Replication Study: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs
Figures
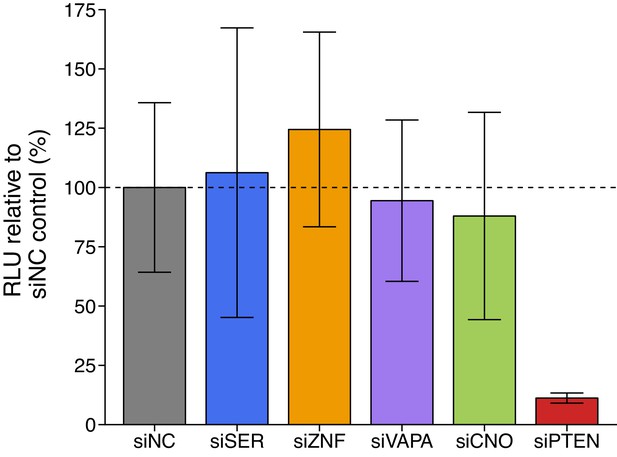
Luciferase activity in DU145 cells co-transfected with siRNA against PTEN ceRNAs and a luciferase-PTEN 3’UTR reporter construct.
DU145 cells were transfected with a luciferase reporter with a fragment of the 3’UTR of PTEN. Cells were also co-transfected with non-targeting control siRNA (siNC) or siRNA plasmids targeting SERINC1 (siSER), ZNF460 (siZNF), VAPA (siVAPA), CNOT6L (siCNO), or PTEN (siPTEN). Cells were harvested 72 hr later for luciferase activity. Relative luminescence unit (RLU) is presented for each condition relative to the siNC condition. Means reported and error bars represent SD from four independent biological repeats. Two-sample t-test of RLU values between siNC and siSER: t(6) = 0.177, uncorrected p=0.866 with a priori Bonferroni adjusted significance threshold of 0.01, Bonferroni corrected p>0.99; siNC and siZNF: t(6) = 0.899, uncorrected p=0.403, Bonferroni corrected p>0.99; siNC and siVAPA: t(6) = 0.225, uncorrected p=0.829, Bonferroni corrected p>0.99; siNC and siCNO: t(6) = 0.426, uncorrected p=0.685, Bonferroni corrected p>0.99; Wilcoxon-Mann-Whitney test of RLU values between siNC and siPTEN: U = 16, uncorrected p=0.029, Bonferroni corrected p=0.143. Additional details for this experiment can be found at https://osf.io/spv4f/.
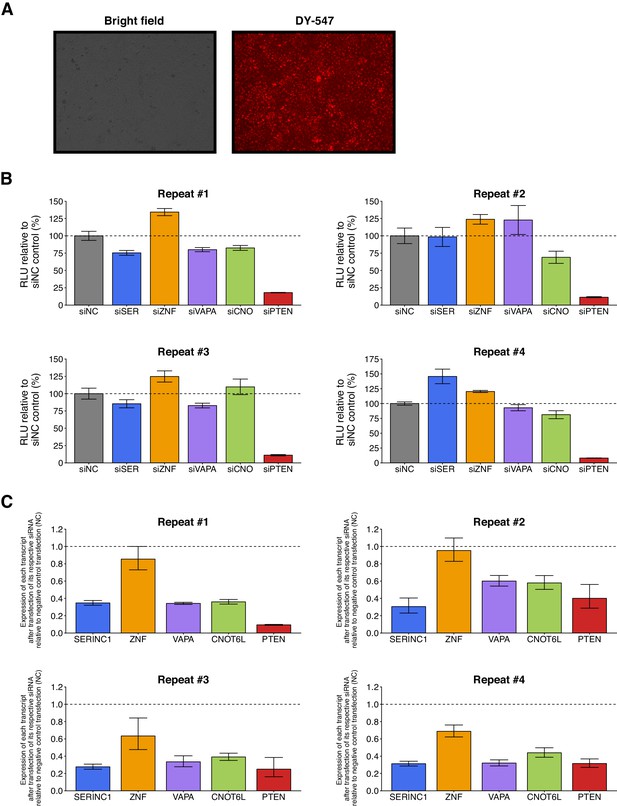
Knockdown efficiency and individual repeats of luciferase-PTEN 3’UTR reporter assay in DU145 cells co-transfected with siRNA against PTEN ceRNAs.
This is the same experiment as Figure 1. (A) Representative microscopy images (10X magnification) of DY-547-labeled siGLO RISC-Free control transfected DU145 cells 48 hr after transfection. Transfection efficiency was estimated to be >90%. (B) Independent biological repeats of luciferase reporter assay. Relative luminescence unit (RLU) is presented for each condition relative to the siNC condition. Means reported and error bars represent SD from three technical replicates. (C) Independent biological repeats of RT-qPCR analysis. Expression of each transcript after transfection of its respective siRNA relative to negative control transfection (siNC) is presented. Transcripts listed on y-axis. Means reported and error bars represent SD from three technical replicates. One-sample t-tests of transcript expression data after transfection of respective siRNA to a constant of 1 (relative value of siNC). SERINC1: t(3) = 45.8, uncorrected p=2.30×10−5, Bonferroni corrected p=1.15×10−4; ZNF460: t(3) = 2.98, uncorrected p=0.059, Bonferroni corrected p=0.294; VAPA: t(3) = 8.94, uncorrected p=0.0030, Bonferroni corrected p=0.015; CNOT6L: t(3) = 11.3, uncorrected p=0.0015, Bonferroni corrected p=0.0074; PTEN: t(3) = 11.2, uncorrected p=0.0015, Bonferroni corrected p=0.0077. Additional details for this experiment can be found at https://osf.io/spv4f/.
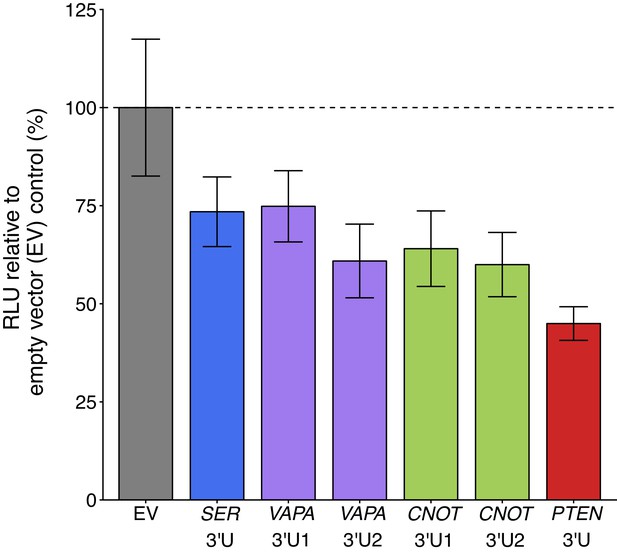
Luciferase activity in DU145 cells co-transfected with 3’UTR of PTEN ceRNAs and a luciferase-PTEN 3’UTR reporter construct.
DU145 cells were transfected with a luciferase reporter with a fragment of the 3’UTR of PTEN. Cells were also co-transfected with empty vector (EV) or plasmids that express the 3’UTR of SERINC1 (SER 3’U), VAPA (VAPA 3’U1 and VAPA 3’U2), CNOT6L (CNOT 3’U1 and CNOT 3’U2), or PTEN (PTEN 3’U). Cells were harvested 72 hr later for luciferase activity. Relative luminescence unit (RLU) is presented for each condition relative to the EV condition. Means reported and error bars represent SD from six independent biological repeats. Two-sample t-test of RLU values between SER 3’U and EV: t(10) = 3.32, uncorrected p=0.0077 with a priori Bonferroni adjusted significance threshold of 0.0083, Bonferroni corrected p=0.046; VAPA 3’U1 and EV: t(10) = 3.13, uncorrected p=0.011, Bonferroni corrected p=0.064; VAPA 3’U2 and EV: t(10) = 4.83, uncorrected p=6.90×10−4, Bonferroni corrected p=0.0041; CNOT 3’U1 and EV: t(10) = 4.42, uncorrected p=0.0013, Bonferroni corrected p=0.0078; CNOT 3’U2 and EV: t(7.1) = 5.09, uncorrected p=0.0014, Bonferroni corrected p=0.0082; PTEN 3’U and EV: t(5.6) = 7.50, uncorrected p=3.99×10−4, Bonferroni corrected p=0.0024. Additional details for this experiment can be found at https://osf.io/mryvq/.
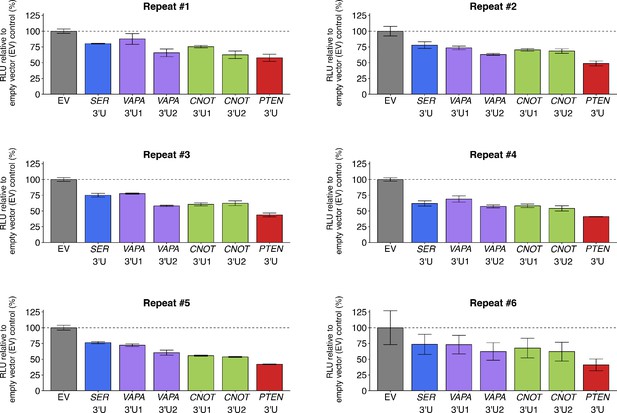
Individual repeats of luciferase-PTEN 3’UTR reporter assay in DU145 cells co-transfected with 3’UTR of PTEN ceRNAs.
This is the same experiment as Figure 2. Independent biological repeats of luciferase reporter assay. Relative luminescence unit (RLU) is presented for each condition relative to the EV condition. Means reported and error bars represent SD from three technical replicates. Additional details for this experiment can be found at https://osf.io/mryvq/.
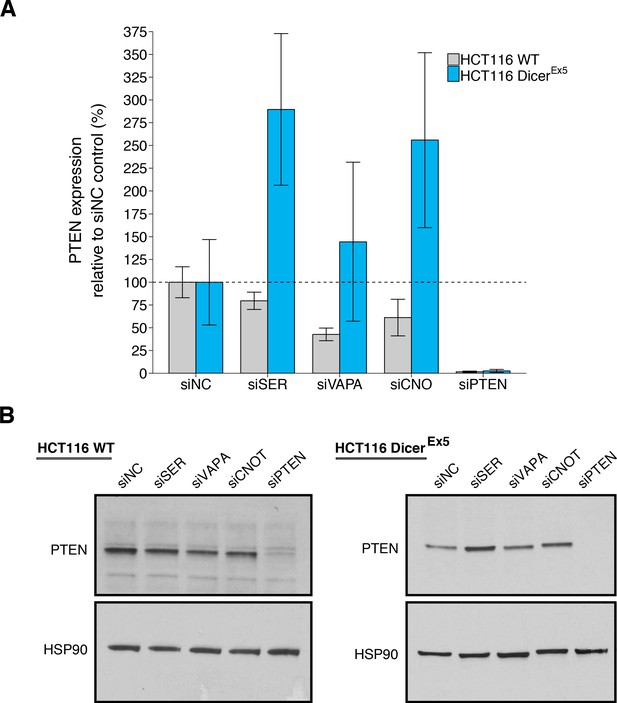
PTEN protein expression in wild-type and DICER mutant HCT116 cells depleted of PTEN ceRNAs.
Wild-type (WT) and DICER mutant (DicerEx5) HCT116 cells were transfected with non-targeting control siRNA (siNC) or siRNA plasmids targeting SERINC1 (siSER), VAPA (siVAPA), CNOT6L (siCNO), or PTEN (siPTEN). Cells were harvested 72 hr later for Western blot analysis. (A) Relative protein expression (PTEN/HSP90) are presented for each condition. Western blot bands were quantified, PTEN levels were normalized to HSP90, with protein expression presented relative to siNC. Means reported and error bars represent SD from three independent biological repeats for wild-type HCT116 cells and four repeats for DicerEx5 HCT116 cells. Analysis of wild-type HCT116 cells: one-way ANOVA (equal variance) on PTEN/HSP90 expression: F(4,10) = 25.4, I=3.18×10−5. Planned contrasts between siNC and siSER: t(10) = 1.94, uncorrected I=0.082 with a priori Bonferroni adjusted significance threshold of 0.0125, Bonferroni corrected p=0.326; siNC and siVAPA: t(10) = 5.44, uncorrected p=2.85×10−4, Bonferroni corrected p=0.0011; siNC and siCNOT: t(10) = 3.69, uncorrected p=0.0042, Bonferroni corrected p=0.017; siNC and siPTEN: t(10) = 9.34, uncorrected p=2.97×10−6, Bonferroni corrected p=1.19×10−5. Analysis of DicerEx5 HCT116 cells: one-way ANOVA (unequal variance) on PTEN/HSP90 expression: F(4,6.0) = 19.3, p=0.0014. Planned comparisons: siNC and siSER: two-sample t-test, t(6) = 3.96, uncorrected p=0.0074 with a priori Bonferroni adjusted significance threshold of 0.0125, Bonferroni corrected p=0.030; siNC and siVAPA: two-sample t-test, t(6) = 0.896, uncorrected p=0.405, Bonferroni corrected p>0.99; siNC and siCNOT: Welch’s t-test, t(4.36) = 2.92, uncorrected p=0.039, Bonferroni corrected p=0.156; siNC and siPTEN: two-sample t-test, t(6) = 4.15, uncorrected p=0.0060, Bonferroni corrected p=0.024. (B) Representative Western blots probed with an anti-PTEN antibody and anti-HSP90 antibody. Additional details for this experiment can be found at https://osf.io/drcbw/.
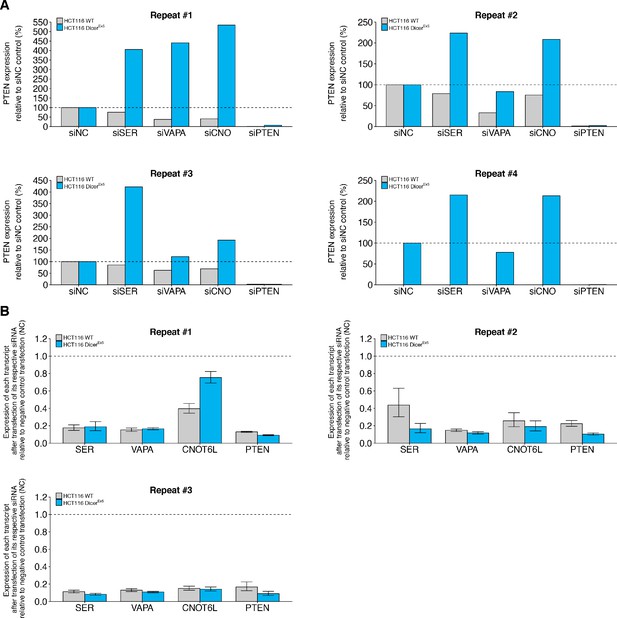
Knockdown efficiency and individual repeats of PTEN protein expression in wild-type and DICER mutant HCT116 cells transfected with siRNA against PTEN ceRNAs.
This is the same experiment as Figure 1. (A) Independent biological repeats of Western blot assay. PTEN/HSP90 protein expression is presented for each condition relative to the siNC condition. (B) Independent biological repeats of RT-qPCR analysis. Expression of each transcript after transfection of its respective siRNA relative to negative control transfection (siNC) is presented. Transcripts listed on y-axis. Means reported and error bars represent SD from three technical replicates. One-sample t-tests of transcript expression data after transfection of respective siRNA to a constant of 1 (relative value of siNC). Wild-type HCT116 cells: SERINC1: t(2) = 8.41, uncorrected p=0.014, Bonferroni corrected p=0.055; VAPA: t(2) = 120, uncorrected p=6.98×10−5, Bonferroni corrected p=2.79×10−4; CNOT6L: t(2) = 10.4, uncorrected p=0.0091, Bonferroni corrected p=0.036; PTEN: t(2) = 30.6, uncorrected p=0.0011, Bonferroni corrected p=0.0043. DicerEx5 HCT116 cells: SERINC1: t(2) = 28.5, uncorrected p=0.0012, Bonferroni corrected p=0.0049; VAPA: t(2) = 48.3, uncorrected p=4.28×10−4, Bonferroni corrected p=0.0017; CNOT6L: t(2) = 3.23, uncorrected p=0.084, Bonferroni corrected p=0.336; PTEN: t(2) = 242, uncorrected p=1.71×10−5, Bonferroni corrected p=6.83×10−5. Additional details for this experiment can be found at https://osf.io/drcbw/.
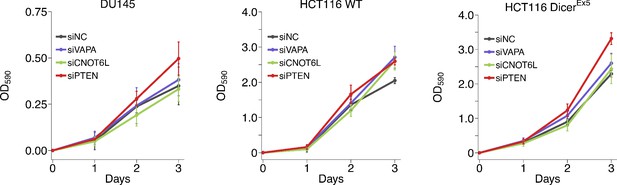
Growth of cells depleted of PTEN ceRNAs.
DU145, wild-type (WT) and DICER mutant (DicerEx5) HCT116 cells were transfected with either a non-targeting control siRNA (siNC) or siRNA plasmids targeting VAPA (siVAPA), CNOT6L (siCNO), or PTEN (siPTEN). Crystal violet proliferation assays were performed each day as indicated starting the day after transfection. Relative OD590 was calculated relative to the average Day 0 values for each condition. Means reported and error bars represent SD from five independent biological repeats for DU145 cells and four times for HCT116 WT and DicerEx5 cells. Analysis on the area under the curve (AUC) for each condition of each biological repeat (reported as dot plot in Figure 4—figure supplement 1A). Analysis results for DU145 cells: one-way ANOVA (equal variance): F(3,16) = 3.27, p=0.049. Planned contrasts between siNC and siVAPA: t(16) = 0.648, uncorrected p=0.526 with a priori Bonferroni adjusted significance threshold of 0.0167, Bonferroni corrected p>0.99; siNC and siCNOT6L: t(16) = 0.950, uncorrected p=0.356, Bonferroni corrected p>0.99; siNC and siPTEN: t(16) = 2.09, uncorrected p=0.053, Bonferroni corrected p=0.158. Analysis of HCT116 cells: two-way ANOVA interaction between DICER status (wild-type or Ex5) and siRNA target: F(3,24) = 0.734, p=0.542; main effect of DICER status: F(1,24) = 1.81, p=0.191; main effect of siRNA target: F(3,24) = 12.1, p=5.20×10−5. Planned contrasts in HCT116 WT cells: siNC and siVAPA: t(24) = 2.02, uncorrected p=0.054 with a priori Bonferroni adjusted significance threshold of 0.0083, Bonferroni corrected p=0.325; siNC and siCNOT6L: t(24) = 0.506, uncorrected p=0.618, Bonferroni corrected p>0.99; siNC and siPTEN: t(24) = 3.03, uncorrected p=0.0057, Bonferroni corrected p=0.034. Planned contrasts in HCT116 DICEREx5 cells: siPTEN and siVAPA: t(24) = 2.43, uncorrected p=0.023, Bonferroni corrected p=0.138; siPTEN and siCNOT6L: t(24) = 4.57, uncorrected p=1.25×10−4, Bonferroni corrected p=7.48×10−4; siNC and siPTEN: t(24) = 4.31, uncorrected p=2.42×10−4, Bonferroni corrected p=0.0015. Additional details for this experiment can be found at https://osf.io/5c7sb/.
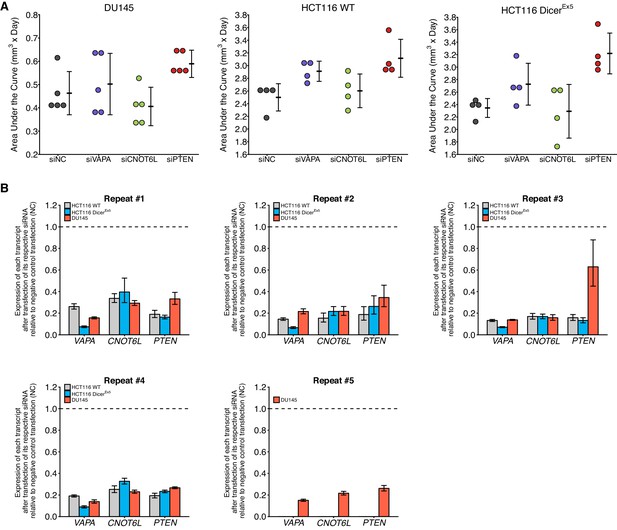
Knockdown efficiency and individual repeats of cell growth assay in cells transfected with siRNA against PTEN ceRNAs.
This is the same experiment as Figure 5. (A) Independent biological repeats of cell growth assay. Dot plot with means reported as crossbars and error bars represent SD. (B) Independent biological repeats of RT-qPCR analysis. Expression of each transcript after transfection of its respective siRNA relative to negative control transfection (siNC) is presented. Transcripts listed on y-axis. Means reported and error bars represent SD from three technical replicates. One-sample t-tests of transcript expression data after transfection of respective siRNA to a constant of 1 (relative value of siNC). DU145 cells: VAPA: t(4) = 58.6, uncorrected p=5.09×10−7, Bonferroni corrected p=1.53×10−6; CNOT6L: t(4) = 36.3, uncorrected p=3.42×10−6, Bonferroni corrected p=1.03×10−5; PTEN: t(4) = 9.05, uncorrected p=8.27×10−4, Bonferroni corrected p=0.0025. Wild-type HCT116 cells: VAPA: t(3) = 18.6, uncorrected p=3.40×10−4, Bonferroni corrected p=0.0010; CNOT6L: t(3) = 108, uncorrected p=1.73×10−6, Bonferroni corrected p=5.19×10−6; PTEN: t(3) = 28.1, uncorrected p=9.90×10−5, Bonferroni corrected p=2.97×10−4. DicerEx5 HCT116 cells: VAPA: t(3) = 14.8, uncorrected p=6.73×10−4, Bonferroni corrected p=0.0020; CNOT6L: t(3) = 27.9, uncorrected p=1.02×10−4, Bonferroni corrected p=3.05×10−4; PTEN: t(3) = 178, uncorrected p=3.89×10−7, Bonferroni corrected p=1.17×10−6. Additional details for this experiment can be found at https://osf.io/5c7sb/.

Meta-analyses of each effect.
Effect size and 95% confidence interval are presented for Tay et al., 2011, this replication study (RP:CB), and a random effects meta-analysis of those two effects. For each effect, Cohen’s d or Glass’ delta, which are standardized differences between the two indicated measurements, is reported. Sample sizes used in Tay et al., 2011 and RP:CB are reported under the study name. (A) These effects are related to the change in luciferase activity between the conditions reported in Figure 1 of this study and Figure 3C of Tay et al., 2011. Meta-analysis p values: siNC and siSER (p=0.374); siNC and siZNF (p=0.233); siNC and siVAPA (p=0.316); siNC and siCNO (p=0.253); siNC and siPTEN (p=0.079). (B) These effects are related to the change in luciferase activity between the conditions reported in Figure 2 of this study and Figure 3D of Tay et al., 2011. Meta-analysis p values: SER 3’U and EV (p=0.881); VAPA 3’U1 and EV (p=0.624); VAPA 3’U2 and EV (p=0.754); CNO 3’U1 and EV (p=0.790); CNO 3’U2 and EV (p=0.716); PTEN 3’U and EV (p=0.766). (C) These effects are related to the differences in PTEN protein expression between the conditions reported in Figure 3 of this study and Figure 3H of Tay et al., 2011. Meta-analysis p values: WT HCT116: siNC and siSER (p=0.091); siNC and siVAPA (p=2.99×10−7); siNC and siCNO (p=0.049); siNC and siPTEN (p=0.0041): DicerEx5 HCT116: siNC and siSER (p=2.78×10−4); siNC and siVAPA (p=0.215); siNC and siCNO (p=3.70×10−4); siNC and siPTEN (p=0.229). (D) These effects are related to the differences in cell growth between the conditions reported in Figure 4 of this study and Figure 5B of Tay et al., 2011. Meta-analysis p values: DU145: siVAPA and siNC (p=0.255); siCNO and siNC (p=0.554); siPTEN and siNC (p=0.082): WT HCT116: siVAPA and siNC (p=0.045); siCNO and siNC (p=0.287); siPTEN and siNC (p=0.082): DicerEx5 HCT116: siVAPA and siPTEN (p=0.075); siCNO and siPTEN (p=0.001); siPTEN and siNC (p=0.048). Additional details for these meta-analyses can be found at https://osf.io/xgrqp/.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens, male) | DU145 | ATCC | cat#:HTB-81; RRID:CVCL_0105 | |
| Cell line (H. sapiens, male) | Wild-type HCT116 cells | Horizon Discovery | cat# HD R02-019; RRID:CVCL_HD76 | |
| Cell line (H. sapiens, male) | DICEREx5 HCT116 cells | Horizon Discovery | cat# HD R02-019; RRID:CVCL_HD76 | |
| Recombinant DNA reagent | psiCHECK-2+PTEN3’UTR | Addgene | plasmid# 50936; RRID:Addgene_50936 | |
| Recombinant DNA reagent | SERINC1 3’UTR | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Recombinant DNA reagent | VAPA 3’UTR1 | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Recombinant DNA reagent | VAPA 3’UTR2 | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Recombinant DNA reagent | CNOT6L 3’UTR1 | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Recombinant DNA reagent | CNOT6L 3’UTR2 | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Recombinant DNA reagent | PTEN 3’UTR | doi:10.1016/j.cell.2011.09.029 | Shared by Dr. Pier Paolo Pandolfi, Beth Israel Deaconess Medical Center | |
| Sequence-based reagent | siGlo RISC-free siRNA | Dharmacon | cat#:D-001600–01 | |
| Sequence-based reagent | siGENOME non-targeting siRNA | Dharmacon | cat#:D-001210–02 | |
| Sequence-based reagent | siGENOME SERINC1 | Dharmacon | cat# M-010725–00 | |
| Sequence-based reagent | siGENOME ZNF460 | Dharmacon | cat# M-032012–01 | |
| Sequence-based reagent | siGENOME VAPA | Dharmacon | cat# M-021382–01 | |
| Sequence-based reagent | siGENOME CNOT6L | Dharmacon | cat# M-016411–01 | |
| Sequence-based reagent | siGENOME PTEN | Dharmacon | M-003023–02 | |
| Sequence-based reagent | TaqMan probe SERINC1 | Thermo Fisher Scientific | Hs00380375_m1 | |
| Sequence-based reagent | TaqMan probe ZNF460 | Thermo Fisher Scientific | Hs01104252_m1 | |
| Sequence-based reagent | TaqMan probe VAPA | Thermo Fisher Scientific | Hs00427749_m1 | |
| Sequence-based reagent | TaqMan probe CNOT6L | Thermo Fisher Scientific | Hs00375913_m1 | |
| Sequence-based reagent | TaqMan probe PTEN | Thermo Fisher Scientific | Hs02621230_s1 | |
| Sequence-based reagent | TaqMan probe PARD3 | Thermo Fisher Scientific | Hs00969077_m1 | |
| Antibody | rabbit anti-PTEN | Cell Signaling Technology | cat#:9559; clone:138G5; RRID:AB_390810 | 1:1000 dilution |
| Antibody | mouse anti-HSP90 | BD Biosciences | cat#:610419; clone:68; RRID:AB_397798 | 1:1000 dilution |
| Antibody | HRP-conjugated donkey anti-rabbit | GE Healthcare | cat#:NA934; RRID:AB_772206 | 1:2000 dilution |
| Antibody | HRP-conjugated rabbit anti-mouse | Abcam | cat#:ab6728; RRID:AB_955440 | 1:2000 dilution |
| Software, algorithm | Veritas Microplate Luminometer software | Turner BioSystems | part#:998–9100; RRID:SCR_018534 | |
| Software, algorithm | StepOne Plus Real-Time PCR software | Applied Biosystems | RRID:SCR_014281 | Version 2.3 |
| Software, algorithm | ImageJ | doi:10.1038/nmeth.2089 | RRID:SCR_003070 | Version 1.50a |
| Software, algorithm | Gen5 software | BioTek Instruments | RRID:SCR_017317 | Version 2.05.5 |
| Software, algorithm | R Project for statistical computing | https://www.r-project.org | RRID:SCR_001905 | Version 3.5.1 |





