Biofilms deform soft surfaces and disrupt epithelia
Figures
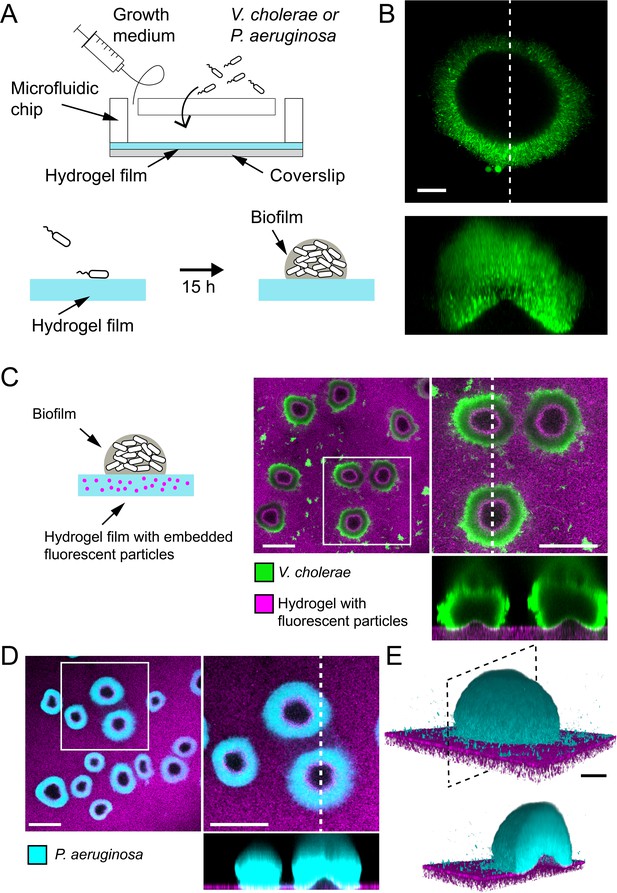
Biofilms deform soft substrates.
(A) Illustration of experimental setup where we generate thin hydrogel films at the bottom surface of microchannels. These devices allow us to study biofilm formation on hydrogels reproducing mechanical properties of host tissues. (B) In-plane and cross-sectional confocal visualizations show that V. cholerae Rg biofilms growing on hydrogels display large gaps at their core. (C) Embedding fluorescence tracer particle in the hydrogel films allow for visualization of deformations. V. cholerae Rg biofilms formed at the surface of the films deform the substrate. (D) P. aeruginosa Rg biofilms similarly deform the soft substrates. Hydrogel elastic modulus: (B and C) E = 12 kPa, (D and E) E = 38 kPa. Scale bars: (C and D) 100 µm, (B and E) 20 µm.
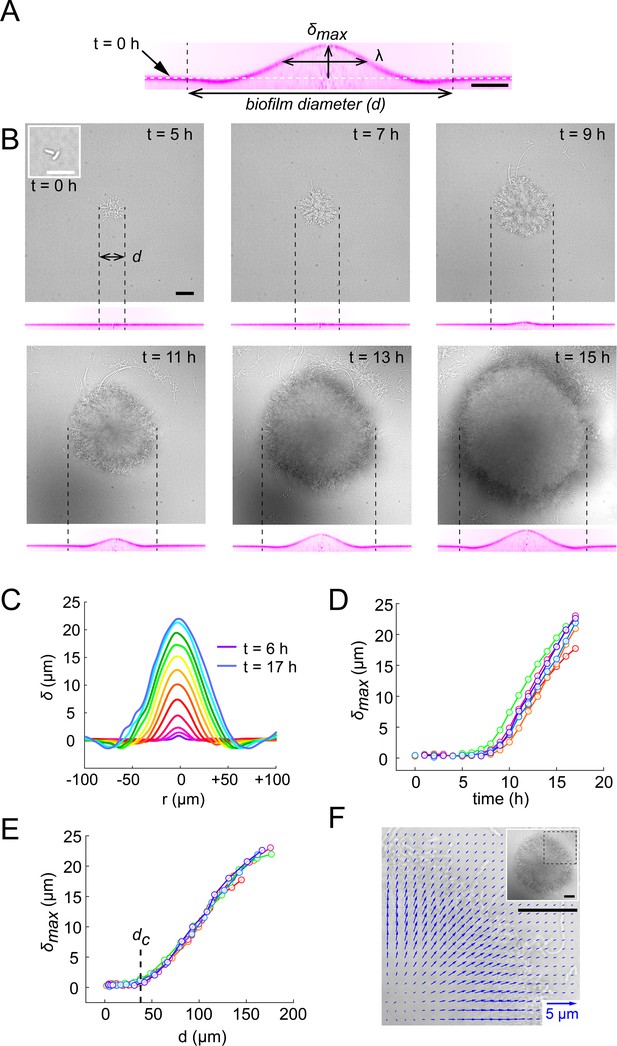
Biofilms deform their substrate by buckling.
(A) Morphological parameters (maximum deformation amplitude) and (half max full width) computed from resliced deformation profiles. Dashed line indicates the baseline position of the gel surface. (B) Timelapse visualization of V. cholerae Rg biofilm growth (brightfield, top) with deformation (reslice, bottom). Dashed lines indicate biofilm position and size on the corresponding hydrogel profile. (C) Superimposition of these profiles shows the rapid deformation and the emergence of a recess at biofilm edges. Each color corresponds to the same biofilm at different times. (D) Time evolution of shows a rapid increase after 6 to 7 hr of growth. (E) The dependence of on biofilm diameter highlights a critical biofilm diameter dc above which deformation occurs. For D and E, each line color corresponds to a different biofilm. (F) Hydrogel strain field computed by digital volume correlation between 11 hr and 12 hr of growth. We superimposed the vector strain field with a brightfield image of the biofilm. For visualization purposes we only display data for the top right quarter of the biofilm shown in inset (dashed lines). E = 38 kPa. Scale bar: 10 µm for inset t = 0 hr in (B), else 20 µm.
-
Figure 2—source data 1
Deformation amplitude and as a function of time and diameter.
- https://cdn.elifesciences.org/articles/56533/elife-56533-fig2-data1-v2.xlsx
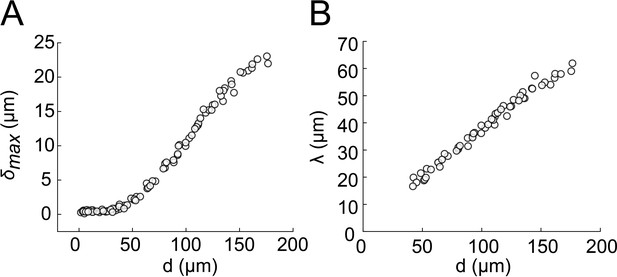
Biofilm diameter-dependence of δmax and λ.
(A) Biofilm diameter-dependence of . (B) Biofilm diameter-dependence of λ. Both and λ linearly scale with the diameter d of the biofilm.
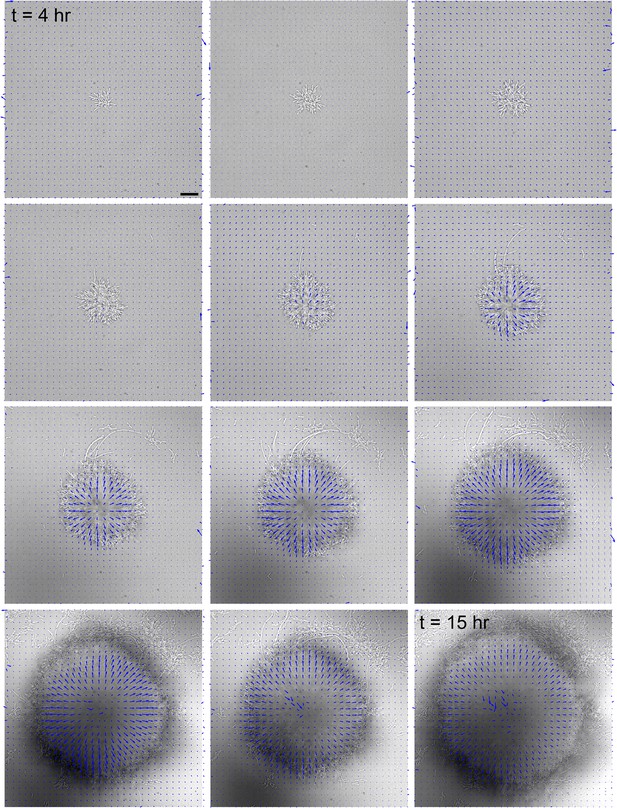
Hydrogel deformation field computed at different growth stages, superimposed with a brightfield image of the biofilm.
Scale bar: 20 µm. The force field at each timestep is normalized by its maximum displacement, thereby showing relative deformations.
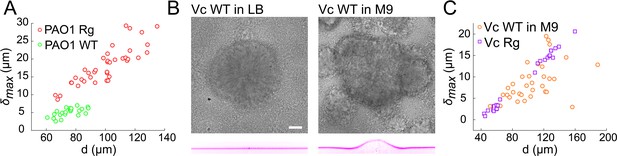
Wild-type and rugose biofilms deform soft-substrates.
(A) Biofilm diameter-dependence of maximum deformation for rugose and smooth variants of P. aeruginosa. (B) Smooth variant of V. cholerae A1552 deforms hydrogels when growing in M9 medium, but not in LB. (C) Biofilm diameter-dependence of maximum deformation for rugose and smooth variants of V. cholerae. Data points correspond to biofilms grown in two microfluidic chambers for PAO1 Rg, PAO1 WT and Vc WT and to biofilms grown in one microfluidic chamber for Vc Rg. E = 38 kPa. Scale bars: 20 µm.
-
Figure 3—source data 1
Deformation amplitude for WT strains.
- https://cdn.elifesciences.org/articles/56533/elife-56533-fig3-data1-v2.xlsx
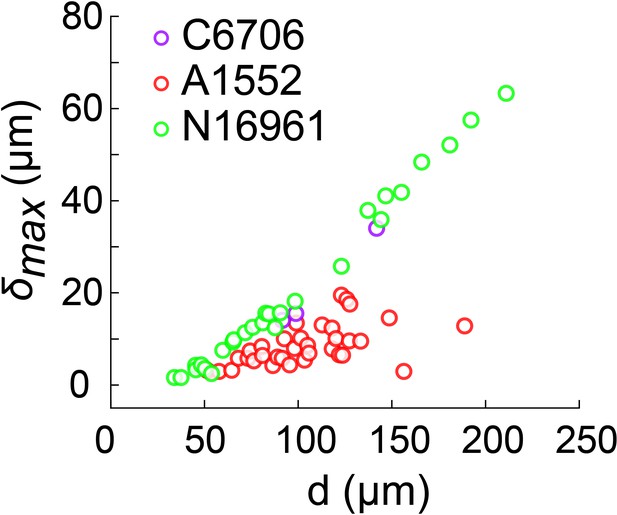
Biofilm diameter-dependence of maximum deformation for the smooth variant of different V. cholerae strains grown in M9.
Data points correspond to different biofilms grown in two microfluidic chambers for A1552 and to biofilms grown in one microfluidic chamber for C6706 and N16961.
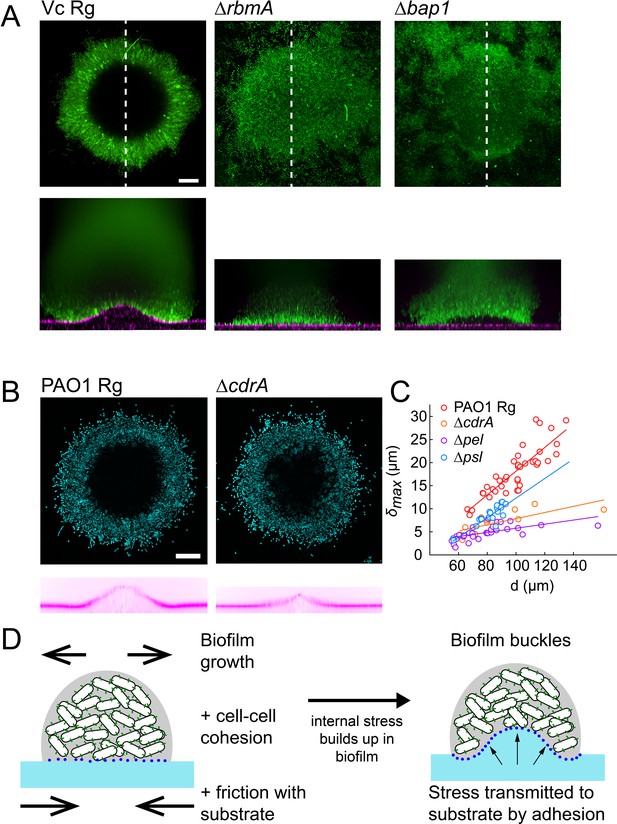
EPS composition drives biofilm and substrate deformations.
(A) Deformations of hydrogel substrates by V. cholerae Rg, rbma- and bap1- biofilms. Biofilms formed by rbma- and bap1- fail to deform the substrate. bap1- biofilms delaminate from the hydrogel surface. (B) Comparison of hydrogel deformations by P. aeruginosa Rg and cdrA- biofilms. (C) Dependence of maximum deformations on P. aeruginosa Rg, cdrA-, pel- and psl- biofilm diameter. All matrix mutants tend to generate weaker deformations compared to Rg. Data points correspond to different biofilms grown in two microfluidic chambers. (D) A model for the mechanism of biofilm deformation of soft substrates. Buildup of mechanical stress in the biofilm induces buckling. Adhesion between the biofilm and the surface transmits buckling-generated stress to the hydrogel, inducing deformations. E = 38 kPa. Scale bars: 20 µm.
-
Figure 4—source data 1
Deformation amplitude for PAO1 matrix mutants.
- https://cdn.elifesciences.org/articles/56533/elife-56533-fig4-data1-v2.xlsx
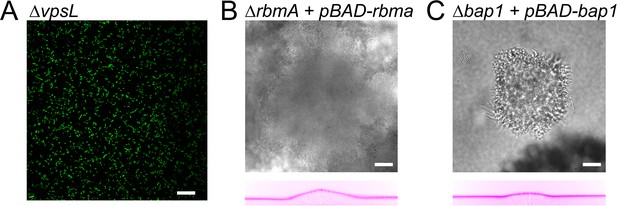
Deformation behaviour for vpsL deletion mutant and complementation strains.
(A) VpsL deletion mutant can not form biofilms. Complementation of (B) V. cholerae rbmA and (C) bap1 deletion mutants (brightfield, top) restore the ability of the biofilm to deform the hydrogel (reslice, bottom). E = 38 kPa. Scale bars: 20 µm.
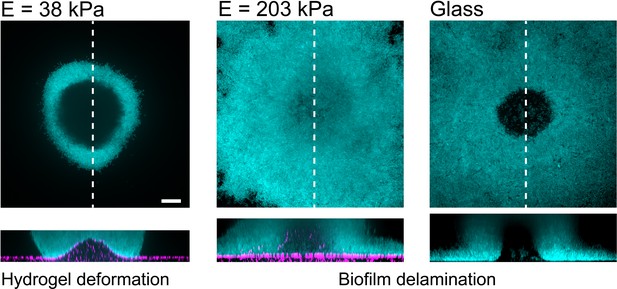
P. aeruginosa biofilms on substrates with different stiffness.
Increasing hydrogel stiffness to 200 kPa induces delamination of biofilms, as observed on glass. Scale bars: 20 µm.
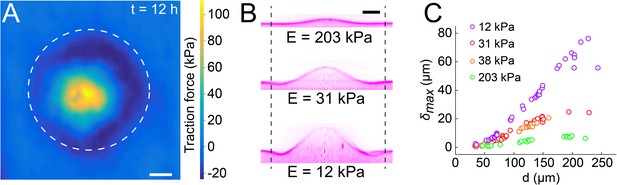
Biofilms generate large traction forces.
(A) Traction force microscopy measurements at the hydrogel-biofilm interface. The dashed line shows the edge of the biofilm. Traction force is largest at the biofilm center, reaching 100 kPa. (B) Deformation profiles generated by V. cholerae Rg biofilms of equal diameters on three hydrogels with different stiffness. (C) Biofilm diameter-dependence of maximum deformation for four different hydrogel composition representing a typical range of tissue stiffnesses. The softest hydrogel can deform up to 80 µm for a biofilm diameter of 220 µm. Data points correspond to different biofilms grown in one microfluidic chamber. Scale bar: 20 µm.
-
Figure 5—source data 1
Deformation amplitude and for substrates of different stiffness.
- https://cdn.elifesciences.org/articles/56533/elife-56533-fig5-data1-v2.xlsx
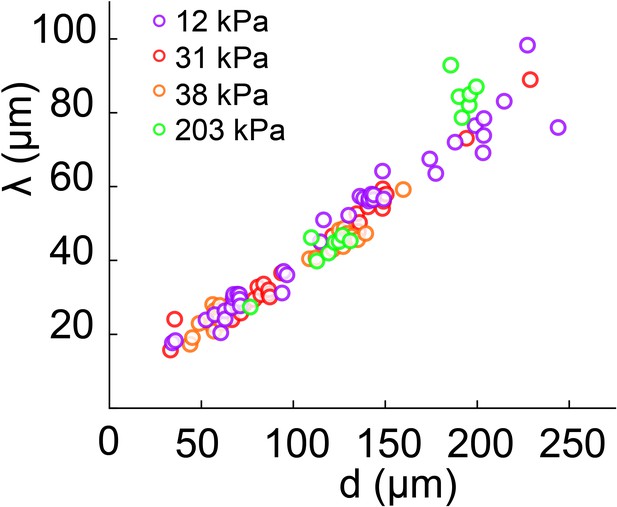
Biofilm diameter-dependence of λ for substrates with different moduli.
λ scales linearly and it is not substrate-stiffness dependent.
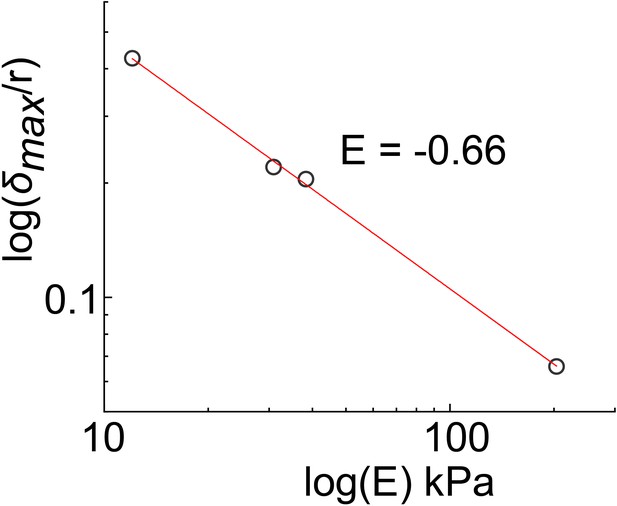
Power-law relationship between deformation and substrate moduli (E).
Values for δmax/r were extrapolated from linear regression of the data points in Figure 5C for r = 50 µm (d = 100 µm).
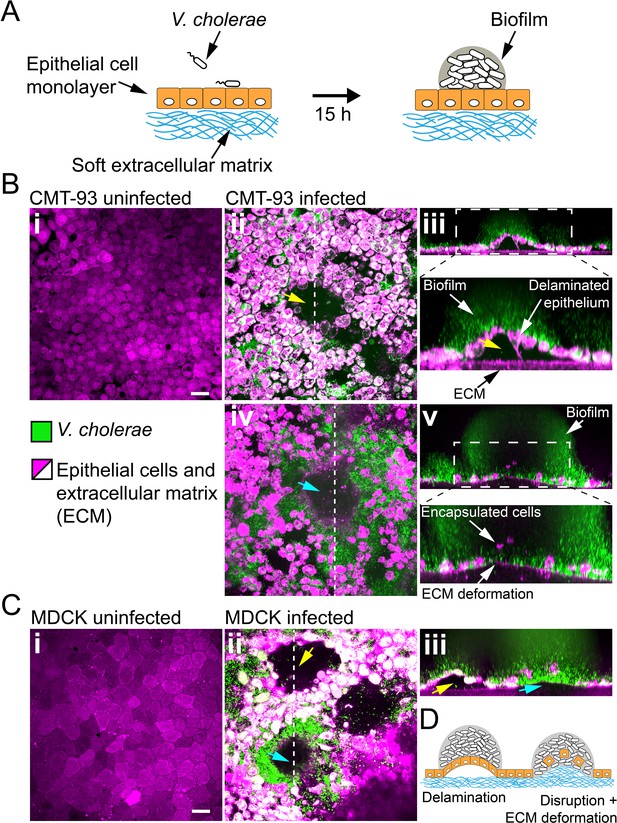
Biofilms deform and disrupt epithelial cell monolayer.
(A) CMT-93 and MDCK cells grow at the surface of a soft ECM into a tight monolayer on which we seed a liquid inoculum of V. cholerae Rg. (B) Confocal images of uninfected (i) and infected (ii-v) monolayers of CMT-93 cells. Yellow arrow indicates gaps in the epithelial monolayer (ii and iii), blue arrow shows deformed ECM (iv). (C) Confocal images of uninfected (i) and infected (ii-iii) monolayers of MDCK cells, also showing delamination and rupture as illustrated in (D). Scale bars: 20 µm.
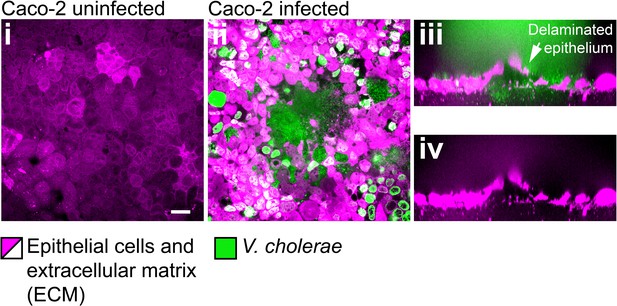
Confocal images of uninfected (i) and infected (ii-iv) monolayers of Caco-2 cells.
Scale bars: 20 µm.
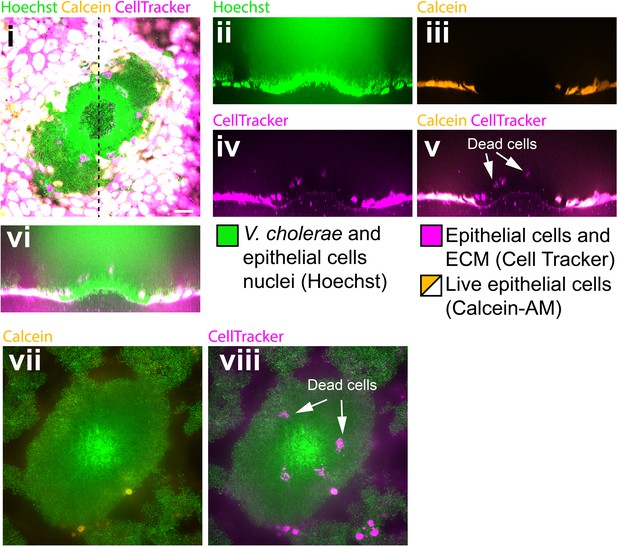
Biofilms perturb the viability of MDCK cell monalyers.
(i) Confocal in plane visualization of a Vc Rg biofilm growing on top of an MDCK cell monolayer stained with CellTracker, Hoechst and Calcein-AM. Cross-section visualization of the infected MDCK monolayer stained with Hoechst (ii), Calcein-AM (iii), Cell Tracker (iv) and merged visualizations (v, vi). (vii, viii) Confocal in plane visualization of the biofilm on a focal plane above (i). Scale bars: 20 µm.
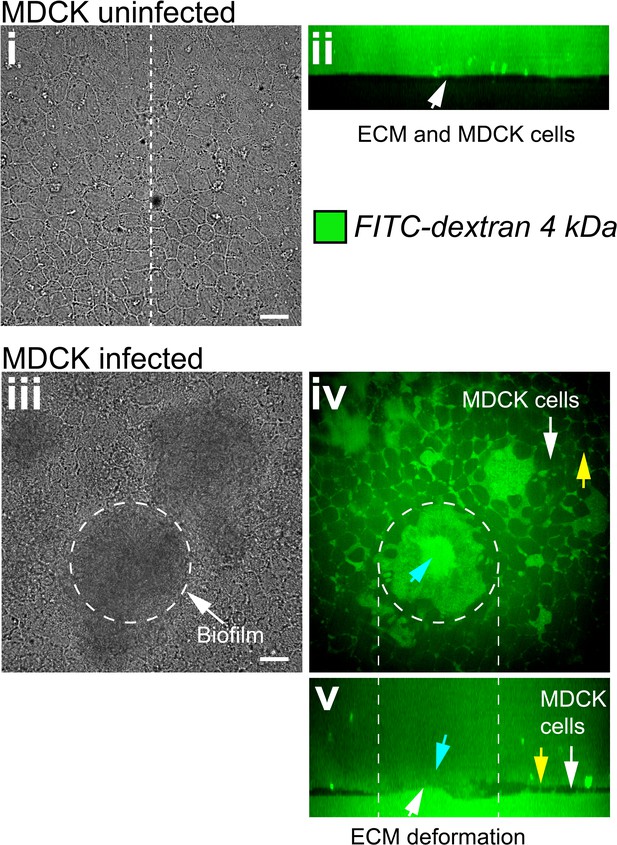
Biofilms increase the permeability of MDCK cell monolayers.
(i) Bright-field in plane image of an uninfected MDCK cell monolayer grown at the surface of a soft ECM. (ii) Confocal cross-section shows that uninfected monolayers are impermeable to FITC labeled Dextran. (iii) Bright-field in plane visualization of an MDCK cell monolayer infected with Vc Rg. The dashed line shows the approximate edge of the biofilm. Confocal in plane (iv) and cross section (v) visualization of FITC-labeled Dextran permeability through the damaged epithelium. Blue arrows show Dextran permeability through the biofilm in direct contact with the ECM, yellow arrows indicate Dextran permeability through epithelial cells junctions. Scale bars: 20 µm.
Videos
Timelapse visualization of V. cholerae Rg biofilm growth (brightfield) and corresponding hydrogel deformation (E = 38 kPa) in the xy and xz planes.
Scale bar 20 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (V. cholerae A1552) | Vc Rg | Yildiz and Schoolnik, 1999 | Rugose variant | |
| Strain, strain background (V. cholerae A1552) | Vc WT | Yildiz and Schoolnik, 1999 | Smooth wild-type variant | |
| Strain, strain background (V. cholerae A1552) | Vc Rg ΔvpsL | This study | In frame deletion of vpsL in rugose background obtained by matings of Vc Rg with S17 harboring deletion plasmid pFY_922 | |
| Strain, strain background (V. cholerae A1552) | Vc Rg ΔrbmA | This study | In frame deletion of rbmA in rugose background obtained by matings of Vc Rg with S17 harboring deletion plasmid pFY_113 | |
| Strain, strain background (V. cholerae A1552) | Vc Rg Δbap1 | This study | In frame deletion of bap1 in rugose background obtained by matings of Vc Rg with S17 harboring deletion plasmid pFY_330 | |
| Strain, strain background (V. cholerae A1552) | Vc Rg ΔrbmA pBADrbmA | This study | Vc Rg ΔrbmA harboring the plasmid pNUT1236 | |
| Strain, strain background (V. cholerae A1552) | Vc Rg Δbap1 pBADbap1 | This study | Vc Rg Δbap1 harboring the plasmid pBAD/Myc-His B | |
| Strain, strain background (V. cholerae N16961) | N16961 | Drescher et al., 2016 | Smooth wild-type variant | |
| Strain, strain background (V. cholerae C6706) | C6706 | Thelin and Taylor, 1996 | Smooth wild-typevariant | |
| Strain, strain background (P. aeruginosa) | PAO1 WT | Hickman et al., 2005 | ||
| Strain, strain background (P. aeruginosa) | PAO1 Rg | Rybtke et al., 2012 | In frame deletions of wspF | |
| Strain, strain background (P. aeruginosa) | PAO1 Rg Δpel | Rybtke et al., 2012 | In frame deletions of wspF, pelA genes | |
| Strain, strain background (P. aeruginosa) | PAO1 Rg Δpsl | Rybtke et al., 2012 | In frame deletions of wspF, pslBCD genes | |
| Strain, strain background (P. aeruginosa) | PAO1 Rg ΔcdrA | Rybtke et al., 2015 | In frame deletions of wspF, cdrA genes | |
| Cell line (Homo sapiens) | Caco-2 | ATCC | HTB-37 RRID:CVCL_0025 | |
| Cell line (Canis) | MDCK | Sigma Aldrich | 84121903-1VL RRID:CVCL_0422 | |
| Cell line (Mus musculus) | CMT-93 | ATCC | RRID:CCL-223 RRID:CVCL_1986 | |
| Recombinant DNA reagent | pFY_113 (plasmid) | Berk et al., 2012 | Plasmid for generation of in-frame rbmA deletion mutants | |
| Recombinant DNA reagent | pFY_330 (plasmid) | Berk et al., 2012 | Plasmid for generation of in-frame bap1 deletion mutants | |
| Recombinant DNA reagent | pFY_922 (plasmid) | Fong et al., 2010 | Plasmid for generation of in-frame vpsL deletion mutants | |
| Recombinant DNA reagent | pNUT1236 (plasmid) | Hartmann et al., 2019 | Arabinose-inducible plasmid containing the coding region of rbmA | |
| Recombinant DNA reagent | pBAD/Myc-His B (plasmid) | Fong and Yildiz, 2007 | Arabinose-inducible plasmid containing the coding region of bap1 | |
| Chemical compound, drug | Lithium phenyl-2,4,6- trimethylbenzoylphosphinate (LAP) | Tokio Chemical Industries | ||
| Chemical compound, drug | PEGDA (MW 3400, 6000, 10000) | Biochempeg | ||
| Chemical compound, drug | PEGDA (MW 700) | Sigma-Aldrich | ||
| Software | Fiji | Fiji | ||
| Software | OriginPro | OriginLab Corporation | ||
| Software | MATLAB | Mathworks | ||
| Software | Imaris | Bitplane | ||
| Algorithm | 3D TFM | Toyjanova et al., 2014 | ||
| Other | SYTO9 stain | Invitrogen | S34854 | 10 µM |
| Other | CellTracker Orange CMRA stain | Invitrogen | C34551 | 10 µM |
| Other | Hoechst stain | Thermo Fischer Scientific | 62249 | 5 µg/ml |
| Other | Calcein-AM stain | Sigma Aldrich | 17783 | 5 µM |
Molecular weight and concentrations of the precursors used for the generation of the hydrogels and resulting elastic modulus.
| Precursor | Concentration wt/vol | Modulus kPa |
|---|---|---|
| PEGDA MW 10000 (Biochempeg) | 10% | 12.1 ± 0.8 |
| PEGDA MW 6000 (Biochempeg) | 10% | 38.3 ± 1.0 |
| PEGDA MW 3400 (Biochempeg) | 10% | 30.9 ± 2.0 |
| PEGDA MW 700 (Sigma-Aldrich) | 15% | 203.3 ± 13.7 |





