 13C of 13C-labeled cells are not significantly different than 12C-labeled cells (B; Welch's t-test p-value=0.2151). In cryolite TS, 13C-labeled cells show a significantly higher Percent 13C than 12C-labeled cells (C; Welch’s t-test p-value=1.348×10−4).
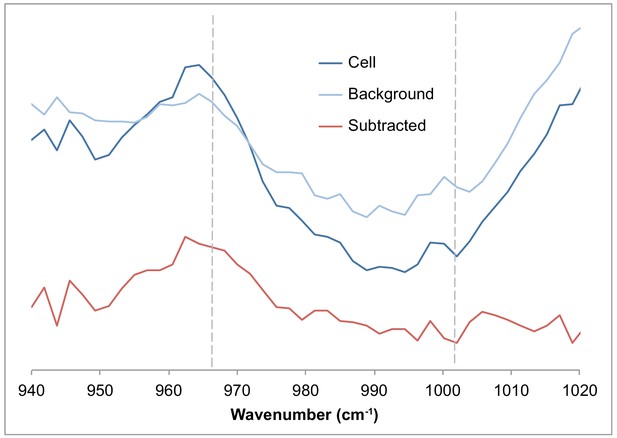
High Nafion background around 965 cm−1 interferes with 13C phenylalanine peak at 966 nm, resulting in unlabeled 12C-rich cells being misclassified as 13C labeled.
Representative single-cell spectrum showing 12C-labeled cell spectrum, a background spectrum taken 20 µm away from the cell and capturing the Raman spectrum of the Nafion matrix itself, and the subtracted spectrum (cell spectrum minus background).
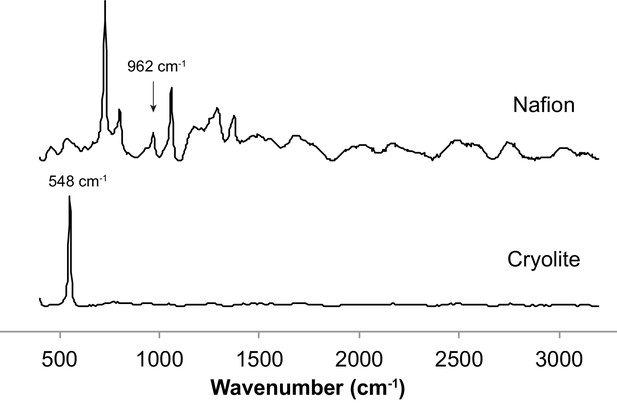
Raman spectra of Nafion and cryolite.
Nafion and cryolite powders were spotted onto an aluminum slide and Raman spectra acquired at 532 nm excitation wavelength. Resultant spectra are shown, with peaks of interest highlighted: 962 cm−1 peak in Nafion interferes with reading phenylalanine peak shift at 966 cm−1 in embedded cells; 548 cm−1 peak in cryolite is the only strong peak in the spectrum.
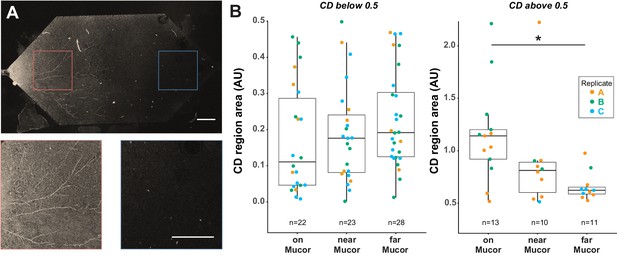
B. subtilis cells on M. fragilis are more metabolically active than cells far from M. fragilis after dry-wet cycle in Nafion-based transparent soil microcosms.
(A) B. subtilis cells grown with dead M. fragilis in Nafion TS microcosm undergo a dry-wet cycle, mimicking the dry-down and wet-up of soils. Cells were exposed to D2O for 16 hr during the wet-up phase. Because M. fragilis spores were trapped within the Nafion matrix on the side where they were inoculated, after they germinated, one side of the microcosm filled with hyphae, while the other side remains empty. Confocal microscopy in GFP channel shows autofluorescent M. fragilis hyphae on one side of microcosm (above, and left inset), and no growth on the other side (right inset). Scale bar is 400 µm in both top micrograph and insets. B. subtilis cells measured were classified as either ‘on’ (cells attached to M. fragilis hyphae), ‘near’ (cells attached to Nafion on M. fragilis-inoculated side of microcosm, within 20 µm radius of nearest hypha), or ‘far’ (cells attached to Nafion on side of microcosm without M. fragilis). ‘Far’ cells are 3 mm or more away from nearest M. fragilis hyphae. (B) Most cells (~60–70 percent) have no detectable activity after a dry-wet cycle, regardless of distance from M. fragilis (left panel, all cells with CD area less than 0.5, indicating no activity detectable by D2O uptake). However, cells that did take up D2O took up more of the label when on M. fragilis than cells far from M. fragilis (right panel, one-way ANOVA of all three categories F-statistic 4.7194, p-value=0.0160; Tukey-Kramer HSD p-value<0.0131 for cells on M. fragilis vs far from M. fragilis, Welch’s t-test p-value=0.002398). Results pooled from three separate biological replicate experiments in three separate TS microcosms, indicated by color.
Videos
Z-stack of fluorescently labeled B. subtilis 3610 through 100 µm of Nafion (fluorescent channels only).
B. subtilis 3610 cells expressing constitutive YFP were inoculated into Nafion microcosms in MSgg, incubated at room temperature (22°C) for 48 hr, and Z-stacks acquired by confocal microscopy. Nafion was stained with sulforhodamine, visible in the RFP channel (green), and AF647 amine was added to the liquid medium, visible in the far-red channel (blue). YPet-producing B. subtilis cells are visible in the YFP channel (red), with some fluorescence also visible in the RFP channel (green) resulting in yellow appearance. Image size is 101.25 × 101.25 µm. Each frame shows a 0.75 µm slice.
Z-stack of fluorescently labeled B. subtilis 3610 through 100 μm of Nafion (fluorescent channels with PMT channel).
Same as in Video 1, but with PMT (non-fluorescent) channel added.
Z-stack of fluorescently labeled B. subtilis 3610 through 100 μm of cryolite (fluorescent channels only).
B. subtilis 3610 cells expressing constitutive YPet were inoculated into cryolite microcosms in MSgg, incubated at room temperature (22°C) for 48 hr, and Z-stacks acquired by confocal microscopy. AF647 amine was added to the liquid medium, visible in the far-red channel (blue). YPet-producing B. subtilis cells are visible in the YFP channel (red), with some fluorescence also visible in the RFP channel (green) resulting in yellow appearance. Image size is 101.25 × 101.25 µm. Each frame shows a 0.75 µm slice.
Z-stack of fluorescently labeled B. subtilis 3610 through 100 μm of cryolite (fluorescent channels with PMT channel).
Same as in Video 3, but with PMT (non-fluorescent) channel added.
Tables
Summary table comparing Nafion and cryolite as transparent porous substrates for applications in soil microbial ecology.
| Criterion | Nafion | Cryolite |
|---|---|---|
| Biocompatible? | Yes | Yes |
| Inert (i.e. not easily metabolizable or decomposable by bacteria or fungi)? | Yes | Yes |
| Submicron resolution through 100 µm of matrix? | Yes | Yes |
| Bacteria visible with fluorescence through 100 µm of matrix? | Somewhat (see text) | Yes |
| Bacteria visible without fluorescence through 100 µm of matrix? | No | Yes |
| Ready to use without pre-treatment? | No | Yes |
| Particles visible under brightfield microscopy? | Yes | No |
| Particles visible under DIC and PMT microscopy? | Yes | Yes |
| Particles autofluorescent? | Yes | No |
| Particles visible after staining with fluorescent dye? | Yes | No |
| Pore water visualizable? | Yes | Yes |
| Commercially available? | Yes | Somewhat (see text) |
| Inexpensive? | No | Yes |
| Pure? | Yes | Somewhat (see text) |
| Desired particle size distributions (between 1 µm and ~ 5 mm) easy to obtain? | Somewhat (see text) | Yes |
| Amenable to drying and rewetting? | Somewhat (see text) | Yes |
| Cation-exchanging? | Yes | Yes |
| Compatible with single-cell D2O tracing by Raman? | Yes | No |
| Compatible with population-level D2O tracing by Raman? | Yes | Yes |
| Compatible with single-cell 13C tracing by Raman? | No | No |
| Compatible with population-level 13C tracing by Raman? | No | Yes |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Bacillus subtilis) | NCIB3610 (abbreviated as 3610); ES3 in Shank Laboratory | Nye et al., 2017 https://doi.org/10.1128/ genomeA.00364-17 | GenBank: CP020102.1 | Available from the Shank lab or Bacillus Genetic Stock Center (BGSCID 3A1) |
| Genetic reagent (Bacillus subtilis 3610) | ES768 | This study; Shank Laboratory | B. subtilis 3610 amyE::PspacC-YPet-camR; available from Shank lab | |
| Genetic reagent (Bacillus subtilis 3610) | ES769 | This study; Shank Laboratory | eps-tasA biofilm gene mutant, B. subtilis 3610 epsA-O::tet tasA::kan amyE::PspacC-YPet-camR; available from Shank lab | |
| Gene (Bacillus subtilis) | epsA-0 | GenBank | BSU_34370 through BSU_34220 | Extracellular polysaccharide biosynthesis operon |
| Gene (Bacillus subtilis) | tasA | GenBank | BSU_24620 | Major component of biofilm matrix |
| Gene (Bacillus subtilis) | YPet | Ethan Garner, Garner Laboratory | Yellow fluorescent protein codon-optimized for B. subtilis | |
| Recombinant DNA reagent | pEA003 | This study; Shank Laboratory | Available from Shank lab | |
| Recombinant DNA reagent | pES037 | This study; Shank Laboratory | Available from Shank lab | |
| Biological sample (Mucor fragilis) | This study; Shank Laboratory | Isolated the strain from a tall fescue plant in the Piedmont region of North Carolina by Fletcher Halliday, University of North Carolina, 2016; available from Shank lab | ||
| Software, algorithm | Fiji/ImageJ | https://imagej.nih.gov/ij/ docs/guide/146-2.html | RRID:SCR_002285 | |
| Software, algorithm | Raman spectrum analysis software | Berry et al., 2015, http://shiny.csb. univie.ac.at:3838/scattr/ | ||
| Other | Nafion | IonPower, Newcastle, DE, USA | POWDion Insoluble −40 + 60 Mesh | https://ion-power.com/product/ powdion-insoluble-4060-mesh/ |
| Other | cryolite | Wilhelm Niemetz Minalerien, Vienna, Austria | Crystalline form from Ivigtut, Greenland |





