Characterization of the kinetic cycle of an ABC transporter by single-molecule and cryo-EM analyses
Figures
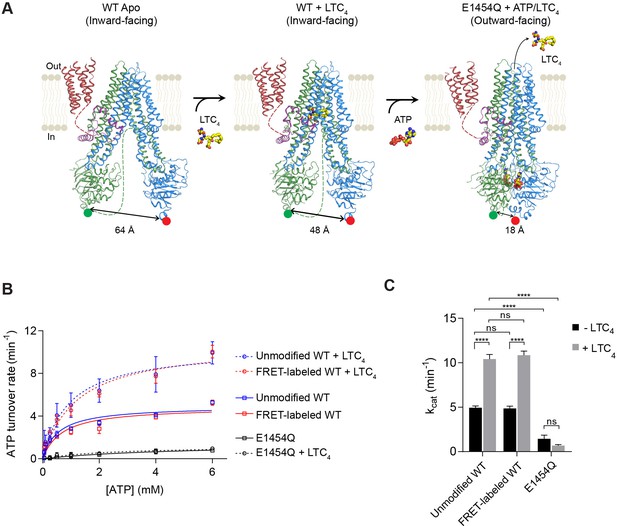
Structure-guided smFRET design for probing MRP1 dynamics.
(A) Structures of bMRP1 captured in ligand-free (left), LTC4-bound (middle), and ATP-bound (right) conformations (PDB accession numbers: 5UJ9, 5UJA, and 6BHU). The positions of tag insertions for site-specific labeling are highlighted in green (FRET donor; peptide sequence substituted following NBD1 residue 867) and red (FRET acceptor; peptide sequence inserted at the C-terminus following NBD2 residue 1530). The distances shown correspond to the separations between residues 867 and 1530 rather than the inter-probe distances. TMD0 is shown in red, the lasso motif in purple, TMD1/NBD1 in green, and TMD2/NBD2 in blue. Flexible linkers not observed in the cryo-EM maps are represented by dotted lines. (B) ATPase activity of unmodified wild-type (WT), FRET-labeled WT, and E1454Q mutant bMRP1, either without or with LTC4 (10 µM). Data are represented as mean ±95% confidence intervals (from 3 to 6 independent measurements) and fitted to Michaelis-Menten equations. (C) Catalytic constants for ATP turnover by unmodified WT, FRET-labeled WT, and E1454Q bMRP1 in the absence and presence of 10 µM LTC4. Data are represented as mean ± SEM. Comparisons were made by one-way ANOVA (****p<0.0001; ns, not significant). The residual ATP turnover seen in the E1454Q sample did not respond to LTC4 stimulation, and was thus most likely due to spontaneous ATP hydrolysis independent of MRP1 activity.
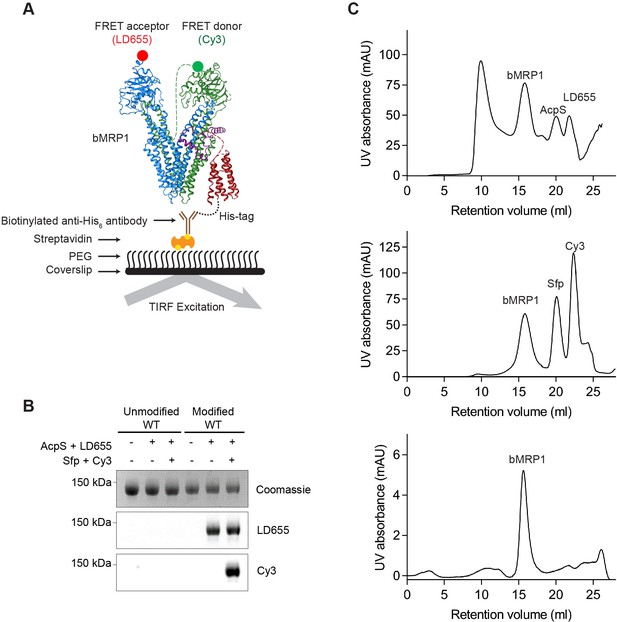
Site-specific labeling and purification of MRP1.
(A) Schematic of the surface immobilization and single-molecule imaging strategy. Labeled bMRP1 molecules were immobilized on a glass surface and imaged at room temperature with total-internal-reflection fluorescence microscopy. (B) SDS-PAGE gel showing fluorescent labeling of bMRP1 with Sfp/AcpS synthases and dye-CoA conjugates. Modified WT stands for MRP1 with inserted peptide sequences for site-specific labeling. (C) Size-exclusion chromatography for generating dual-labeled bMRP1. The AcpS synthase and excess free LD655 dye were removed by gel filtration (top), followed by the removal of Sfp synthase and excess free Cy3 dye (middle). The bottom graph depicts the final dual-labeled sample used in single-molecule experiments.
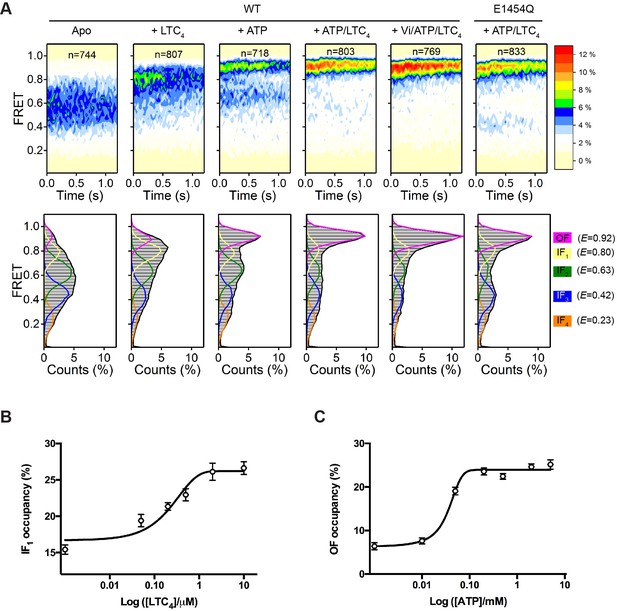
Conformational landscapes of MRP1 revealed by smFRET.
(A) Contour plots (top) and histograms (bottom) of FRET distributions obtained with WT bMRP1 in the following conditions (from left to right): apo, + LTC4 (10 µM), + ATP (5 mM), + ATP/LTC4 (5 mM/10 µM), + Vi/ATP/LTC4 (1 mM/5 mM/10 µM). Shown in the right column are data for the E1454Q mutant in the presence of ATP/LTC4 (5 mM/10 µM). The time-dependent changes in the contour plots were due to fluorophore photobleaching, which depopulated FRET-active molecules over time. Time points after photobleaching were excluded from subsequent analysis. The histograms represent the cumulative FRET distributions over the entire 1.25 s time window. Overlaid on the histograms are fitted distributions by the five-state model with mean FRET values of 0.92 (magenta, OF), 0.80 (yellow, IF1), 0.63 (green, IF2), 0.42 (blue, IF3), and 0.23 (orange, IF4). n denotes the number of molecules analyzed. (B) Relative occupancy of the IF1 state in the presence of increasing concentrations of LTC4. Data are fitted to a dose-response function with the Hill equation, yielding an EC50 of 0.32 ± 0.17 µM. (C) Relative occupancy of the OF state in the presence of increasing concentrations of ATP. Data are fitted to a dose-response function with the Hill equation, yielding an EC50 of 0.05 ± 0.02 mM. Data are represented as mean ± SEM.
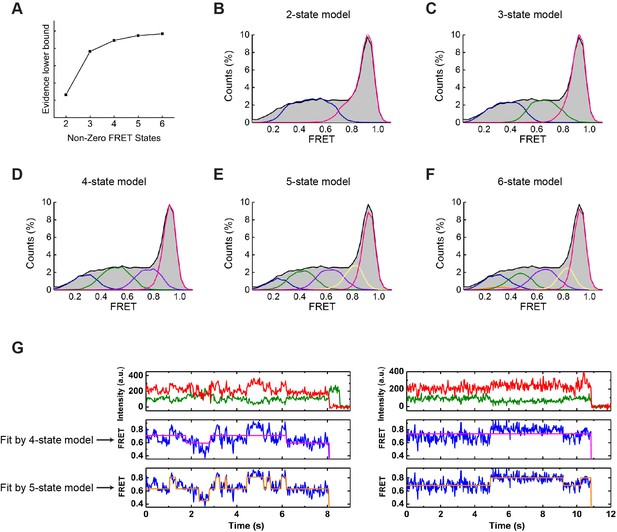
Determination of model parameters for idealizing smFRET trajectories.
(A) Evidence lower bound determined by ebFRET, which was used to determine the number of non-zero FRET states and provide initial estimates of model parameters. (B–F) Models with two (B), three (C), four (D), five (E), and six (F) non-zero FRET states were implemented in SPARTAN using the initial parameters generated by ebFRET. FRET trajectories were then idealized through segmental k-means optimization of each model. The raw FRET histogram (gray, from WT MRP1 with 5 mM ATP and 10 µM LTC4) is overlaid with histograms of the assigned states (shown in colors). (G) Example single-molecule trajectories fitted by four non-zero FRET states (magenta line) versus five non-zero FRET states (orange line). The four-state model fails to recognize obvious FRET transitions.
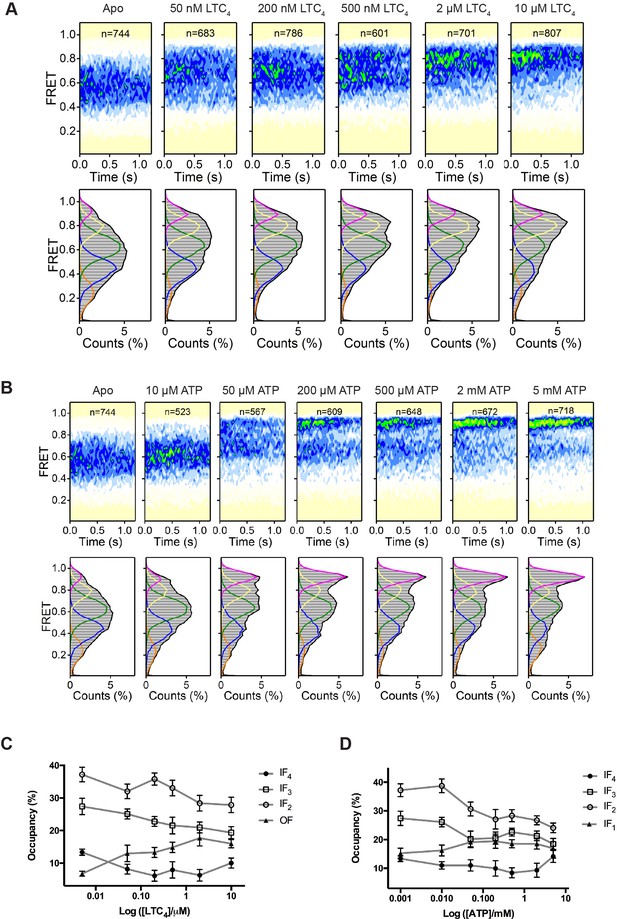
Dependence of the conformational distribution of MRP1 on substrate and ATP.
(A) FRET contour plots (top) and histograms (bottom) obtained at varying concentrations of LTC4. n denotes the number of molecules analyzed for each condition. (B) FRET contour plots and histograms obtained at varying concentrations of ATP. (C) Relative occupancies of states other than IF1 at different LTC4 concentrations. IF1 occupancy is shown in Figure 2B. (D) Relative occupancies of all IF states at different ATP concentrations. OF occupancy is shown in Figure 2C. Data are represented as mean ± SEM.
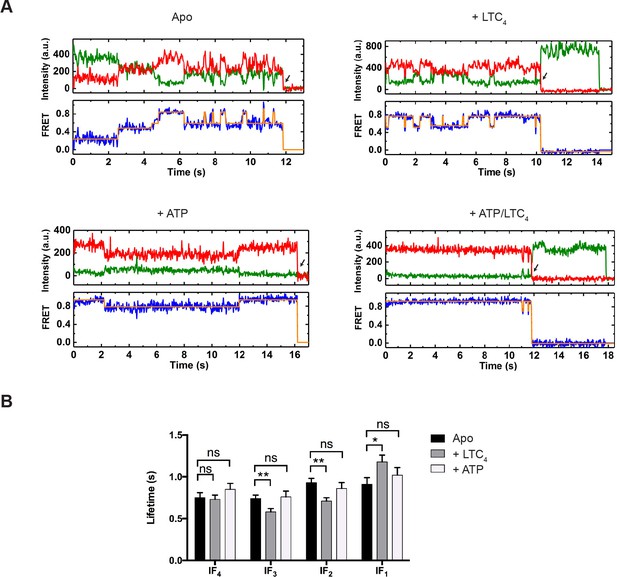
Real-time dynamics of MRP1 under steady state.
(A) Representative single-molecule donor (green) and acceptor (red) fluorescence trajectories, and the corresponding FRET trajectories (blue) obtained at a frame rate of 25 ms. Idealized FRET states are overlaid in orange lines. Arrows indicate fluorophore photobleaching events, after which the data were excluded from further analysis. The following conditions were analyzed: apo, + LTC4 (10 µM), + ATP (5 mM), and + ATP/LTC4 (5 mM/10 µM). (B) Average lifetimes of each IF state under different conditions. Data are represented as mean ± SEM.
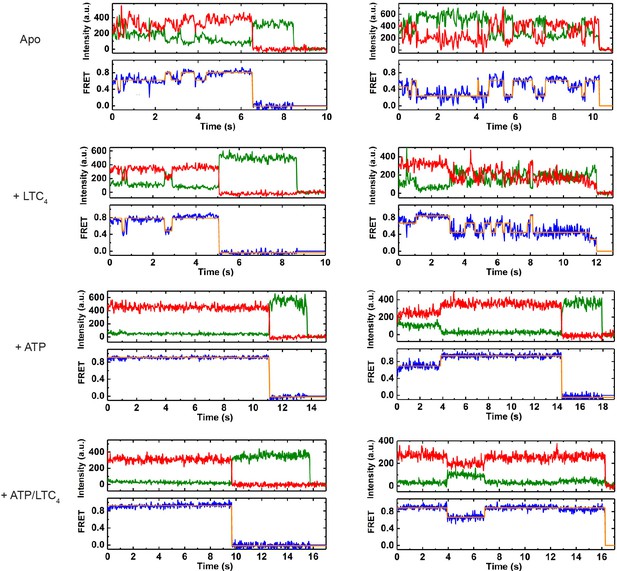
Additional representative fluorescence and FRET trajectories obtained at 25 ms time resolution for the indicated conditions.
Idealized FRET states are overlaid in orange.
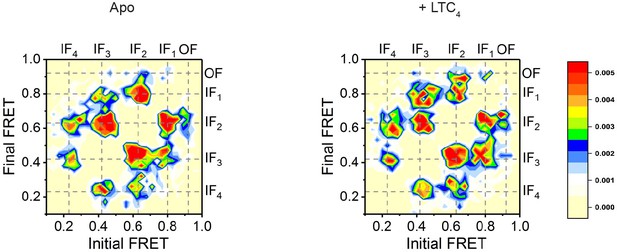
Transition density plots displaying the distributions of FRET values before (x-axis) and after (y-axis) each transition identified in the idealized FRET traces.
Dashed lines represent the mean values for the five discrete FRET states.
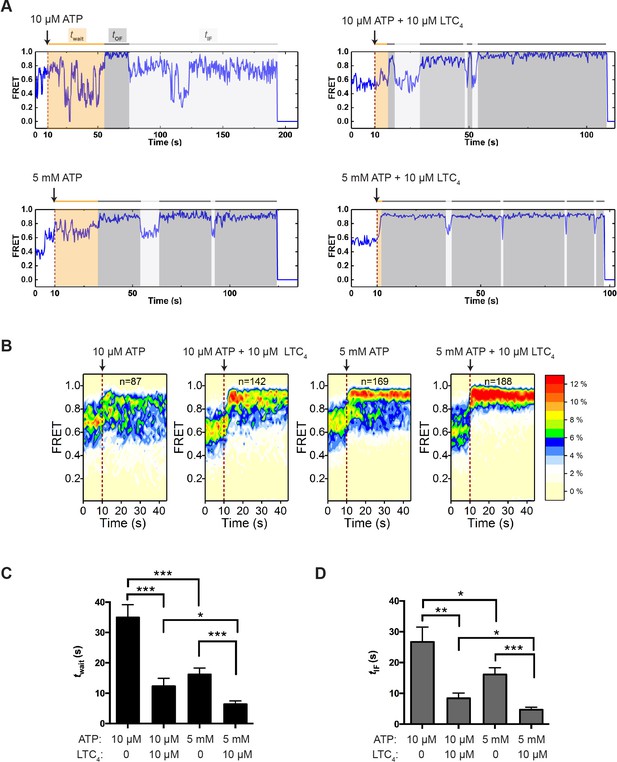
Transitions between IF and OF states from perturbation experiments.
(A) Representative smFRET trajectories of active molecules from the perturbation experiments obtained at a frame rate of 300 ms. A limiting (10 µM) or saturating (5 mM) concentration of ATP with or without LTC4 (10 µM) was injected into the imaging chamber at 10 s (dashed lines). The wait time until the onset of the first OF state (twait) and the lifetime of the subsequent OF states (tOF) and IF states (tIF) are shaded in orange, dark gray, and light gray, respectively. (B) Contour plots of smFRET trajectories for active molecules under each perturbation condition. n denotes the number of active molecules analyzed. (C) Average twait for different perturbation conditions. (D) Average tIF for different perturbation conditions. The values have been corrected for the photobleaching rate. Data are represented as mean ± SEM.
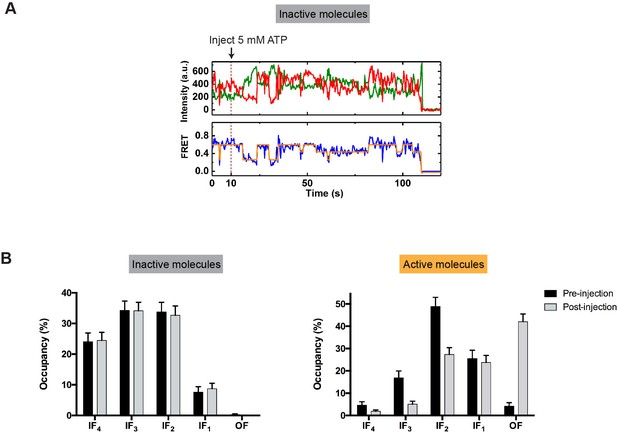
Differentiation between inactive and active molecules.
(A) Representative fluorescence and FRET trajectories of the inactive molecules. These molecules transitioned among distinct IF states but never visited the OF state before photobleaching. (B) Occupancy of each FRET state before and after the injection of 5 mM ATP for the inactive (left) and active (right) population. Data are represented as mean ± SEM.
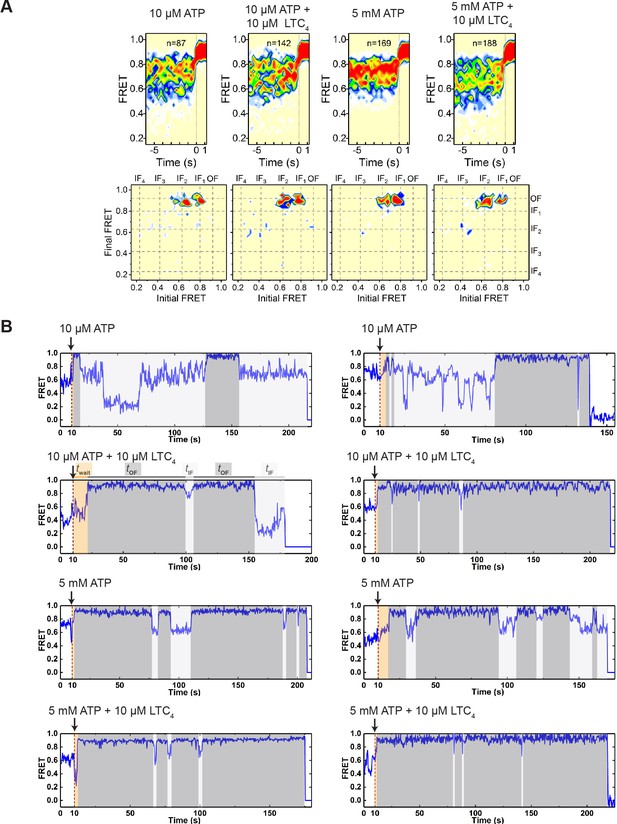
Characteristics of IF-to-OF transitions under different conditions.
(A) Contour plots (top) and corresponding transition density plots (bottom) of smFRET trajectories aligned at the appearance of the first OF state from the perturbation experiments in which 10 µM ATP, 10 µM ATP + 10 µM LTC4, 5 mM ATP, or 5 mM ATP + 10 µM LTC4 (from left to right) was injected into the imaging chamber during data acquisition. (B) Additional representative smFRET trajectories for the perturbation experiments. Dashed lines indicate the time of injection. The wait time until the onset of the first OF state (twait) and the lifetimes of the subsequent OF states (tOF) and IF states (tIF) are shaded in orange, dark gray and light gray, respectively.
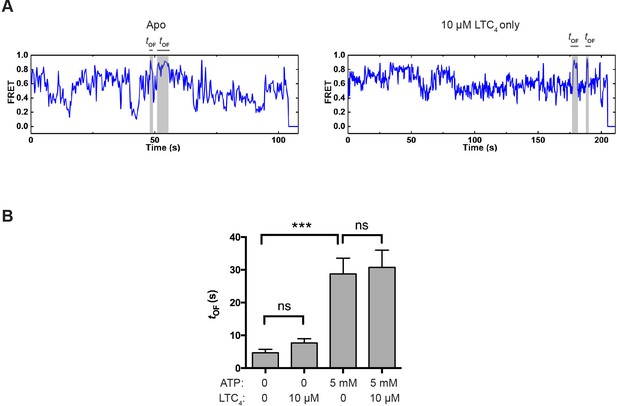
OF state lifetime under different conditions.
(A) Example smFRET trajectories for apo and LTC4-only conditions showing spontaneous transitions into the OF state independent of ATP. (B) Average lifetime of the OF states under different conditions. The values have been corrected for the photobleaching rate. Data are represented as mean ± SEM.
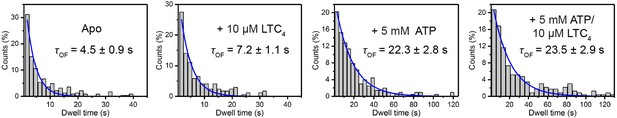
Histograms of OF state lifetimes for different conditions and their fit to single-exponential decay functions.
The corresponding time constants (τOF) are reported.
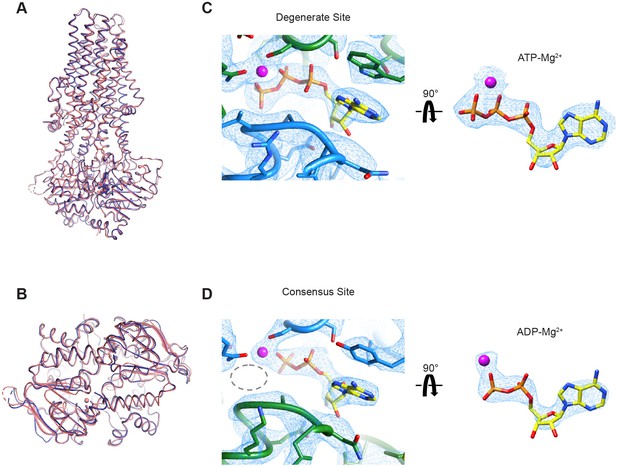
Cryo-EM structure of wild-type MRP1 in its post-hydrolytic state.
(A) The structure of WT bMRP1 in the presence of saturating ATP and LTC4 shown in blue, overlaid with the structure of the non-hydrolyzing bMRP1-E1454Q mutant in the presence of saturating ATP and LTC4 (PDB 6BHU) shown in salmon. The structures are shown in cartoon representation viewed from within the plane of the membrane with ATP/ADP shown as sticks and Mg2+ shown as spheres. (B) The same structural overlay as in (A), rotated 90° to view the NBD dimer from the cytoplasmic side. (C) Cryo-EM density for the degenerate ATPase site (left) and the ATP-Mg2+ from the degenerate site alone (right, rotated 90°). NBD1 is shown in green, NBD2 in blue, ATP as yellow sticks colored by heteroatom, and Mg2+ as a magenta sphere. (D) Cryo-EM density for the consensus ATPase site (left) and the ADP-Mg2+ from the consensus site alone (right, rotated 90°). In the left panel, the position of the missing γ-phosphate is demarcated with a gray dotted oval. The color code is the same as in (C).
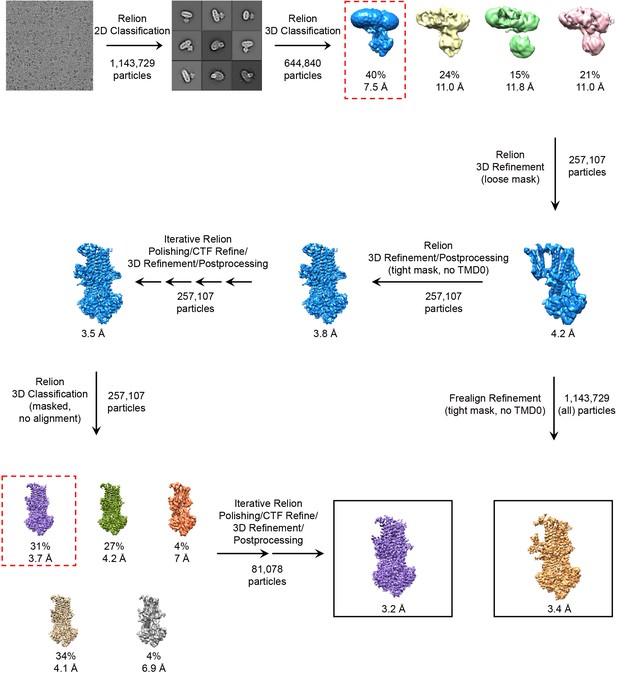
Cryo-EM data processing workflow.
The number of particles used in each step is indicated next to the arrow associated with that step. After initial classification and refinement in RELION, two separate routes were taken to generate each of the final maps. To generate the 3.2 Å map from a subset of particles, the data were further processed in RELION. To generate the 3.4 Å map from all particles, the data were refined using Frealign.
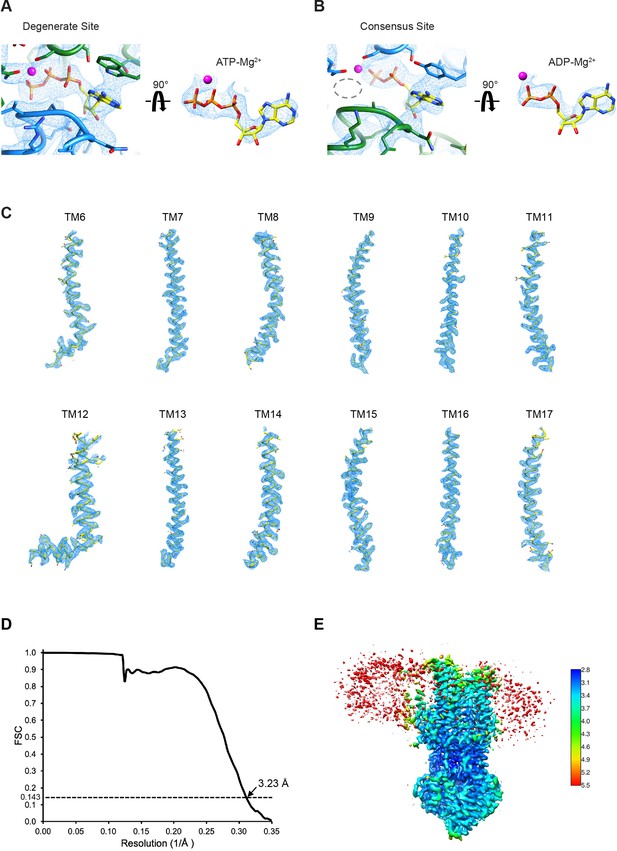
Quality of the cryo-EM maps.
(A) Cryo-EM density in the degenerate ATPase site from the Frealign map using all particles, displayed in the same way as Figure 5C. (B) Cryo-EM density in the consensus ATPase site from the Frealign map using all particles, displayed in the same way as Figure 5D. (C) Cryo-EM density for the transmembrane helices from the RELION map using a subset of the particles. (D) Fourier shell correlation (FSC) curve for the two half maps used in the final reconstruction of the RELION map using a subset of the particles. (E) Local resolution of the RELION map using a subset of the particles. Local resolution was estimated using cryoSPARC.
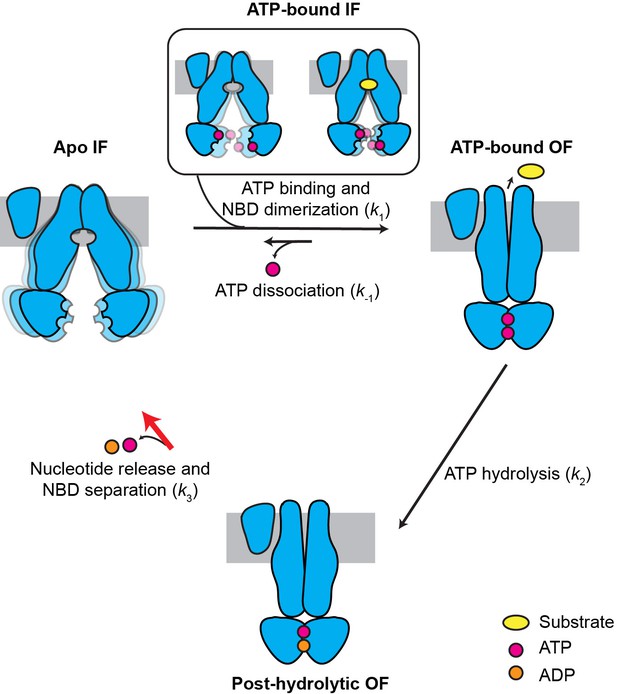
Kinetic model for the transport cycle of MRP1.
MRP1 is intrinsically dynamic, transitioning between multiple IF conformations both in the absence and presence of ATP. Under physiological conditions, ATP rapidly binds to the IF state, promoting NBD dimerization and formation of the OF state. LTC4 accelerates the IF-to-OF transition (k1) but not the other transitions, yielding a faster overall ATPase turnover rate. The reverse isomerization (k-1) resulting from ATP dissociation occurs at a much slower rate than the forward reaction. ATP hydrolysis in the consensus site (k2) is fast and results in an asymmetric post-hydrolytic OF state with ATP in the degenerate site and ADP in the consensus site. This represents the predominant conformation determined by cryo-EM under active turnover conditions. The entire transport cycle is limited by the rate of dissociation of the NBD dimer (k3) after ATP hydrolysis.
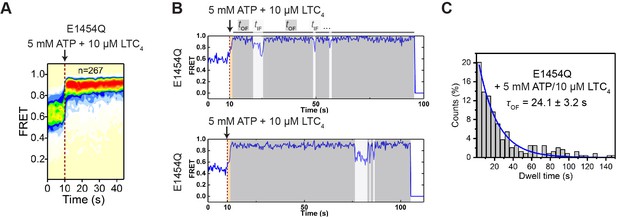
Perturbation experiments with E1454Q mutant MRP1.
(A) FRET contour plot for the perturbation experiment in which 5 mM ATP and 10 µM LTC4 were delivered to bMRP1-E1454Q at 10 s. (B) Representative smFRET trajectories for the above experiment. (C) Histogram of the OF state lifetimes extracted from the above experiment and its fit to a single-exponential decay function (blue line).
Tables
Kinetics of the transitions between IF and OF conformations
| Condition | tIF (s) | tOF (s) |
|---|---|---|
| WT, 5 mM ATP | 16.1 ± 2.2 | 28.8 ± 4.8 |
| WT, 5 mM ATP + 10 µM LTC4 | 4.7 ± 0.8 | 30.8 ± 5.2 |
| E1454Q, 5 mM ATP + 10 µM LTC4 | 7.7 ± 1.5 | 31.7 ± 5.5 |
-
Shown are the average lifetimes (mean ± SEM) of the composite IF state and the OF state for WT and E1454Q MRP1 with indicated ATP and substrate concentrations. The effect of dye photobleaching on the apparent IF/OF lifetime has been corrected for.
Summary of EM data and structure refinement statistics
| Data collection | ||
|---|---|---|
| Microscope | Titan krios (FEI) | |
| Voltage (kV) | 300 | |
| Detector | K2 Summit (Gatan) | |
| Pixel size (Å) | 1.03 | |
| Defocus range (μm) | 0.7 to 2.4 | |
| Movies | 3604 | |
| Frames/movie | 50 | |
| Dose rate (electrons/pixel/s) | 8.0 | |
| Total dose (electrons/Å2) | 75 | |
| Number of particles | 1,143,729 | |
| Model composition | ||
| Non-hydrogen atoms | 9684 | |
| Protein residues | 1210 | |
| Lipids/Detergents/Ligands | 3 CHS/1 ATP/1 ADP/2 Mg2+ | |
| Refinement | ||
| Resolution (Å) | 3.23 | |
| Rwork | 0.265 | |
| Rfree | 0.276 | |
| RMS deviations | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 1.297 | |
| Validation | ||
| Molprobity score | 1.11 | |
| Clashscore, all atoms | 1.07 | |
| Favored rotamers (%) | 97.7 | |
| Ramachandran plot (%) | ||
| Favored | 95.7 | |
| Allowed | 4.3 | |
| Outliers | 0.0 | |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Biotinylated 6x-His Tag monoclonal antibody | Invitrogen | Cat# MA1-21315-BTIN | Molar ratio of 2:1 (antibody:bMRP1) |
| Cell line | Sf9 | ATCC | CRL-1711 | |
| Cell line | HEK293S GnTI- | ATCC | CRL-3022 | |
| Recombinant DNA reagent | bovine MRP1 in pUC57 vector | Bio Basic | Codon-optimized | |
| Recombinant DNA reagent | bovine MRP1 in a modified pEG BacMam vector | Johnson and Chen, 2017 | Suitable for expression in mammalian cells | |
| Recombinant DNA reagent | bovine MRP1 with S6/A1 peptides for site-specific labeling | This paper | ||
| Recombinant DNA reagent | bovine MRP1 E1454Q with S6/A1 peptides for site-specific labeling | This paper | ||
| Recombinant DNA reagent | Sfp pet29b C-terminal His Tag | (Worthington and Burkart, 2006) | Addgene Plasmid# 75015 | |
| Recombinant DNA reagent | pET15b-ACPs (from S. pneumoniae) | Gift from Michael Johnson | Addgene Plasmid# 63687 | |
| Chemical compound, drug | Cy3 maleimide mono-reactive dye | GE Healthcare | Cat# PA23031 | |
| Chemical compound, drug | LD655 maleimide mono-reactive dye | Lumidyne Technologies | Cat# LD655-MAL | |
| Chemical compound, drug | Trolox | Sigma-Aldrich | Cat# 238813 | |
| Chemical compound, drug | 4-Nitrobenzyl alcohol (NBA) | Sigma-Aldrich | Cat# N12821 | |
| Chemical compound, drug | Cyclooctatetraene (COT) | Sigma-Aldrich | Cat# 138924 | |
| Chemical compound, drug | 3,4-Dihydroxybenzoic acid (PCA) | Sigma-Aldrich | Cat# 37580 | |
| Chemical compound, drug | Digitonin | Sigma-Aldrich | Cat# D141 | |
| Peptide, recombinant protein | Protocatechuate 3,4-Dioxygenase (PCD) | Sigma-Aldrich | Cat# P8279 | |
| Peptide, recombinant protein | Leukotriene C4 | Cayman Chemical | Cat# 20210 | |
| Commercial assay, kit | NHS-activated Sepharose 4 Fast Flow resin | GE Healthcare | Cat# 17-0430-01 | |
| Commercial assay, kit | Superose 6, 10/300 GL | GE Healthcare | Cat# 17-5172-01 | |
| Software, algorithm | SPARTAN | (Juette et al., 2016) | https://www.scottcblanchardlab.com/software | |
| Software, algorithm | ebFRET | (van de Meent et al., 2014) | http://ebfret.github.io | |
| Software, algorithm | MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html | |
| Software, algorithm | Origin | OriginLab | https://www.originlab.com | |
| Software, algorithm | GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | RELION 1.4 | (Scheres, 2012) | https://www2.mrc-lmb.cam.ac.uk/relion | |
| Software, algorithm | Frealign | (Grigorieff, 2016) | https://grigoriefflab.janelia.org/frealign | |
| Software, algorithm | Coot | (Emsley and Cowtan, 2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot | |
| Software, algorithm | PHENIX | (Adams et al., 2010) | https://www.phenix-online.org | |
| Software, algorithm | REFMAC | (Brown et al., 2015) | https://www.ccp4.ac.uk/html/refmac5.html | |
| Software, algorithm | MolProbity | (Chen et al., 2010) | https://molprobity.biochem.duke.edu | |
| Software, algorithm | Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera | |
| Software, algorithm | PyMOL | PyMOL | https://www.pymol.org | |
| Software, algorithm | cryoSPARC | (Punjani et al., 2017) | https://cryosparc.com | |
| Other | R1.2/1.3 400 mesh Au holey carbon grids | Quantifoil | Cat# 1210627 | 1 µg/mL |





