Assigning mitochondrial localization of dual localized proteins using a yeast Bi-Genomic Mitochondrial-Split-GFP
Figures
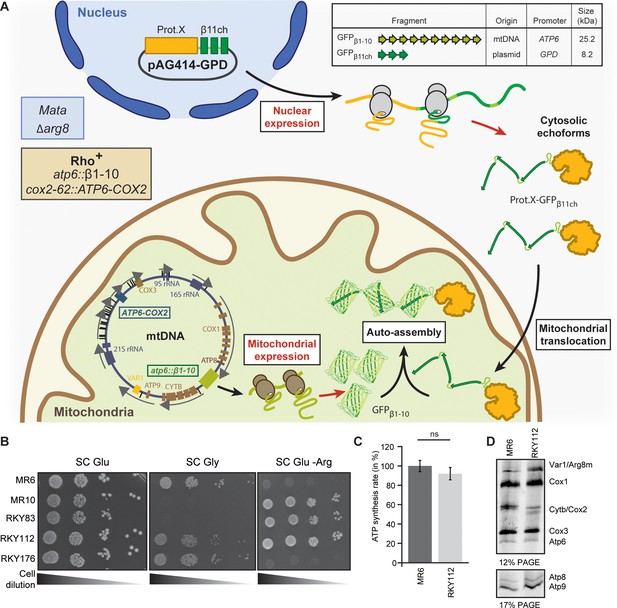
Engineering of the BiG Mito-Split-GFP system in S. cerevisiae.
(A) Principle of the Split-GFP system. When present in the same subcellular compartment, two fragments of GFP namely GFPβ1-10 and GFPβ11ch can auto-assemble to form a fluorescent BiG Mito-Split-GFP chaplet (three reconstituted GFPs). GFPβ1-10 sequence encoding the first ten beta strands of GFP has been integrated into the mitochondrial genome under the control of the ATP6 promoter. GFPβ11ch consists of a tandemly fused form of the eleventh beta strand of GFP and is expressed from a plasmid under the control of a strong GPD promoter (pGPD). The molecular weight of the tag is indicated. (B) Growth assay on permissive SC Glu plates, respiratory plates (SC Gly), and restrictive media lacking arginine (SC Glu -Arg) of the different strains used in the study (N = 2). All generated strains are derivative from MR6. (C) ATP synthesis rates of the MR6 and RKY112 strains presented as the percent of the wild type control strain (N = 2). P-value was 0.7456 (not significant). 95% confidence interval was −273.4 to 229.9, R squared = 0.064 (D) Mitochondrial translation products in the MR6 and RKY112 strains (N = 2). Cells were grown in rich galactose medium. Pulse-chase of radiolabeled [35S]methionine + [35S]cysteine was performed by a 20 min incubation in the presence of cycloheximide. Total cellular extracts were separated by SDS PAGE in two different polyacrylamide gels prepared with a 30:0.8 ratio of acrylamide and bis-acrylamide. Upper gel: 12% polyacrylamide gel containing 4 M urea and 25% glycerol. Lower gel: 17.5% polyacrylamide gel. Gels were dried and exposed to X-ray film. The representative gels are shown.
-
Figure 1—source data 1
Respiratory competency and translation of mtDNA-encoded respiratory subunits of the strains used in this study.
Growth assay on permissive SC Glu plates, respiratory plates (SC Gly), and restrictive media lacking arginine (SC Glu -Arg) of the different strains used in the study (related to Figure 1B). Mitochondrial translation products in the MR6 and RKY112 strains (N = 2) monitored by pulse-chase labeling with radiolabeled [35S]methionine and [35S]cysteine (related to Figure 1D).
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig1-data1-v1.docx
-
Figure 1—source data 2
Statistics of the comparison of ATP synthesis rates between RKY112 and MR6 strains (related to Figure 1C).
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig1-data2-v1.docx

Optimized sequence and secondary structure of the GFPβ1-10 and GFPβ11ch that were used in this study (related to Figure 1).
(A) The amino acid sequence and numbering of the residues of wild type GFPβ1-10 are shown. The β-strands are schematized as blue arrows. The amino acid residues of wild type GFP that were mutated to generate the Folding Reporter GFP are in green. The six amino acids of Folding reporter GFP that were then mutated to build the Superfolder GFP are in red (Pédelacq et al., 2006) and the seven amino acid residues of Superfolder GFP that were mutated to generate GFPβ1-10 OPT are indicated in orange (Cabantous et al., 2005a). (B) The amino acid sequence of the GFPβ11ch is shown and the numbering corresponds to the aa residues of the β11-strand of wild type GFP. The three consecutive β11 strands are schematized as green arrows and the three mutations that were introduced into each β11 strand (GFP11M3) are in purple (Cabantous et al., 2005a). The linker sequences are colored gray.
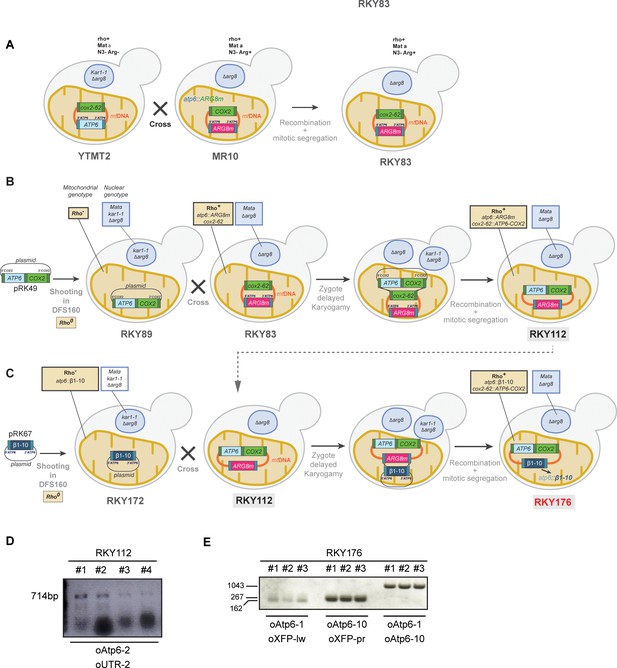
Engineering of the strains and verification of the correct integration of ATP6 under the control of COX2 gene UTRs or GFPβ1-10 under the control of ATP6 gene UTRs (related to Figure 1).
(A) Construction of the RKY83 strain. (B–C) Construction of RKY112 and RKY176, a strain that expresses GFPβ1-10 from the mitochondrial genome. Detailed description can be found in the Materials and methods section. (D) Total DNA prepared from the RKY112 clones 1 to 4 was used as templates for amplification of the 3’part of ATP6 and the 3’UTR region of COX2 (N = 4). (E) Total DNA prepared from or RKY176 clones 1 to 3 was used as templates for amplification of the 3’ and the 5’ ATP6 gene UTRs/GFPβ1-10 regions (N = 3). The oligonucleotides used for each reaction and products lengths are indicated (Supplementary file 1).
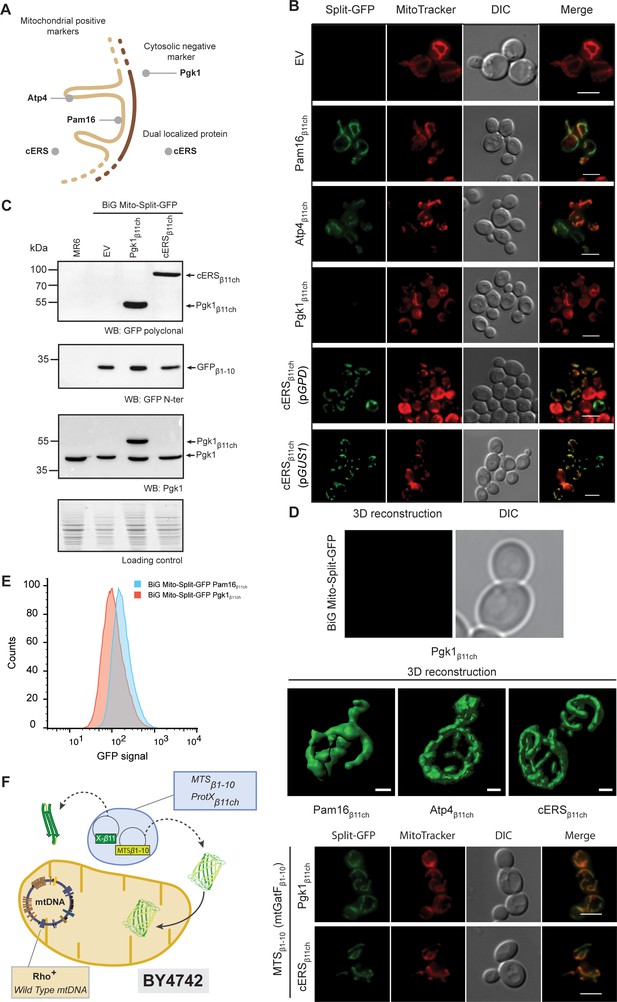
The reconstitution and fluorescence emission of the BiG Mito-Split-GFP is confined to mitochondria and exclusively generated by mitochondrial proteins.
(A) Schematic of the spatial localization of proteins used as positive mitochondrial control proteins (Atp4, Pam16), negative cytosolic control protein (Pgk1) and as dual localized protein (cERS) in S. cerevisiae. (B) Empty pAG414pGPDβ11ch vector (EV) or pAG414pGPDβ11ch vectors expressing each of the four GFPβ11ch-tagged proteins used as markers in our study were transformed into the BiG Mito-Split-GFP strain. cERSβ11ch was either expressed under the dependence of the GPD (pGPD) or its own promoter (pGUS1) from a centromeric plasmid. GFP reconstitution upon mitochondrial import was followed by epifluorescence microscopy (N = 3). (C) Immunodetection of the GFPβ1-10, cERSβ11ch and Pgk1β11ch fusion protein in whole cell extract from the transformed BiG Mito-Split-GFP strain using anti-GFP and -Pgk1 antibodies, confirming expression of Pgk1β11ch. Loading control: stain-free. The representative gels are shown. (D) The strains described in the legend of panel (B) were used for three-dimensional reconstitution of yeast mitochondrial network (N = 1). Z-Stack images from Pam16β11ch, Atp4β11ch, cERSβ11ch and Pgk1β11ch were taken using an Airyscan microscope. Scale bar: 1 µm. (E) Flow cytometry measurements of total GFP fluorescence of the BiG Mito-Split-GFP strain stably expressing Pgk1β11ch or Pam16β11ch (N = 3). (F) The mitochondrial GatF protein was fused to the GFPβ1-10 fragment (mtGatF β1-10), thereby targeting the ten first GFP beta-strands to mitochondria after being transcribed in the nucleus and translated in the cytoplasm. This construct was co-expressed with either cERSβ11ch or Pgk1β11ch. The GFP reconstitution was monitored by epifluorescence microscopy. Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm. Representative fields are shown.
-
Figure 2—source data 1
Micrographs of the BiG Mito-Split-GFP expressing Pgk1β11ch, cERSβ11ch, Pam16β11ch, (related to Figure 2B).
The micrograph of the BiG Mito-Split-GFP expressing Pgk1β11ch which is magnified in Figure 2B is presented here with adjusted or enhanced contrast settings. A new panel of the BiG Mito-Split-GFP expressing Pgk1β11ch was added with enhanced or adjusted contrast settings.
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig2-data1-v1.docx
-
Figure 2—source data 2
Confirmation of the expression of the GFPβ1-10, cERSβ11ch and Pgk1β11ch fusion proteins in whole cell extract from the transformed BiG Mito-Split-GFP strains (Related to Figure 2C).
Antibodies used for immunoblotting are indicated below WBs. Loading control corresponds to the gel stained with the stain-free procedure.
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig2-data2-v1.docx
-
Figure 2—source data 3
Flow cytometry measurements of total GFP fluorescence of the three biological replicates of the BiG Mito-Split-GFP strain stably expressing Pgk1β11ch or Pam16β11ch (related to Figure 2F).
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig2-data3-v1.docx
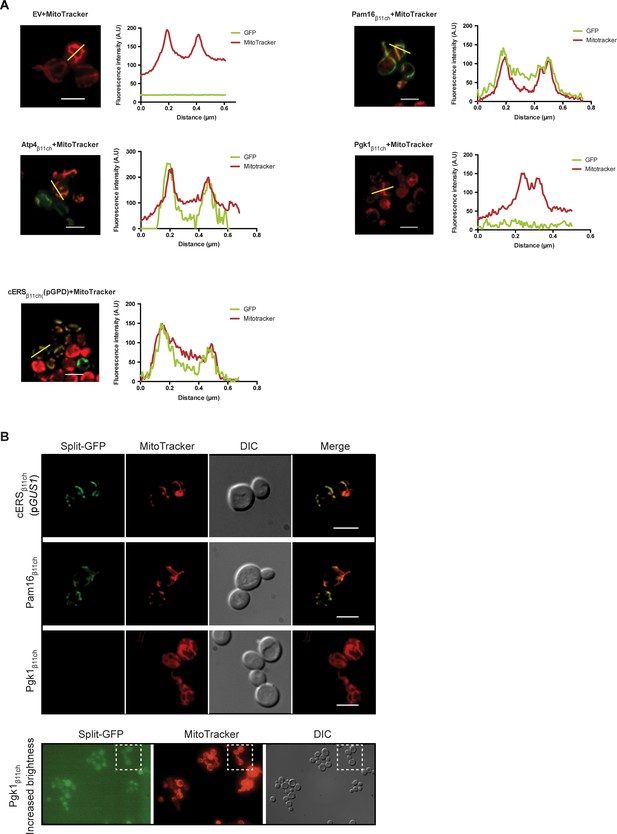
Mitochondrial relocation of mitochondrial proteins or echoforms tagged with GFPβ11 (related to Figure 2).
(A) Colocalization measurement of the reconstituted GFP (β11+ β1-10) with MitoTracker Red CMXRos-stained mitochondria on merged micrographs shown in Figure 2B. Fluorescent signals were measured along the yellow line with the ImageJ software. (B) Fluorescence microscopy analysis of the BiG Mito-Split-GFP strains bearing integrated into the TRP1 locus of GUS1 (cERS), PAM16 or PGK1 genes fused to GFPβ11ch. The cERSβ11ch is expressed from the own promoter (GUS1) while Pam16β11ch and Pgk1β11ch are expressed from GPD promoter. The last panel (Pgk1β11ch Increased brightness) shows the full field from which the Pgk1β11ch micrograph of the upper panel was taken from, with enhanced brightness, thereby illustrating the absence of any faint mitochondrial fluorescence. Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm.
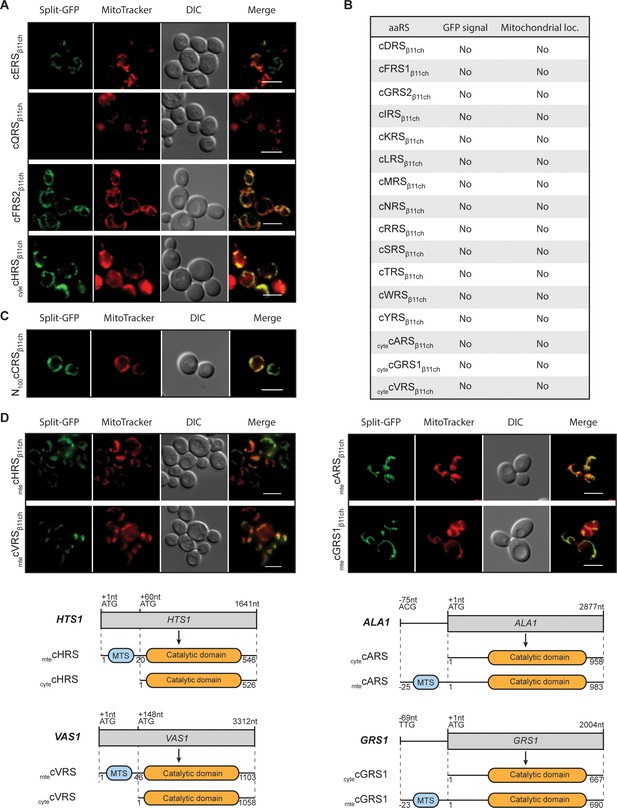
Identification and visualization of mitochondrial echoforms of yeast cytosolic aaRSs using the BiG Mito-Split-GFP strategy.
Fluorescence microscopy analyses of BiG Mito-Split-GFP strain transformed with pAG414pGPDβ11ch expressing yeast caaRSs (also see Table S3). Genes encoding 18 out of the 20 yeast caaRS, including those encoding the α- and β-subunits of the cytosolic α2β2 FRS (cFRS2), and the cGRS2 pseudogene, as well as the four encoding the cytosolic echoforms of cGRS1 (cytecGRS1), cARS (cytecARS), cHRS (cytecHRS) and cVRS (cytecVRS) were cloned in the pAG414pGPDβ11ch and expressed in the BiG Mito-Split-GFP strain (N = 2). (A) From the set of caaRSs tested, only cERS, cQRS, cFRS2 and cytecHRS micrographs are shown. (B) Table summarizing the GFP emission and mitochondrial localization of the caaRSs not shown in A). The corresponding micrographs are shown in Fig. S4A. (C) Fluorescence microscopy analysis of the BiG Mito-Split-GFP strain expressing the first 100 amino acids of the N-ter region of the cCRS fused to GFPβ11ch (N = 2). (D) Fluorescence microscopy analyses of BiG Mito-Split-GFP strain transformed with pAG414pGPDβ11ch expressing the mitochondrial echoforms mtecGRS1, mtecARS, mtecHRS and mtecVRS. Schematics of cARS, cGRS1, cHRS and cVRS echoforms expression in yeast. Expression can be initiated upstream of the initiator ATG+1 (mtecARS at ACG-75 and mtecGRS1 at TTG-69) but the synthesis of this echoform can also be initiated at the ATG+1. In this case, the expression of the cytosolic echoform is initiated downstream (cytecHTS at ATG+60 and cytecVRS at ATG+148). Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm. Representative fields are shown.
-
Figure 3—source data 1
Confirmation, by WB, of the expression of the 18 full-length aaRSβ11ch and N100cCRSβ11ch in whole cell extracts from the transformed BiG Mito-Split-GFP strains (Related to Figure 3).
Antibodies used for immunoblotting are indicated below WBs. Loading controls correspond to gels stained with the stain-free procedure.
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig3-data1-v1.docx
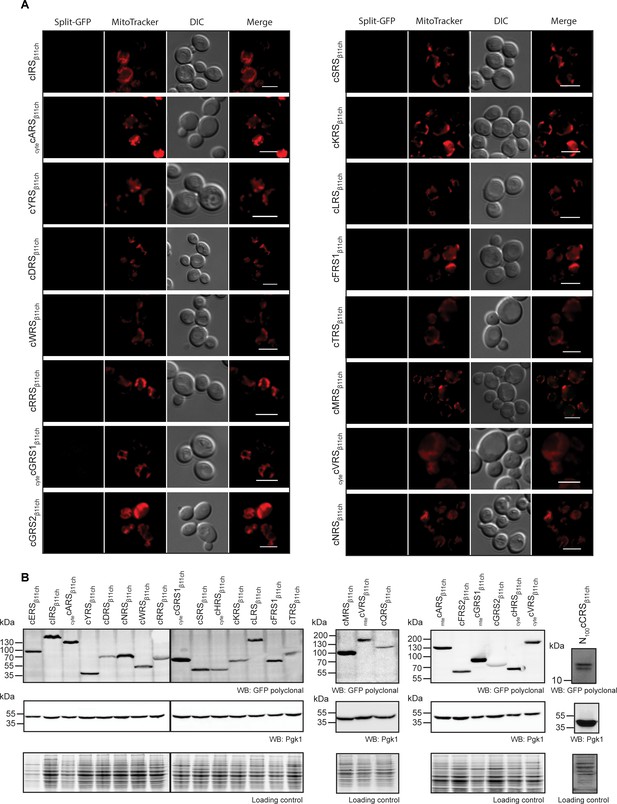
Screening of caaRSs and expression level of each GFPβ11ch-tagged proteins (related to Figure 3).
(A) Micrographs of all the other caaRSs tested in Figure 3A. Representative panels from two independent experiments are shown. Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm. Representative fields are shown. (B) Immunodetection of all the GFPβ11ch-tagged aaRSs expressed in the BiG Mito-Split-GFP strain. aaRSβ11ch were detected by anti-GFP antibodies. Equal loading was verified by anti-Pgk1 antibodies and by stain-free technology (Loading control). caaRS: cytosolic aaRS, cytecaaRS: cytosolic echoform of the caaRS, mtecaaRS: mitochondrial echoform of the caaRS.
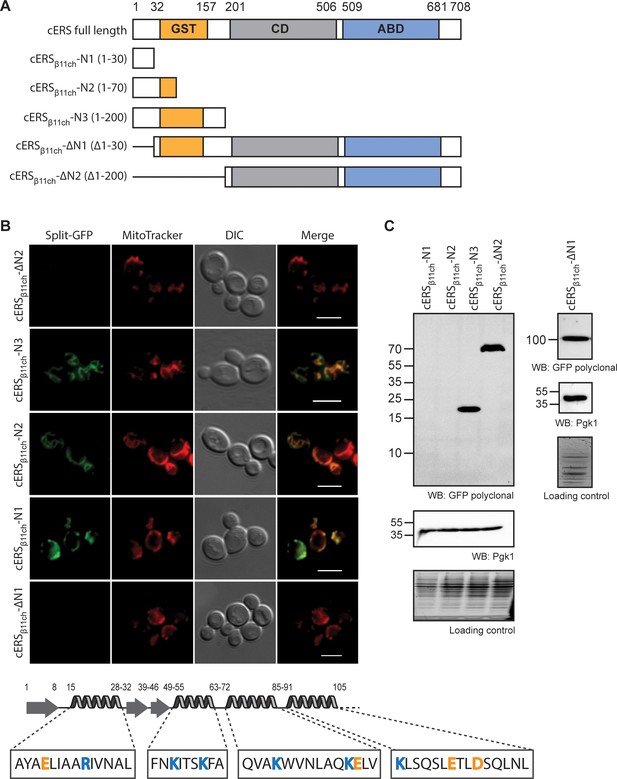
The BiG Mito-Split-GFP is a suitable tool to delimit regions containing non-canonical MTSs.
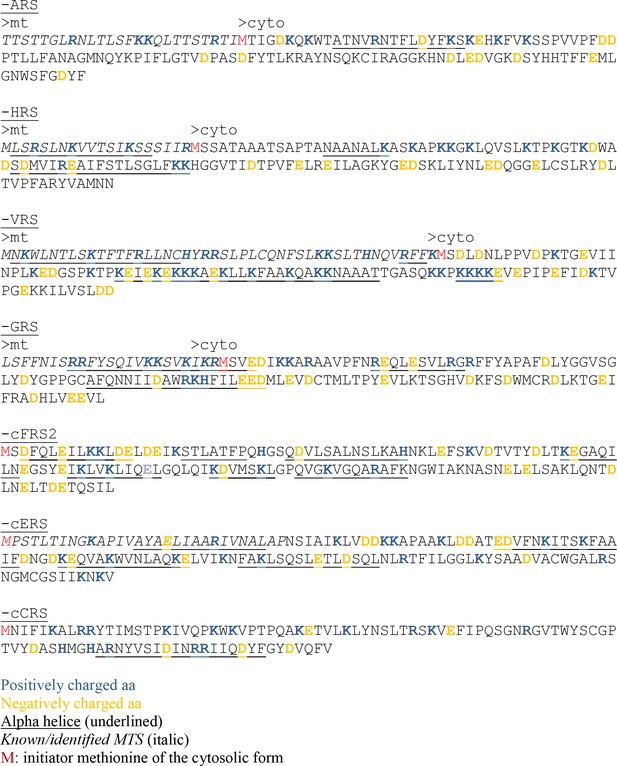
(A) Schematic representation of the cERS fragments fused to GFPβ11ch. Orange boxes correspond to the GST-like domain necessary for Arc1 interaction (GST), the grey boxes represent the catalytic domain (CD), and the blue box, the tRNA-binding domain generally named anti-codon binding domain (ABD). Numbering above corresponds to cERS amino acids residues. (B) Fluorescence microscopy analyses of the BiG Mito-Split-GFP strain expressing the cERS variants shown on A. Mitochondria were stained with MitoTracker Red CMXRos; scale bar: 5 µm. The secondary structure (according to Simader et al., 2006) of the smallest peptide that still contains the non-conventional MTS of cERS is described together with the amino acid sequence of each helices. Positively and negatively charged amino acids are shown in orange and blue respectively. (C) Immunodetection of the cERS variants in BiG Mito-Split-GFP whole cell extracts using anti-GFP antibodies. Quantity of proteins loaded in each lane was estimated using anti-Pgk1 antibodies or by the stain-free procedure. The bands corresponding to the mutants N1 and N2 could not be detected. The representative fields or gel are shown.
-
Figure 4—source data 1
Immunodetection of the cERS variants in BiG Mito-Split-GFP whole cell extracts using anti-GFP antibodies (related to Figure 4C).
Antibodies used for immunoblotting are indicated below WBs. Loading controls correspond to gels stained with the stain-free procedure.
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig4-data1-v1.docx
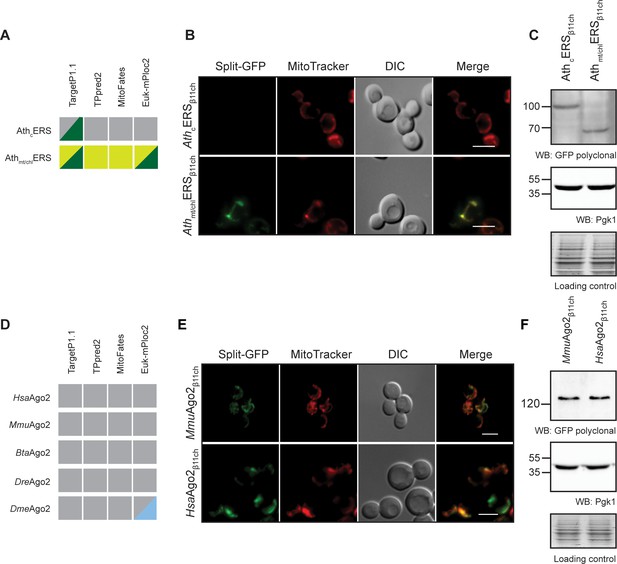
The BiG Mito-Split-GFP can be used to study mitochondrial importability of mammalian and plant proteins.
(A, D) Prediction of MTS and mitochondrial localization of (A) two ERS from Arabidopsis thaliana (AthcERS and Athmt/chlERS) and (D) five eukaryotic Ago2 proteins [HsaAgo2 (Protein argonaute-2 isoform X2 [Homo sapiens] NCBI sequence ID: XP_011515267.1), MmuAgo2 (protein argonaute-2 Mus musculus NCBI sequence ID: NP_694818.3.), BtaAgo2 (Bos Taurus), DreAgo2 (Danio rerio), DmeAgo2 (Drosophila melanogaster). MTS were predicted using TPpred2.0 (http://tppred2.biocomp.unibo.it/tppred2), TargetP1.1 (http://cbs.dtu.dk/services/TargetP/), MitoFates (http://mitf.cbrc.jp/MitoFates/cgibin/top.cgi) and the EukmPloc2 website (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/). Grey boxes indicate prediction of a cytosolic localization, light and dark green indicate prediction of mitochondrial or chloroplastic localization respectively. Blue boxes indicate prediction of nuclear localization. (B, E) Fluorescence microscopy analyses of the BiG Mito-Split-GFP strain expressing the GFPβ11ch-tagged AthcERS and Athmt/chlERS (N = 2) (B) andMmuAgo2, HsaAgo2 (N = 2) (E). Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm. Representative fields are shown. (C, E) Protein expression was checked by WB with anti-GFP antibodies and equal amount of loaded protein was controlled using anti-Pgk1 antibodies and by the stain-free technology (Loading control: stain-free). The representative gels are shown.
-
Figure 5—source data 1
Confirmation, by WB, of the expression of AthERSβ11ch and mouse and human Ago2β11ch in whole cell extract from the transformed BiG Mito-Split-GFP strains (Related to Figure 5C and F).
Antibodies used for immunoblotting are indicated below WBs. Loading controls correspond to gels stained with the stain-free procedure.
- https://cdn.elifesciences.org/articles/56649/elife-56649-fig5-data1-v1.docx
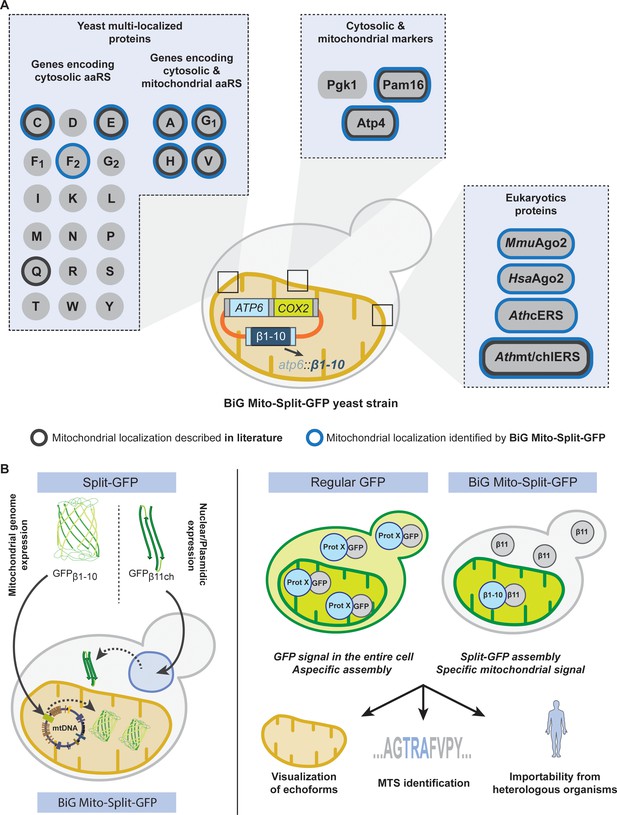
Schematic of the BiG Mito-Split-GFP system and its applications.
(A) Using our engineered strain, we could show the dual localization of echoforms in the aaRS family of proteins and foster its power by studying localization of heterologous proteins originating from plants, mice and human. (B) The BiG Mito-Split-GFP strain was generated by integrating the sequence encoding the first 10 beta barrel segments into yeast mitochondrial DNA, and by either expressing any protein of interest fused to the 11th GFP segment from a plasmid or by integration in yeast nuclear DNA. As opposed to regular GFP-tagging where visualizing an echoform ultimately results in a GFP signal diffusing in the entire cell, our BiG Mito-Split-GFP system abolishes the fluorescence originating from cytosolic echoform to only display a specific mitochondrial signal. Further applications range from high-throughput experiments to identify relocating proteins involved in mitochondria homeostasis or metabolism, to identify non-conventional MTSs or seek for mitochondrial localization of heterologous proteins.
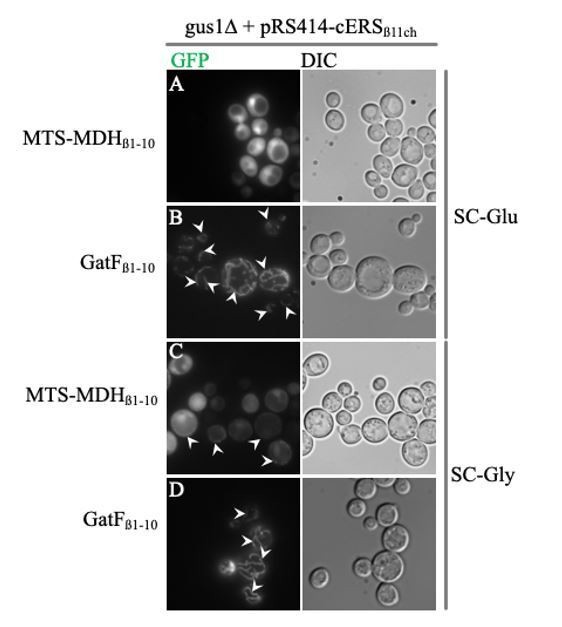
Efficiency of mitochondria-specific GFP labeling induced by MTS-MDHβ1-10 compared to that of GatFβ1-10.
Saccharomyces cerevisiae gus1∆ strain complemented with the pRS414 plasmid expressing cERSβ11ch was transformed with a pRSX expressing either MTS-MDHβ1-10 (A and C) or GatFβ1-10 (B and D) and grown either on SC-Glu (A and B) or SC-Gly (C and D). Epifluorescence micrographs were taken with an AXIO Observer d1 (Carl Zeiss) epifluorescence microscope using a 100 × plan apochromatic objective (Carl Zeiss) and processed with the Image J software. Arrowheads: mitochondria.
Tables
Genotypes of yeast strains used or generated for this study.
| Strain | Nuclear genotype | mtDNA | Source |
|---|---|---|---|
| MR6 | MATa ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+ | Rak et al., 2007 |
| DFS160 | MATα leu2∆ ura3-52 ade2-101 arg8::URA3 kar1-1 | ρo | Steele et al., 1996 |
| NB40-3C | MATa lys2 leu2-3,112 ura3-52 his3∆HindIII arg8::hisG | ρ+ cox2-62 | Steele et al., 1996 |
| MR10 | MATa ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 CAN1 arg8::hisG | ρ+ atp6::ARG8m | Rak et al., 2007 |
| SDC30 | MATα leu2∆ ura3-52 ade2-101 arg8::URA3 kar1-1 | ρ-COX2 ATP6 | Rak et al., 2007 |
| YTMT2 | MATα leu2∆ ura3-52 ade2-101 arg8::URA3 kar1-1 | ρ+cox2-62 | This study |
| RKY83 | MATa ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 arg8::HIS3 | ρ+cox2-62 atp6::ARG8m | This study |
| RKY89 | MATα leu2∆ ura3-52 ade2-101 arg8::URA3 kar1-1 | ρ-S5`UTRCOX2 ATP6 3`UTRCOX2 COX2 | This study |
| RKY112 | MATa ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 arg8::HIS3 | ρ+ atp6::ARG8m 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| RKY172 | MATα leu2∆ ura3-52 ade2-101 arg8::URA3 kar1-1 | ρ-S atp6::GFPβ1-10 COX2 | This study |
| RKY176 | MATa ade2-1 his3-11,15 trp1-1 leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+atp6::GFPβ1-10 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| BiG Mito- Split-GFP | MATa his3-11,15 trp1-1 leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+atp6::GFPβ1-10 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| BiG Mito- Split- GFP+PAM16β11ch | MATa his3-11,15 trp1-1::PAM16β11ch leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+atp6::GFPβ1-10 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| BiG Mito- Split- GFP+PGK1β11ch | MATa his3-11,15 trp1-1::PGK1β11ch leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+atp6::GFPβ1-10 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| BiG Mito- Split- GFP+GUS1β11ch | MATa his3-11,15 trp1-1:: GUS1β11ch leu2-3,112 ura3-1 CAN1 arg8::HIS3 | ρ+atp6::GFPβ1-10 5`UTRCOX2ATP6 3`UTRCOX2 | This study |
| BY 4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | ρ+ | Winston et al., 1995 |
List of genes encoding S. cerevisiae cytosolic and mitochondrial aminoacyl-tRNA synthetases and their cytosolic or mitochondrial echoforms
| Gene coding for | ||||
|---|---|---|---|---|
| aaRSs forms | aaRS echoforms | |||
| aaRS | cytosolic (c) | mitochondrial (mt) | cytosolic (cyte) | mitochondrial (mte) |
| IRS | ILS1 | ISM1 | - | - |
| GRS | GRS1/GRS2 | - | GRS1 | GRS1 −23 |
| SRS | SES1 | DIA4 | - | - |
| KRS | KRS1 | MSK1 | - | - |
| RRS | RRS1 | MSR1 | - | - |
| ERS | GUS1 | MSE1 | GUS1 | GUS1 |
| VRS | VAS1 | - | VAS1∆46 | VAS1 |
| YRS | TYS1 | MSY1 | - | - |
| MRS | MES1 | MSM1 | - | - |
| NRS | DED81 | SLM5 | - | - |
| PRS | YHR020W | AIM10 | - | - |
| TRS | THS1 | MST1 | - | - |
| DRS | DPS1 | MSD1 | - | - |
| FRS | FRS1 (β)/FRS2 (a) | MSF1 (a) | - | - |
| CRS | CRS1 | - | - | - |
| WRS | WRS1 | MSW1 | - | - |
| QRS | GLN4 | - | - | - |
| ARS | ALA1 | - | ALA1 | ALA1 −25 |
| LRS | CDC60 | NAM2 | - | - |
| HRS | HTS1 | - | HTS1∆20 | HTS1 |
-
The Saccharomyces Genome Database standard gene names are used. The amino acid (aa) one-letter code is used for the aminoacyl-tRNA synthetase aa specificity and (-) means that the gene encoding the corresponding aaRS is missing. Two genes encode the cytosolic phenylalanyl-tRNA synthetase (cFRS) since the enzyme is an α2β2 hetero-tetramer. For echoforms, the position of the alternative initiation start codon is indicated and corresponds to the nomenclature described in Figure 3; briefly, (- number) means that the start codon of the mteaaRS is located (number) aa upstream the one that starts translation of the corresponding cyteaaRS while (∆number) means that the start codon of the cyteaaRS is located (number) aa downstream the one that starts translation of the corresponding mteaaRS.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (S. cerevisiae) | BiG Mito- Split-GFP | This study | RKY176 strain with ADE2 gene (ρ+atp6::GFPβ1-105`UTRCOX2 ATP6 3`UTRCOX2) | |
| Genetic reagent (S. cerevisiae) | BiG Mito- Split-GFP+Pgk1β11ch | This study | RKY176 strain (PGK1:: β11ch::TRP1) | |
| Genetic reagent (S. cerevisiae) | BiG Mito- Split-GFP+PAM16β11ch | This study | RKY176 strain (PAM16:: β11ch::TRP1) | |
| Genetic reagent (S. cerevisiae) | BiG Mito- Split-GFP+cERSβ11ch | This study | RKY176 strain (GUS1:: β11ch::TRP1) | |
| Antibody | Anti-GFP (Mouse polyclonal) | Sigma | Cat# G1544 | WB (1:5000) Called GFP N-ter in Figure 2C recognizes GFPβ1–10 |
| Antibody | Anti-GFP (Mouse monoclonal IgG1κ clones 7.1 and 13.1) | Roche | Cat# 11814460001 | WB (1:5000) Called GFP polyclonal in Figure 2C recognizes GFPβ11 |
| Antibody | Anti-Pgk1 (Mouse monoclonal IgG1, clone 22C5D8) | Molecular Probes | Cat# 459250 | WB (1:5000) |
| Recombinant DNA reagent | pAG414-pGPD- β11ch (plasmid) | This study | Template vector used for all constructs. Cloning done by Gibson assembly | |
| Chemical compound, drug | MitoTracker Red CMXRos | ThermoFisher | Cat# M7512 | Mitochondria staining |
| Chemical compound, drug | 0.5% (v/v) 2,2,2-Trichloroethanol | Sigma | Cat# T54801 | Used to detect total protein loading in SDS-PAGE, referred to Loading control |
Additional files
-
Supplementary file 1
Sequence of the BamHI-EcoRI DNA fragment of GFPβ1-10 flanked by the regulatory sequences of ATP6 gene Regulatory sequences of ATP6 are underlined, 5’-BamHI and 3’-EcoRI sites are in italicized bold characters.
The GFPβ1-10 sequence is in gray background and has been codon-optimized to be expressed by S. cerevisiae mitochondrial translation machinery.
- https://cdn.elifesciences.org/articles/56649/elife-56649-supp1-v1.docx
-
Supplementary file 2
Primers used in the study to verify integration of ectopic ATP6 or GFPβ1-10 in mtDNA.
The use of each oligo is described in the Materials and methods section.
- https://cdn.elifesciences.org/articles/56649/elife-56649-supp2-v1.docx
-
Supplementary file 3
Primers used for PCR amplifications of genes fused to GFPβ11ch sequence.
The primers in black and blue were used for Gateway and Gibson cloning methods respectively (see Material and methods section).
- https://cdn.elifesciences.org/articles/56649/elife-56649-supp3-v1.docx
-
Supplementary file 4
List of expression plasmids generated for this study.
- https://cdn.elifesciences.org/articles/56649/elife-56649-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56649/elife-56649-transrepform-v1.docx