Acute disruption of the synaptic vesicle membrane protein synaptotagmin 1 using knockoff in mouse hippocampal neurons
Figures
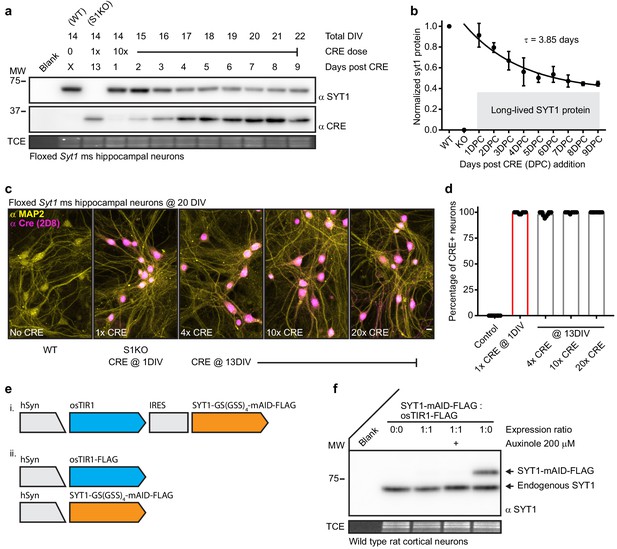
Synaptotagmin 1 is a difficult to disrupt, long-lived protein.
(a) Representative anti-SYT1 and anti-CRE immunoblot of WT, Syt1 KO (generated using 1x CRE lentivirus at day 1), and neurons transduced with 10x CRE lentivirus at 13 DIV. Total DIV, CRE dose, and incubation time post CRE transduction are labeled at the top of the blot. Trichloroethanol (TCE) in-gel fluorescence served as a loading control for each immunoblot in this, and all subsequent, figures. (b) Cleavage was quantified via densitometry of the SYT1 signals in panel (a), data were fitted with a single exponential function (R2 = 0.7583), yielding a τ for SYT1 turnover of 3.85 days. The plateau represents a significant population of long-lived SYT1 that was not turned-over. Mean +/- SEM from three trials. (c) Representative confocal fluorescent ICC images from mouse hippocampal neurons at 20 DIV. Images of WT, S1KO, and neurons transduced with 4x, 10x, and 20x CRE lentivirus at 13 DIV, stained with anti-MAP2 (yellow) and anti-CRE (magenta) antibodies. (d) Percentage of CRE positive MAP2 positive soma from indicated conditions (n = 10 for each condition from three independent trials). (e) Schematics of auxin-induced degron expression vectors. (f) Representative anti-SYT1 immunoblot from cultures transduced with vectors shown in e ii; leak prevented detectable expression of the fusion protein, even in the presence of the osTIR inhibitor, auxinole.
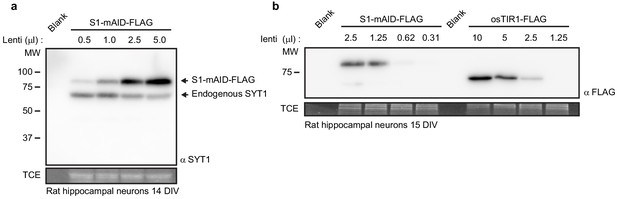
Characterizing expression levels of AID components in neurons.
(a) Representative anti-SYT1 immunoblot of endogenous and Auxin Induced Degron (AID) modified SYT1 from rat hippocampal neurons at 14 DIV. (b) Representative anti-FLAG immunoblot of the SYT1-mAID fusion protein and osTIR1 (both tagged with 1x FLAG) from rat hippocampal neurons at 15 DIV. The anti-FLAG blot was used to determine ~1:1 expression of osTIR1 and the SYT1-mAID fusion protein.
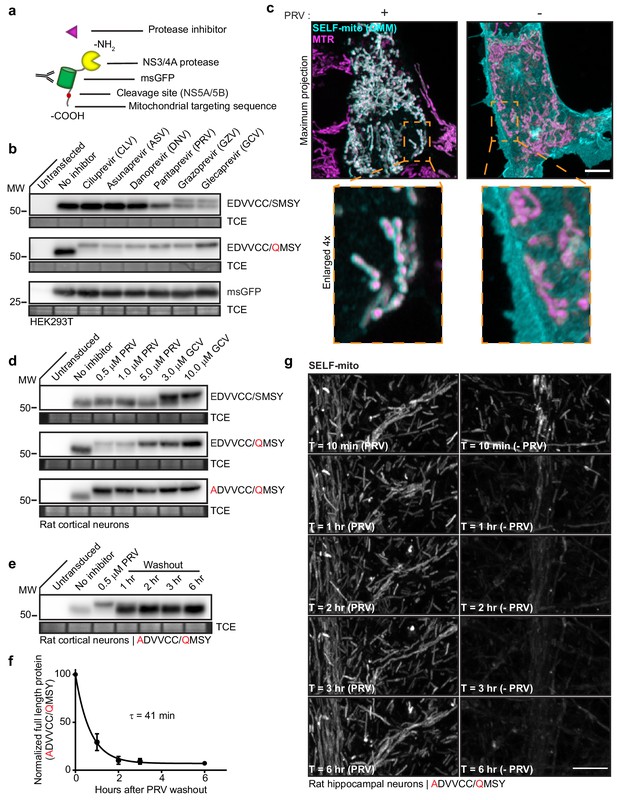
Knockoff development using a model substrate.
(a) Schematic of the model substrate (SELF-mito) used to develop knockoff. (b) Representative anti-GFP immunoblots of HEK293T cells expressing model substrates in the presence of the indicated inhibitors. The upper band represents full-length substrate protein; the lower band is the cleavage product. The negative control was untagged msGFP. (c) Super-resolution fluorescent images of HEK293T cells expressing SELF-mito with the modified cleavage sequence (EDVVCC/QMSY) counterstained with MitotrackerRed (MTR), +/- 5 µM PRV. Maximum projection and enlarged inset examples are shown. (d) Knockoff in neurons required further attenuation of the cleavage site. Representative anti-GFP immunoblots from rat cortical neurons expressing substrates bearing the natural NS5A/5B cleavage sequence (EDVVCC/SMSY), modified sequence (EDVVCC/QMSY), and further attenuated sequence (ADVVCC/QMSY) in the presence of inhibitors. GCV was the most effective inhibitor of substrate cleavage. (e) Representative anti-GFP immunoblot from rat cortical neurons expressing the attenuated (ADVVCC/QMSY) model substrate showing the time-course of cleavage after washout of 0.5 µM PRV. (f) Self-cleavage time-course of the attenuated (ADVVCC/QMSY) model knockoff protein after inhibitor washout. Cleavage in (e) was quantified by densitometry and the data fitted with a single exponential function to yield a τ = 41 min (R2 = 0.9675). Mean +/- SEM from three independent trials are shown. (g) Representative super-resolution fluorescence images of attenuated (ADVVCC/QMSY) substrate (SELF-mito) self-cleaving in a dendrite from a rat hippocampal neuron. (see another example Video 1). Scale bars, 5 µm.
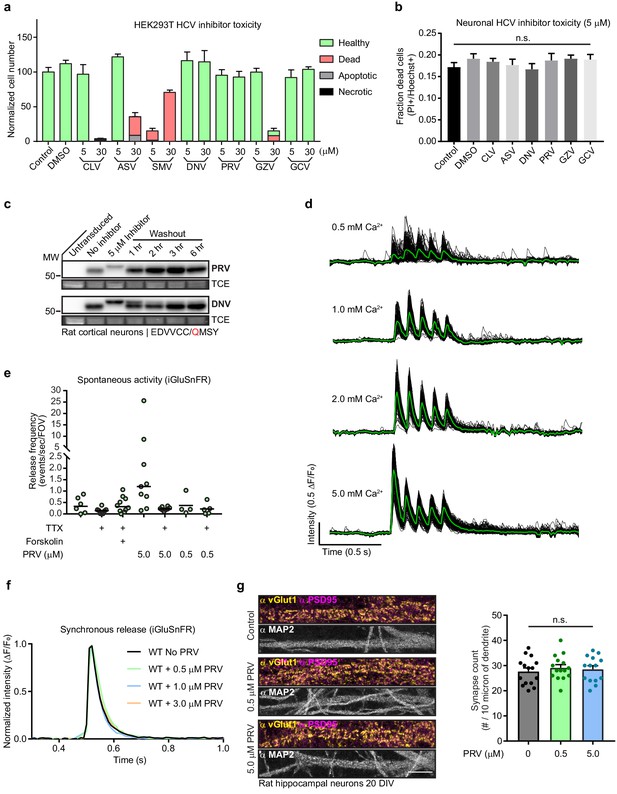
Knockoff development using a model substrate.
(a) Inhibitor toxicity measured in HEK293T cells using a standard dead cell apoptosis assay. Healthy cells are not fluorescent (green), dead cells are propidium iodide (PI) and annexin V (AV) positive (red). Apoptotic cells are only AV positive (grey), while necrotic cells are only PI positive (black). Hoescht nuclear counterstain was to determine the total number of cells. Values are mean +/- SEM, n = 7–8 for each condition from three independent trials. (b) Inhibitor toxicity measured in primary cultures of rat cortical neurons using PI (Dead) and Hoechst (Total) stains. Values are mean +/- SEM, n = 10 fro each condition from two independent trials. (c) Representative anti-GFP immunoblots showing time-course of cleavage for DNV and PRV washout with the modified EDVVCC/QMSY model. PRV has the fastest apparent dissociation from the protease, as the model is almost completely cleaved within 1 hr following washout. (d) Representative, individual ROI (black) and average (green) iGluSnFR traces showing the Ca2+-dependence of release in WT rat hippocampal neurons transduced with iGluSnFR. The examples shown were from a single FOV for each condition. (e) Quantification of miniature glutamate transients (mGT) in WT rat hippocampal neurons at 15 DIV using iGluSnFR (Marvin et al., 2013) in 2 mM extracellular Ca2+. One µM TTX was used to block action potentials where indicated; in some experiments, forskolin (10 µM) was also included to increase mGT frequency. Addition of PRV drastically increased the rate of spontaneous glutamate release events; TTX blocked this increase. A low dose of PRV (0.5 µM) did not influence spontaneous release. Two separate trials were conducted; bars represent means and individual dots represent events/s from a FOV (n) imaged for one minute, n = 4 to 10. (f) Normalized, average traces (n = 5 to 6) of evoked glutamate release from a single action potential in WT neurons, and WT neurons treated with 0.5, 1, or 3 µM PRV, in 1 mM extracellular Ca2+. (g) Representative confocal images of 20 DIV rat hippocampal neurons stained with anti-PSD95, anti-vGLUT1, and anti-MAP2. Neurons grown in vehicle, 0.5 µM, or 5.0 µM PRV. Quantitation of synapse number to the right, control (grey) 0.5 µM (green), or 5.0 µM PRV (blue).
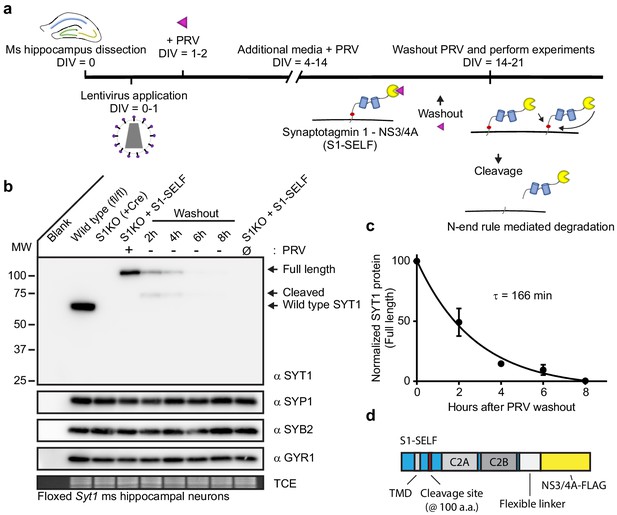
Rapid, efficient, targeted degradation of synaptotagmin 1.
(a) Illustration of the SYT1 knockoff protocol. At 14–21 DIV, inhibitor was removed from neuronal cultures and experiments were performed. Following inhibitor washout, cleavage reactions can occur in cis or trans, and the resulting cleavage product is degraded via the N-end rule. (b) Representative anti-SYT1, anti-synaptophysin 1 (SYP1), anti-synaptobrevin 2 (SYB2), anti-synaptogyrin 1 (GYR1) immunoblots of WT, SYT1 KO (generated using a CRE virus), and KO neurons expressing S1-SELF, in mouse hippocampal neurons at 14 DIV. (Ø) denotes a condition in which cultures have never been exposed to PRV. (c) Self-cleavage time course of S1-SELF, upon PRV washout. Cleavage was quantified via densitometry of the SYT1 immunoblots in panel b, and plotted. Mean +/- SEM from three independent trials are shown, and the time constant was determined by fitting the data with a single exponential function (R2 = 0.9498). The τ for S1-SELF cleavage was 2 hr 45 min. (d) Schematic of S1-SELF; domains are labeled.
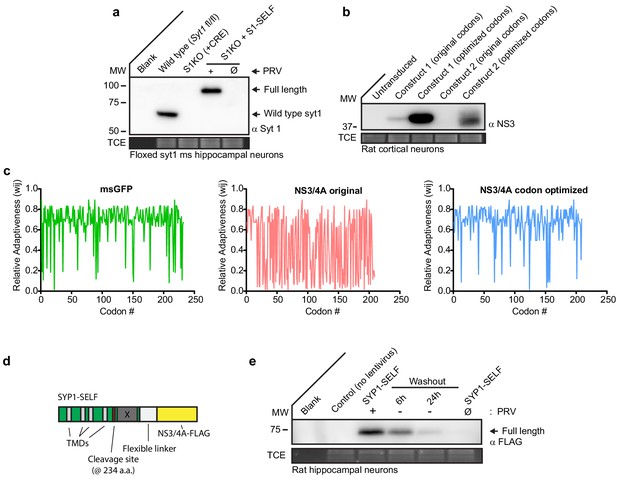
NS3/4A codon optimization and application to the carboxy terminus of synaptophysin.
(a) Representative anti-SYT1 immunoblot of mouse hippocampal neurons at 14 DIV showing: 1) CRE excision of Syt1, 2) stable expression of SYT1-SELF + 0.5 µM PRV, and 3) complete cleavage and degradation of SYT1-SELF in the absence of PRV. (Ø) denotes a condition in which cultures have never been exposed to PRV. These experiments follow the protocol that is detailed in Figure 3a. (b) Immunoblot of msGFP comparing expression levels of two different NS3/4A containing constructs using original codons and optimized codons (construct 1: mito-NS3/4A, construct 2: ER-NS3/4A). Constructs were identically prepped and transduced, using lentivirus, into rat cortical neurons. (c) Relative adaptiveness (wij) of codons from msGFP, original NS3/4A, and NS3/4A codon optimized transcript. msGFP CAI-value = 0.630283. Note how this high value contrasts with the original NS3/4A transcript. NS3/4A original CAI-value = 0.276799 and NS3/4A codon optimized CAI-value = 0.621645. (d) Diagram of the synaptophysin self-cleaving construct, denoted SYP1-SELF. The protease cleavage site was inserted between the fourth TMD and the carboxy termini YG(P/Q) repeats, denoted in grey and with an X. (e) Representative anti-Flag immunoblot showing time-course of cleavage for SYP1-SELF in 0.5 µM PRV.
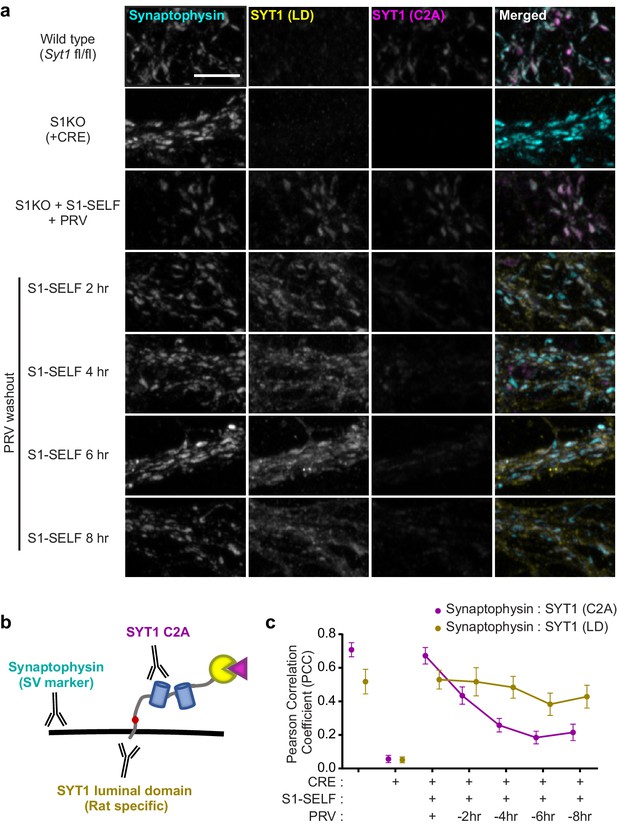
Distinct fates of the S1-SELF cleavage products.
(a) Representative super-resolution fluorescent ICC images from mouse hippocampal neurons at 15 DIV. Images of WT, S1KO, and neurons expressing S1-SELF, stained with anti-synaptophysin (cyan), anti-SYT1 luminal domain (LD) (yellow), and anti-SYT1 C2A domain (C2A) (magenta) antibodies; in the last column, all three signals were merged. Note the SYT1 LD antibody recognizes the rat luminal domain with much higher affinity than the mouse; therefore, in the WT example, only a trace signal is detected. All images were acquired using the same microscope settings, and all samples were prepared in parallel. Scale bar, 5 µm. (b) Illustration of S1-SELF, and the antibodies used for ICC. (c) Pearson’s correlation coefficient (PCC) plot, measured using JaCoP for ImageJ (Bolte and Cordelières, 2006). As S1-SELF is cleaved, the PCC of the synaptophysin to SYT1 C2A signal decreases (Mean PCC +/- SEM are plotted, n = 10 for each condition from three independent trials).
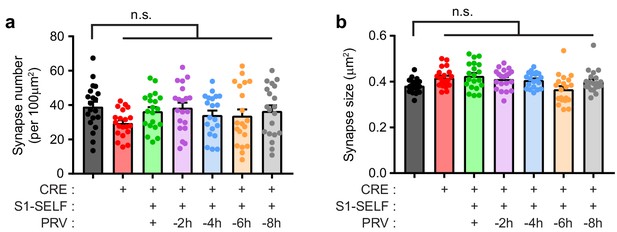
Synaptic number and size are not altered during knockoff of SYT1.
(a) Synapse number in WT, S1KO, and neurons expressing S1-SELF during knockoff. No statistical differences were observed between WT and any other group. (b) Synapse size in WT, S1KO, and neurons expressing S1-SELF during knockoff; again, statistical differences were not observed. Values are mean +/- SEM, n = 20 from three independent trials.
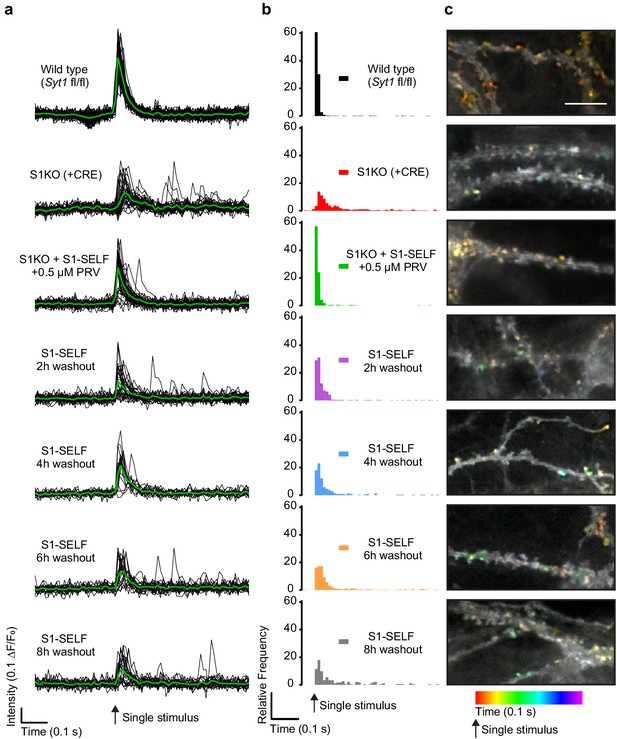
Loss of S1-SELF via knockoff disrupts synchronous release.
(a) Representative, individual ROI (black) and average (green) iGluSnFR traces from a single-field stimulus, recorded optically at 100 Hz. The examples shown are from a single FOV for each condition. (b) Histograms of iGluSnFR (ΔF/F0) peaks plotted using 10 ms bins. Peaks were binned over the entire 1.5 s of recording; a 0.5 s epoch, just before and after the stimulus, is shown. Samples were color-coded as follows: WT (black), Syt1 KO (+CRE) (red), S1KO + S1-SELF + 0.5 µM PRV (green); the S1-SELF PRV washout samples were: 2 (purple), 4 (blue), 6 (orange), and 8 hr (grey). The histograms include all combined data from four independent trials, with 10 to 16 FOVs for each group. Comparisons between all conditions, and statistical analysis, are provided in Figure 5—source data 1. (c) Representative images showing temporally color-coded max projections (time projection) of iGluSnFR ΔF/F0 peaks 100 ms after a single stimulus. Temporal color code is from red (time of stimulation) to purple (100 ms after the stimulus). Scale bar, 5 µm.
-
Figure 5—source data 1
Loss of SYT1 via knockoff disrupts synchronous release.
Table summarizing the Kruskal-Wallis test and Dunn’s multiple comparison test for the histograms shown in Figure 5b.
- https://cdn.elifesciences.org/articles/56469/elife-56469-fig5-data1-v1.docx
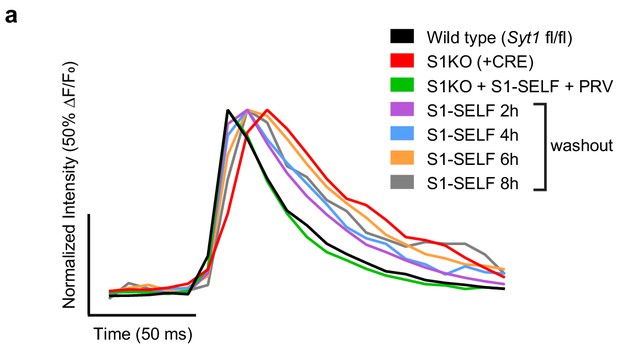
Average normalized synchronous glutamate release measured optically during knockoff of SYT1.
(a) Average, normalized, traces of iGluSnFR ΔF/F0 from Figure 5. Samples are color coded as follows: WT (black), Syt1 KO (+CRE) (red), S1KO + S1-SELF + 0.5 µM PRV (green), S1-SELF 2 hr washout (purple), S1-SELF 4 hr washout (blue), S1-SELF 6 hr washout (orange), and S1-SELF 8 hr washout (grey). Values from four independent trials, with 10 to 16 field of views for each group.
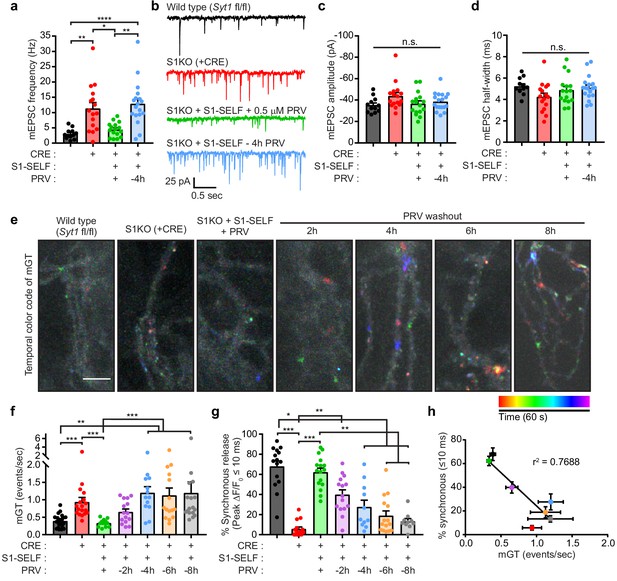
Acute knockoff of SYT1 unclamps spontaneous release.
(a) mEPSC rates in WT (black), S1KO (red), and neurons expressing S1-SELF (+ PRV green and −4 hr PRV blue). Values are mean +/- SEM, 12 to 18 neurons per condition (n). Comparisons between conditions and statistical analysis, are provided in Figure 6—source data 1. (b) Representative traces from (a). (c) Average mEPSC amplitude. (d) Average mEPSC 10–90% half-width. No statistical differences between groups in (c) or (d) were observed. (e) Representative images showing temporally color-coded max projections (time projection) of iGluSnFR ΔF peaks. mGT denotes miniature glutamate release events. Temporal color code is from red (start of image acquisition) to purple (60 s after image acquisition start). Scale bar, 5 µm. (f) mGT rates in WT, S1KO, and neurons expressing S1-SELF, recorded optically with iGluSnFR. Values are mean +/- SEM from three independent trials, with 13 to 21 field of views for each group. Comparisons between conditions and statistical analysis, are provided in Figure 6—source data 2. (g) Percentage of iGluSnFR ΔF/F0 peaks within 10 ms following a single stimulus (data are from Figure 5). Values are mean +/- SEM from four independent trials, with 10 to 16 field of views for each group. Comparisons between conditions and statistical analysis, are provided in Figure 6—source data 3. (h) Inverse correlation between synchronous release (≤10 ms) and spontaneous (mGT) release in WT, S1KO, and S1-SELF neurons. Data were fitted using a linear regression; r2 = 0.7671, p-value=0.0097.
-
Figure 6—source data 1
Acute knockoff of SYT1 unclamps spontaneous release as determined electrophysiologically.
Table summarizing the Kruskal-Wallis test and Dunn’s multiple comparison test for data in Figure 6a.
- https://cdn.elifesciences.org/articles/56469/elife-56469-fig6-data1-v1.docx
-
Figure 6—source data 2
Acute knockoff of SYT1 unclamps spontaneous release via iGluSnFR.
Table summarizing the Kruskal-Wallis test and Dunn’s multiple comparison test for data in Figure 6f.
- https://cdn.elifesciences.org/articles/56469/elife-56469-fig6-data2-v1.docx
-
Figure 6—source data 3
Acute knockoff of SYT1 decreases synchronous release (increases asynchronous release).
Table summarizing the Kruskal-Wallis test and Dunn’s multiple comparison test for histogram data in Figure 6g.
- https://cdn.elifesciences.org/articles/56469/elife-56469-fig6-data3-v1.docx
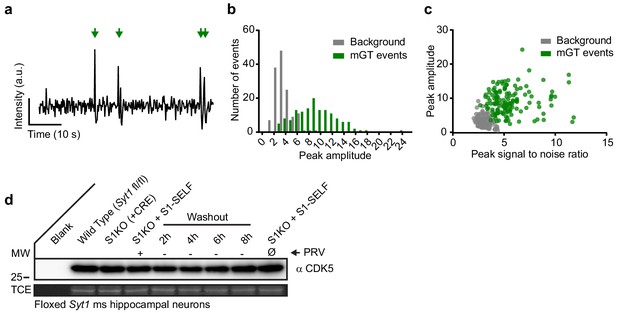
Identification of spontaneous glutamate transients and probing homeostatic plasticity markers during knockoff.
(a) Example trace of background-subtracted, iGluSnFR ΔF spontaneous events, from cultured hippocampal neurons expressing S1-SELF, 6 hr after washout of PRV; four spontaneous events were detected (green arrows). (b) Histogram of background ROI peak ΔF (grey) compared to manually identified, ROI ΔF peaks (green) of spontaneous events. Background peaks are centered at 3 a.u., while mGT events are centered at 9 a.u., n = 141 for each group. (c) The peak amplitude of mGT events (green, panel C) and random ROIs (grey), plotted against the peak signal-to-noise ratio at each ROI. This graph indicates that low-intensity events were identified because the background noise, for those specific events, was also low, n = 141 for each group. (d) Representative anti-CDK5 immunoblot of WT, Syt1 KO (generated using a CRE virus), and KO neurons expressing S1-SELF. (Ø) PRV means that the culture has never seen inhibitor (PRV). (Ø) denotes a condition in which cultures have never been exposed to PRV.
Videos
Knockoff development using a model substrate a, Images of SELF-mito being cleaved in a dendrite from a rat hippocampal neuron.
Images acquired using fast Airyscan mode on a Zeiss 880 LSM, with 10 min imaging intervals for six total hours. Video from Figure 2g. After cleavage, the model (msGFP) is liberated from the mitochondrial membrane. Scale bar, 5 µm.
Acute knockoff of SYT1 unclamps spontaneous release a, Example image sequence of background-subtracted, iGluSnFR ΔF spontaneous events, from S1-SELF 6 hr after washout neuron; four spontaneous events were detected (green arrows).
The arrow indicates the position of the ROI. The image sequence and associated data are from the ROI indicated in Figure 6—figure supplement 1a. Scale bar, 5 µm.
Acute knockoff of SYT1 unclamps spontaneous release a, Larger area from the same image sequence in Video 2.
Numerous spontaneous release events are seen throughout the FOV. Scale bar, 5 µm.
Additional files
-
Supplementary file 1
Knockoff development using HEK293T cells and model substrates.
Table summarizing commercially available HCV inhibitors and their properties.
- https://cdn.elifesciences.org/articles/56469/elife-56469-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56469/elife-56469-transrepform-v1.docx





