A divergent cyclin/cyclin-dependent kinase complex controls the atypical replication of a malaria parasite during gametogony and transmission
Figures
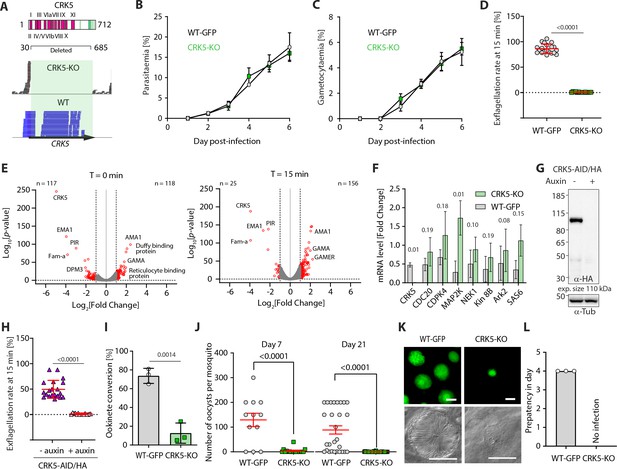
CRK5 is a key regulator of gametogony and sporogony during mosquito transmission.
(A) Schematic representation of CRK5 showing the eleven regions that constitute the catalytic domain in pink, and the Plasmodium-specific C-terminal extension that is phosphorylated at eight sites (green bars). Amino acids 31 to 685 were replaced by the selection marker in the CRK5-KO line. The lower panel shows the sequence reads mapping on crk5 in the KO and WT lines, confirming deletion of the gene in the KO. (B) Deletion of crk5 has no effect on the growth of asexual blood stage parasites (error bars show standard deviation from the mean; 3 and 2 independent infections for the CRK5-KO and WT lines, respectively). (C) Deletion of crk5 has no effect on gametocytaemia (error bars show standard deviation from the mean; 3 and 2 independent infections for the CRK5-KO and WT lines, respectively). (D) Deletion of crk5 leads to a strong defect in exflagellation as measured by the percentage of active exflagellation centres per microgametocyte (error bars show standard deviation from the mean; technical replicates from three independent infections; two-way ANOVA). (E) Volcano plots show limited transcriptional changes between the CRK5-KO and wild type (WT) parasite lines, comparing non-activated gametocytes (T = 0 min) with gametocytes 15 min after activation (T = 15 min). (F) The levels of mRNA for multiple critical regulators of gametogony do not change significantly in CRK5-KO compared to WT parasites, despite a trend for up-regulation in the mutant (error bars show standard deviation from the mean; two independent biological replicates, two-tailed unpaired t-test). (G) Depletion of CRK5-AID/HA protein upon addition of auxin to mature purified gametocytes, as measured by western blotting; α-tubulin serves as a loading control. (H) Depletion of CRK5-AID/HA upon auxin treatment leads to a profound defect in exflagellation (error bars show standard deviation from the mean; technical replicates from three independent infections; two-way ANOVA). (I) Effect of crk5 deletion on ookinete formation as a measure of fertile male gamete formation. Conversion is reported as the percentage of female gametes forming ookinetes (error bars show standard deviation from the mean; three independent infections, two-tailed unpaired t-test). (J) crk5 deletion leads to a dramatic reduction in oocyst formation when mature gametocytes are fed to mosquitoes. Midguts were dissected and GFP-positive oocysts were counted at 7 and 21 days post-infection (error bars show standard deviation from the mean; three independent infections; two tailed unpaired t-test). (K) The residual CRK5-KO oocysts show abnormal development; scale bar = 20 µm. (L) No viable CRK5-KO sporozoite develop. Mosquitoes were infected and 21 days later allowed to feed on naive mice. For the WT line, blood stage parasites were readily observed after a 4 day prepatent period, while no mice became infected with CRK5-KO parasites (error bars show standard deviation from the mean; three independent infections).
-
Figure 1—source data 1
CRK5 is a key regulator of gametogony and sporogony during mosquito transmission.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Panel G, anti-HA western blot.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig1-data2-v1.pdf
-
Figure 1—source data 3
Panel G, anti-αtubulin western blot.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig1-data3-v1.pdf
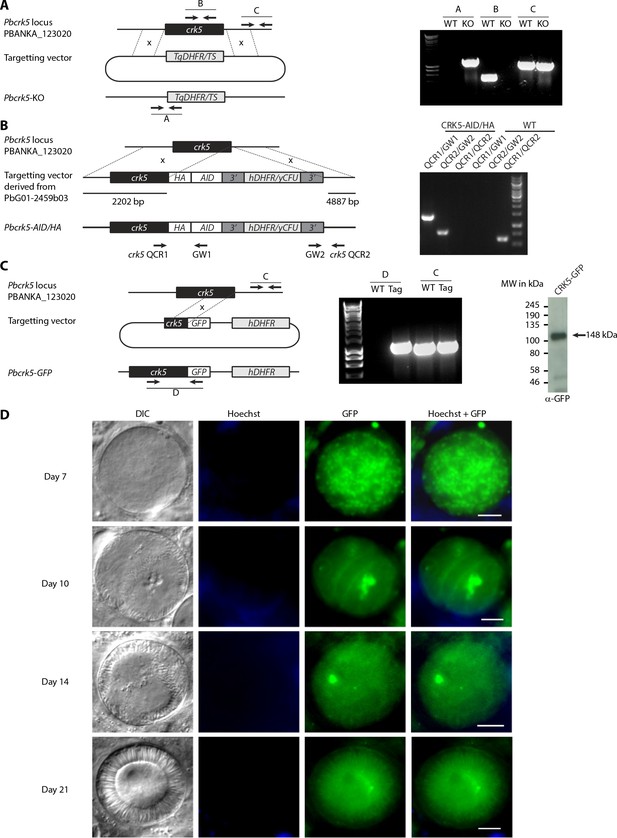
Generation of CRK5-KO and CRK5-AID/HA transgenic lines and CRK5-GFP expression in oocysts.
(A-C) Genetic modification vectors and genotyping data for CRK5-KO (A), CRK5-AID/HA (B), and CRK5-GFP (C). Oligonucleotides used for PCR genotyping are indicated and agarose gels for corresponding PCR products from genotyping reactions are shown. (C) Western blot detection of CRK5-GFP. (D) CRK5-GFP expression and localisation in midgut oocysts at day 7, 14, 17, and 21 post-infection. Scale bar = 10 µm.
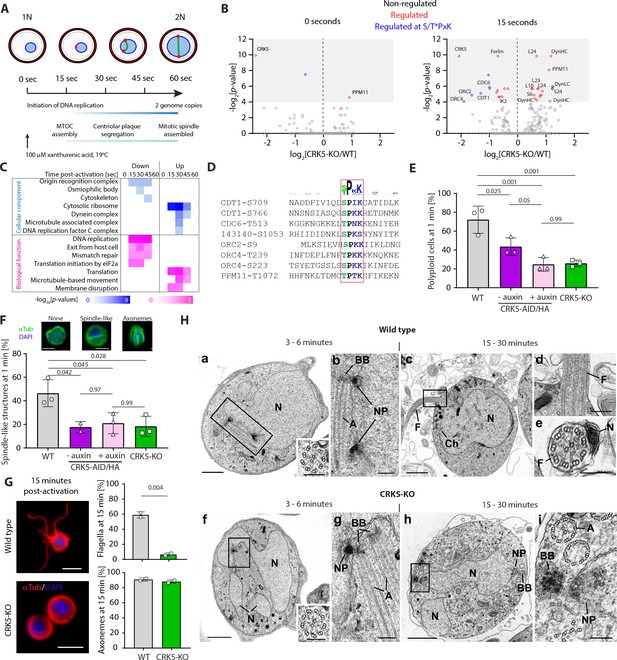
CRK5 shows functional similarities to canonical CDKs and directly regulates DNA replication during gametogony.
(A) Schematic of the first mitosis following activation of male gametogony. Red dots represent the nuclear poles and the green line the mitotic-spindle. Time points selected for a high-time resolution phosphoproteome survey of CRK5-KO parasites are indicated. B. Volcano plots showing the extent of differentially phosphorylated peptides in non-activated and activated gametocytes after 15 s, comparing CRK5-KO and WT parasite lines. Significantly regulated sites (p-value<0.05) with a fold change >2 are highlighted in red, and in blue when corresponding to the CDK S/T*PxK motif. C. GO term enrichment analysis for down- and up-regulated phosphopeptides in CRK5-KO gametocytes during the first minute of gametogony. D. Alignment of eight down-regulated phosphopeptides showing the classical S/T*PxK CDK motif. E. Reduction in the number of male gametocytes replicating their DNA upon crk5 deletion or CRK5-AID/HA depletion. Proportion of male gametocytes undergoing DNA replication was determined at 1 min pa (post-activation) and is expressed as a percentage of polyploid (>1N) cells (error bars show standard deviation from the mean; three independent infections; two-way ANOVA). F. Reduction in the number of male gametocytes with spindle-like structures upon crk5 deletion or CRK5-AID/HA depletion, as assessed by α-tubulin staining 1 min pa (error bars show standard deviation from the mean; three independent infections; two-way ANOVA). Inset pictures show representative typical tubulin distribution patterns (green), observed in WT microgametocytes counter stained with DAPI (blue); scale bar = 2 µm. G. CRK5-KO gametocytes, fifteen min after activation, have fully assembled axonemes that do not initiate flagellar motility, in contrast to their WT counterparts (error bars show standard deviation from the mean; two independent infections; two tailed unpaired t-test). Scale bar = 5 µm. H. Electron micrographs of male gametogony in wild type parasites at 3–6 min (a, b) and 15–30 min (c, d, e) and CRK5-KO mutant parasites at 3–6 min (f, g) and 15–30 min (h, i). Bars represent 1 µm (a, c, f, h) and 100 nm in other images. (a) Low power micrograph an early gametocyte showing the large nucleus (N) and two nuclear poles. Insert: detail of a cross-sectioned axoneme. (b) Detail of the enclosed area showing the nuclear poles (NP) with an adjacent basal body (BB) and elongated axoneme (A). (c) Late microgametocyte undergoing exflagellation showing the nucleus (N) with some clumped heterochromatin (Ch) and a flagellum (F) protruding from the surface. (d) Detail of the enclosed area showing the point of exit for the microgamete flagellum (F). (e) Cross-section through a free microgamete showing the 9+two flagellum (F) and electron dense nucleus (N). (f) Low power of an early microgametocyte showing an irregular lobated nucleus (N) and the presence of a nuclear pole and axonemes unusually centrally located in the cytoplasm. Insert: detail of a cross-sectioned axoneme. (g) Detail of the enclose area showing the nuclear pole (NP) closely associated with a basal body (BB) and longitudinally running axonemes. (h) At a late time point, the parasite still resembles an early stage with lobated nucleus (N) which displays nuclear poles (NP) and basal bodies (BB). (i) Detail of the enclosed area showing the nuclear pole (NP) and associated cross section basal body (BB). Note the normal and abnormal axonemes (A).
-
Figure 2—source data 1
CRK5 shows functional similarities to canonical CDKs and directly regulates DNA replication during gametogony.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig2-data1-v1.xlsx
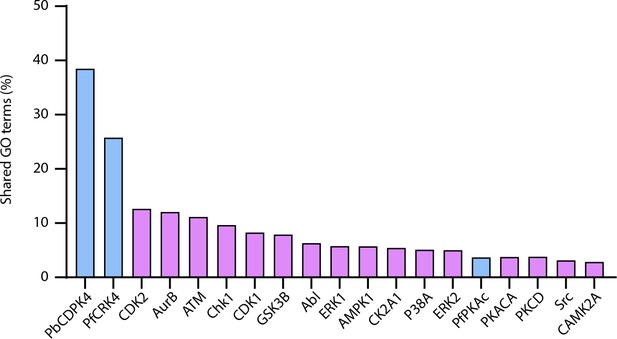
Percentage of GO terms shared between CRK5 and a set of Plasmodium and human kinases.
For CRK5 GO terms, proteins with a > 2 fold change (p-value<0.05) in the CRK5-KO were retained.
-
Figure 2—figure supplement 1—source data 1
Percentage of GO terms shared between CRK5 and a set of Plasmodium and human kinases.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig2-figsupp1-data1-v1.xlsx
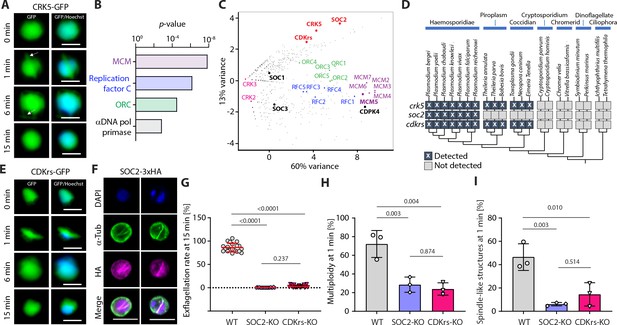
CRK5 interacts with a divergent cyclin and a conserved CDK regulatory subunit.
(A) Localisation of CRK5-GFP during male gametogony. The protein colocalises with Hoechst DNA-binding dye and shows a mitotic spindle distribution during M-phases. Scale bar = 5 µm. (B) GO term enrichment analysis of proteins co-immunoprecipitated with CRK5 from gametocytes. Bonferroni corrected p-values are indicated. (C) Spectral count values as identified by mass spectrometry for proteins co-purifying with CRK5, CDKrs, SOC2, CDPK4, SOC1, SOC3 and MCM5 (highlighted in bold) following immunoprecipitation, and displayed in first and second principal components. Proteomic sets (including CRKs, cyclins, MCMs, ORCs and RepC complex proteins) are additionally highlighted. (D) Phylogenetic distribution of the CSC complex components detected in alveolate genomes. Dark boxes with X denote the presence of genes. (E) Localisation of CDKrs-GFP during male gametogony. The protein colocalises with Hoechst dye and shows a mitotic spindle distribution during M-phases. Scale bar = 5 µm. (F) Localisation of SOC2-3xHA by immunofluorescence in activated male gametocytes. The protein colocalises with DAPI and shows overlap with mitotic spindles stained by α-tubulin antibodies during M-phase. Scale bar = 5 µm. (G) Deletions of soc2 or cdkrs lead to a profound defect in exflagellation (error bars show standard deviation from the mean; three independent infections; two-way ANOVA). (H) Reduction in the number of male gametocytes replicating their DNA in parasites with soc2 or cdkrs deletions. The proportion of male gametocytes undergoing DNA replication was determined at 1 min pa and is expressed as the percentage of cells that are polyploid (>1N) (error bars show standard deviation from the mean; three independent infections; two-way ANOVA). (I) Reduction in the number of male gametocytes with spindle-like structures following soc2 or cdkrs deletion, as assessed by α-tubulin staining 1 min pa (error bars show standard deviation from the mean; three independent infections; two-way ANOVA).
-
Figure 3—source data 1
CRK5 interacts with a divergent cyclin and a conserved CDK regulatory subunit.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig3-data1-v1.xlsx
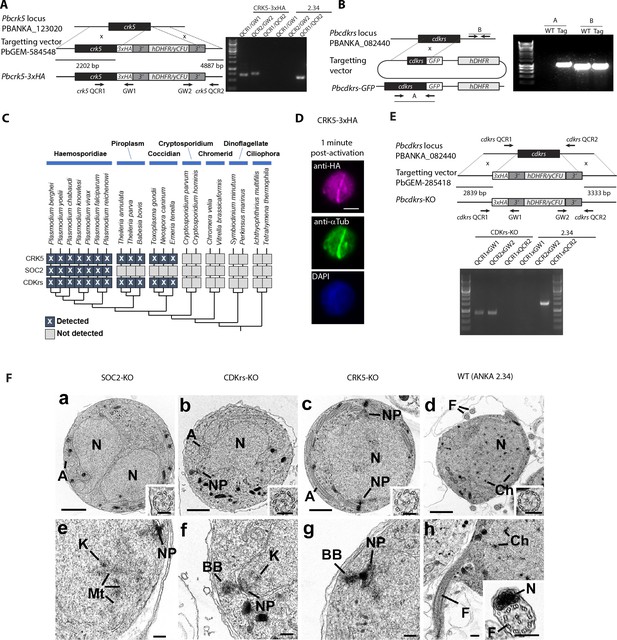
Identification and characterisation of the CSC complex components.
(A-B) Genetic modification vectors and genotyping data for CRK5-3xHA (A) and CDKrs-GFP (B). Oligonucleotides used for PCR genotyping are indicated and agarose gels for corresponding PCR products from genotyping reactions are shown. (C) Phylogenetic distribution of the CSC complex components and of Plasmodium CRKs in selected alveolate genomes (see methods). Dark boxes with X denote the presence of genes. (D) Localisation of CRK5-3xHA in 1 min activated gametocytes. Scale bar = 5 µm. (E) Genetic modification vectors and genotyping data for CDKrs-KO. F. Electron micrographs of the male gametocytes of the SOC2-KO (a, e), CDKrs-KO (b, f), CRK5-KO (c, g) and wild type (d,h) parasites at 15 min pa. The three KO parasites consist of predominately early stages (95–100%) while the WT showed a number of late stages (~40%) with exflagellation and free microgametes. Bars represent 1 µm in a-d and 100 nm in all other micrographs. (a to c) SOC2-KO, CDKrs, and CRK5 male gametocytes showing the early stage of development with a lobed nucleus (N) and cytoplasmic axonemes (A). Inserts. Detail of cross-sectioned axonemes. (d) WT showing a late stage in development with nucleus (N) containing small clumps of condensed chromatin (Ch). Note the free microgamete flagella (F). Insert. Cross-sectioned axoneme. (e) Detail of a similar stage parasite to that in (a) showing a nuclear pole (NP) but note the disorganised arrangement of spindle microtubules (Mt) and attached kinetochores (K). (f) Detail of the nucleus in (b) showing the nuclear pole (NP) with a closely associated basal body (BB) in the cytoplasm. K – Kinetochore. (g) Detail of a similar stage to that in (c) showing the nuclear pole (NP) and associated basal body (BB). (h) Detail of a similar parasite to that in (d) undergoing exflagellation showing the flagellum (F) protruding from the microgametocyte. Ch – condensed chromatin. Insert. Cross section through a free microgamete showing the flagellum (F) and electron dense nucleus (N).
-
Figure 3—figure supplement 1—source data 1
DNA sequences used in panel D.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig3-figsupp1-data1-v1.txt
-
Figure 3—figure supplement 1—source data 2
Accession numbers of genes analysed in panel D.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig3-figsupp1-data2-v1.xlsx
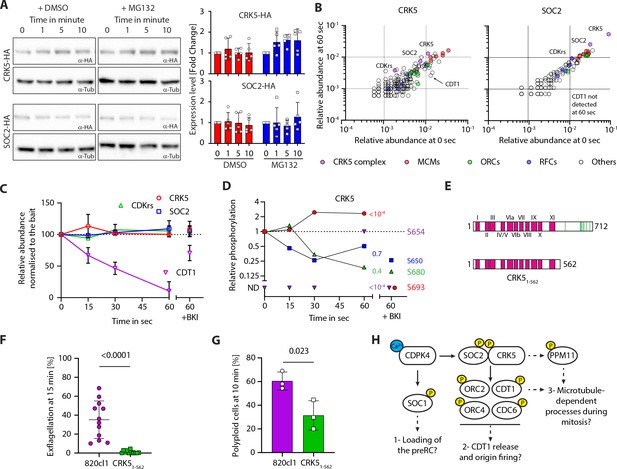
The CSC complex is stable but dynamically phosphorylated during gametogony.
(A) Western blot analysis of CRK5-3xHA and SOC2-3xHA over the course of gametogony shows that the level of each protein is stable in absence or presence of the proteasome inhibitor MG132; α-tubulin serves as a loading control. Quantification from five independent biological samples is shown to the right (error bars show standard deviation from the mean). (B) Dot blots showing the relative abundance of proteins immunoprecipitated (% of all spectra) from gametocytes at 0 or 60 s activation, with CRK5-3xHA (left panel) and SOC2-3xHA (right panel). No major changes were observed between the two conditions; CDT1 is however recovered less in both IPs at 60 s. (C) Relative abundance of CRK5, SOC2, CDKrs and CDT1 in SOC2-3xHA and CRK5-3xHA IPs over the first min of gametogony and in gametocytes pre-treated with the CDPK4 inhibitor BKI-1294 (Ojo et al., 2014) (1 µM). Abundance are relative to the bait and normalised to the 0 s time point (error bars show standard deviations from the mean; data are extracted from single SOC2-3xHA and CRK5-3xHA immunoprecipitations). (D) Phosphorylation profile of immunoprecipitated CRK5 during the first minute of gametogony in presence or absence of the CDPK4 inhibitor BKI-1294 (1 µM). ND = Not detected. (E) Schematic representation of the CRK51-562 mutant generated in this study. (F) CRK51-562 gametocytes show a strong defect in exflagellation (error bars show standard deviation from the mean; three independent infections; two-tailed t-test). (G) Reduction in the number of male gametocytes with a crk5 3’ deletion replicating their DNA. The proportion of male gametocytes undergoing DNA replication was determined at 10 min pa and is expressed as a percentage of cells that are polyploid (>1N) (error bars show standard deviation from the mean; three independent infections; two-tailed t-test). (H) Possible signalling cascades regulating entry and progression through the first replicative cycle during P. berghei gametogony. Early calcium mobilisation upon gametocyte activation leads to CDPK4-dependent phosphorylation of SOC1 and the CSC complex. SOC1 may be involved in the loading of the pre-replicative complex (preRC) (Fang et al., 2017). Here, we propose that CDPK4-dependent phosphorylation of SOC2 and/or CRK5 activates the kinase activity to phosphorylate components of the pre-replicative complex allowing initiation of DNA replication. Indirect regulation of dyneins through phosphatases such as PPM11 could possibly regulate progression into M-phase. Yellow circles represent phosphorylation.
-
Figure 4—source data 1
Panel A, CRK5-HA western blot.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig4-data1-v1.pdf
-
Figure 4—source data 2
Panel B, SOC2-HA western blot.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig4-data2-v1.pdf
-
Figure 4—source data 3
The CSC complex is stable but dynamically phosphorylated during gametogony.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig4-data3-v1.xlsx
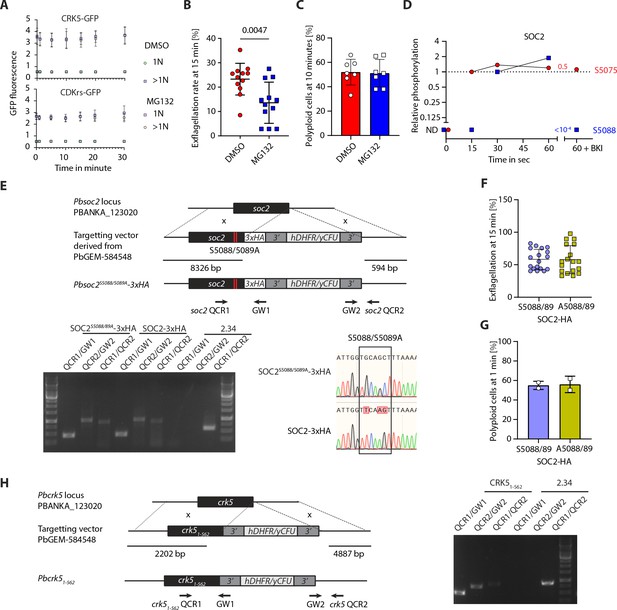
Study of the CSC complex regulation.
(A) Flow cytometry analysis of CRK5-GFP and CDKrs-GFP fluorescence over the course of gametogony in haploid or multiploid (>1N) cells in the presence or absence of 1 µM of the proteasome inhibitor MG132 (error bars show standard deviation from the mean; 3 or four independent infections; two-way ANOVA). (B) Pre-treatment of gametocytes with 1 µM MG132 leads to reduced exflagellation (error bars show standard deviation from the mean; technical replicates from three independent infections; unpaired two-tailed t-test). (C) Treatment with 1 µM MG132 has no effect on the number of male gametocytes replicating their DNA. The proportion of male gametocytes undergoing DNA replication was determined at 10 min pa and is expressed as a percentage of cells that are polyploid (>1N) (error bars show standard deviation from the mean; technical replicates from three independent infections; unpaired two-tailed t-test). (D) SOC2 phosphorylation levels show that S5088 is phosphorylated in a CDPK4-dependent manner 15 s after activation. ND = not detected. BKI = CDPK4 inhibitor. (E) Genetic modification vector and genotyping data for SOC2S5088/5089A mutagenesis. Oligonucleotides used for PCR genotyping are indicated and agarose gels for corresponding PCR products from genotyping reactions are shown. (F) S5088/5089A substitution in SOC2-3xHA does not impair exflagellation (error bars show standard deviation from the mean; technical replicates from two independent infections; two-way ANOVA). (G) There is no reduction in the number of male gametocytes replicating their DNA in SOC2S5088/5089A-3xHA gametocytes. The proportion of male gametocytes undergoing DNA replication was determined at 1 min pa and is expressed as a percentage of cells that are polyploid (>1N) (error bars show standard deviation from the mean; from two independent infections; two-way ANOVA). (H) Genetic modification vector and genotyping data for CRK5 mutagenesis. Oligonucleotides used for PCR genotyping are indicated and agarose gels for corresponding PCR products from genotyping reactions are shown.
-
Figure 4—figure supplement 1—source data 1
Study of the CSC complex regulation.
- https://cdn.elifesciences.org/articles/56474/elife-56474-fig4-figsupp1-data1-v1.xlsx
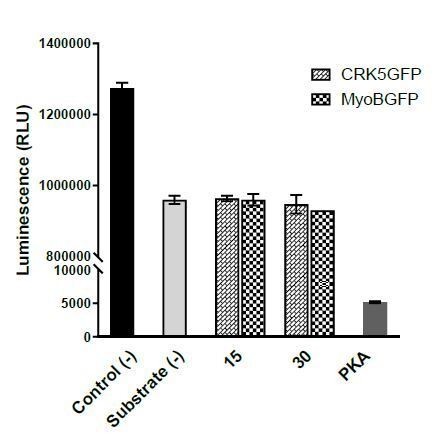
CRK5 Kinase activity assay using Kinase-Glo Assay kit.
Kinase reactions containing 40 mM Tris-HCl (pH 7.5), 20 mM MgCl2 were performed at 10 μM ATP, 50 μM Histone with different dilutions of kinases for 1 hour. X-axis represents: Control (-) – No kinase, Substrate (-) – No substrate, 15 and 30 – quantity of CRK5-GFP and MyoB-GFP beads (μl), PKA – 10 units of Protein kinase A. Error bars represent standard deviations from the mean; two independent biological replicates.

Expression of CRK5, CDKrs and SOC2 in E. coli BL21 cells.
Overnight IPTG-induced expression of CRK5, CDKrs or SOC2 in E. coli BL21 (DE3) cells at 16ᵒC. Cell pellets were resuspended in 60 ml of lysis buffer (50 mM Tris pH 7.4/ 200 mM NaCl/ 3 mM 2-ME – fresh) and lysed by two passages through a French Press at 1,000 psi, followed by sonication for 20 sec, 20 % duty cycle, power 5. Cell debris and membranes were pelleted by centrifugation (30'000 g, 35 min, 4 °C). Soluble (supernatant) and non-soluble (pellet) fractions were denatured and analysed by SDS-PAGE. Expected MWs are: CRK5: 85 kDa, CDKrs: 35 kDa, and SOC2 158 kDa. Please also note that lower MW are observed for CRK5 and SOC2.
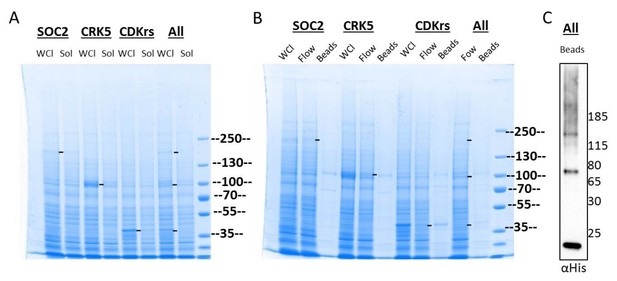
Expression and tentative purification of SOC2, CRK5 and CDKrs in Sf9 cells.
(A) Expression of SOC2, CRK5 and CDKrs N-terminally fused to the mellitin signal peptide, as well as 10xHis and 2xStreptavidin tags individually or in co-expression (all). Cell pellets were resuspended in 60 ml of lysis buffer (50 mM Tris pH 7.4/ 200 mM NaCl/ 3 mM 2-ME – fresh) and lysed by two passages through the French Press at 1000 psi, followed by sonication for 20 sec, 20 % duty cycle, power 5. Cell debris and membranes were pelleted by centrifugation (30'000 g, 35 min, 4°C). Soluble (Sol) and non-soluble (WCL, whole cell lysate) fractions were denatured and analysed by SDS-PAGE. (B) The supernatant containing soluble proteins was applied to a 5 ml Strep-TACTIN column and washed with 40 ml lysis buffer (Flow). WCL, flow and proteins retained on beads were analysed by SDS-PAGE stained with coomassie. C. Western blot analysis of proteins bound to beads.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BigEasy-TSA | Lucigen | Cat# 60224–1 | Electrocompetent cells |
| Transfected constructs (P. berghei) | PlasmoGEM vectors | https://plasmogem.sanger.ac.uk/ | Materials and method ssection and supplementary figures | |
| Cell line (P. berghei) | ANKA 2.34 | Billker et al., 2004 | ||
| Cell line (P. berghei) | 507cl1 | Janse et al., 2006b | ||
| Cell line (P. berghei) | 820cl1 | Mair et al., 2010 | ||
| Cell line (P. berghei) | 615 | Philip and Waters, 2015 | ||
| Cell line (P. berghei) | 507cl1 CRK5-KO | This study | Available from the Nottingham laboratory | |
| Cell line (P. berghei) | 615 CRK5-AID/HA | This study | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 2.34 CRK5-GFP | This study | Available from the Nottingham laboratory | |
| Cell line (P. berghei) | 2.34 CRK5-3xHA | This study | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 2.34 CDKrs-GFP | This study | Available from the Nottingham laboratory | |
| Cell line (P. berghei) | 820cl1 CDKrs-KO | This study | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 820cl1 SOC2-KO | Fang et al., 2017 | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 820cl1 SOC2-3xHA | Fang et al., 2017 | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 820cl1 SOC2S5088/89A-3xHA | This study | Available from the Geneva laboratory | |
| Cell line (P. berghei) | 820cl1 CRK51-562 | This study | Available from the Geneva laboratory | |
| Antibody | Anti-HA High Affinity from rat IgG1 (3F10) | Sigma | Cat# 0000011867423001 | IF(1:1000), WB (1:1000), IP (1:1000) |
| Antibody | Anti-c-Myc antibody produced in rabbit | Sigma | Cat# C3956 | WB (1:1000) |
| Antibody | Anti-α-Tubulin antibody from mouse (DM1A) | Sigma | Cat# T6199 | IF(1:1000), WB (1:1000) |
| Antibody | Goat anti-Rat IgG (H+L), Alexa Fluor 594 | Thermofisher | Cat# A-11007 | IF(1:1000) |
| Antibody | Goat anti-Mouse IgG (H+L), Alexa Fluor 488 | Thermofisher | Cat# A-11029 | IF(1:1000) |
| Antibody | Goat anti-Rabbit IgG (H+L), Alexa Fluor 488 | Thermofisher | Cat# A-11034 | IF(1:1000) |
| Sequence-based reagent | PCR primers | Microsynth | Materials and methodssection and Supplementary file 5 | |
| Chemical compound, drug | BKI-1294 | Ojo et al., 2014 | CDPK4 inhibitor (1 μM) |
Additional files
-
Supplementary file 1
RNAseq data.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp1-v1.xls
-
Supplementary file 2
Peptides detected in WT and CRK5-KO gametocytes.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp2-v1.xlsx
-
Supplementary file 3
Phosphopeptides detected in WT and CRK5-KO gametocytes.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp3-v1.xlsx
-
Supplementary file 4
Spectral count values of proteins identified in co-immunoprecipitates by mass-spectrometry.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp4-v1.xlsx
-
Supplementary file 5
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp5-v1.xlsx
-
Supplementary file 6
Correspondence between TMT labels and samples.
- https://cdn.elifesciences.org/articles/56474/elife-56474-supp6-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56474/elife-56474-transrepform-v1.docx





