Super-resolution microscopy reveals majorly mono- and dimeric presenilin1/γ-secretase at the cell surface
Figures
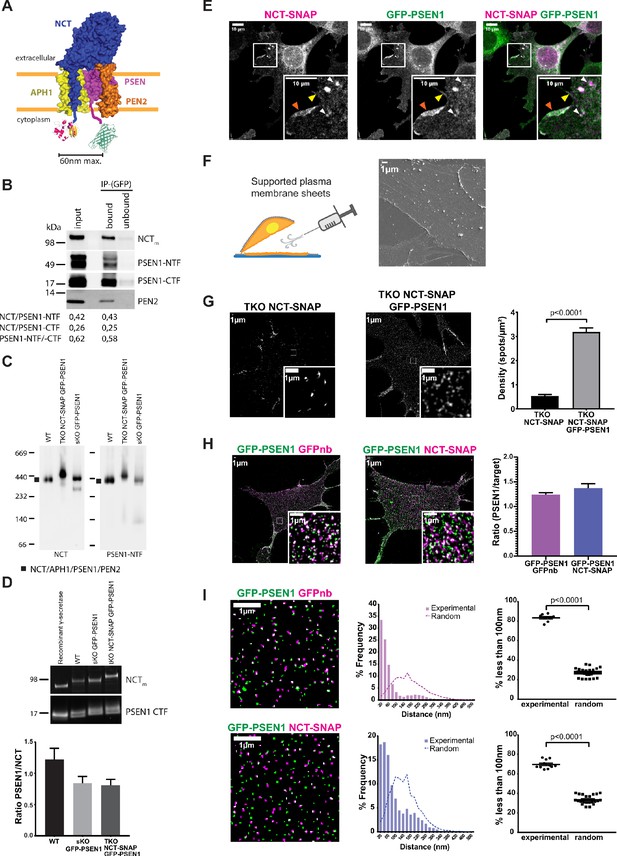
Visualizing the stoichiometry of γ-secretase subunits.
(A) Scheme of double-tagged γ-secretase. Both tags face the cytoplasm with the SNAP-tag and a linker fused to the C-terminus of NCT, and GFP with a linker to the N-terminus of PSEN1. Maximum distance between both tags is theoretically ~60 nm. (B) Co-immunoprecipitation of CHAPSO extracts of SPION isolated PM fractions using anti-GFP antibodies. Western blot shows an almost complete depletion of the extract for γ-secretase components in a ratio (NCT/PSEN1-NTF (0.43), NCT/PSEN1-CTF (0.25) and PSEN1-NTF /-CTF (0.58)) similar to those from input (0.42, 0.26, 0.62, respectively). (C) Blue native PAGE and western blot analysis of PM fractions of WT, NCT-SNAP/GFP-PSEN1 rescued tKO and GFP-PSEN1 rescued sKO MEFs. All samples show a band of around 440 kDa corresponding to the full γ-secretase complex (Fraering et al., 2004). The faint lower band corresponds to NCT/PSEN1-NTF/APH1A as DDM extraction slightly affects the stability of the complex as previously reported (Spasic et al., 2007; Fraering et al., 2004). (D) Quantitative western blot of SPION isolated PM fractions normalized to known amounts of purified recombinant γ-secretase. The PSEN1/NCT ratio is close to one for all samples. n = 4 replicates (E) Confocal microscopy of SiR-labelled NCT-SNAP/GFP-PSEN1 rescued tKO MEFs showing co-localization at the cell surface (yellow arrowhead), including membrane ruffles (orange arrowhead), and LAMPI-positive vesicles (white arrowheads). Scale bar = 10 µm (F) (Left panel) Schematic representation of supported PM sheet preparation. (Right panel) Scanning Electron Microscopy (SEM) of PM sheets of GFP-PSEN1 rescued sKO MEFs showing the basal PM attached to the coverslip. Some cytoskeleton can be seen still attached. Scale bar = 1 µm (G) SIM on PM sheets of NCT-SNAP and NCT-SNAP/GFP-PSEN1 rescued tKO MEFs showing a dramatically reduced NCT-SNAP spot density when tKO MEFs are only rescued with NCT-SNAP (mean ± SEM, NCT-SNAP n = 8 cells; NCT-SNAP/GFP-PSEN1 n = 14 cells). Scale bar = 1 µm (H) SIM image of PM sheets of NCT-SNAP/GFP-PSEN1 rescued tKO MEFs labeled with GFPnb-Atto647n (left panel) or SNAP-SiR (middle panel) showing similar spot densities in both channels (right panel) (mean ± SEM, GFPnb n = 8 cells; NCT n = 11 cells). (I) Masks of SIM PM sheets of NCT-SNAP/GFP-PSEN1 rescued tKO MEFs with either GFPnb-Atto647n (upper panels) or SNAP-SiR (lower panels). Scale bar = 1 µm. Histograms show the distribution of nearest-neighbor distances of either GFPnb to PSEN1 or NCT to PSEN1 spot centroids. (Left panels) Dot plot summarizing nearest-neighbor distances below 100 nm. Random distances were calculated from unpaired experimental data. Each dot represents one cell. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, GFPnb n = 8 cells; NCT n = 11 cells). See Figure 1—source data 1.
-
Figure 1—source data 1
Source Data for Nearest Neighbor Analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig1-data1-v1.zip
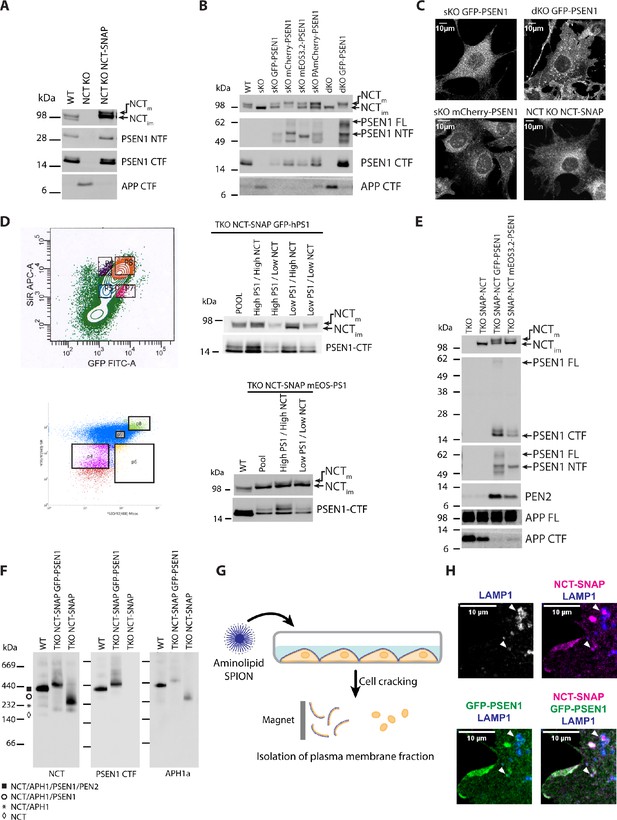
Biochemical characterization of MEF rescued cell lines.
(A) Western blot of total cell lysates of WT, NCT KO and NCT-SNAP rescue NCT KO MEFs showing maturation of NCT, endoproteolysis of PSEN1 and processing of APP-CTF after reintroducing NCT. (B) Western blot of total cell lysates of WT, PSEN1 sKO and PSEN1 and 2 dKO MEFs and the corresponding rescued MEFs with different tagged-PSEN1 constructs showing maturation of NCT, endoproteolysis of exogenous tagged PSEN1 and processing of APP-CTF after reintroducing the corresponding fluorescently tagged PSEN1 subunit. (C) Representative confocal microscopy of tagged-PSEN1 or NCT-SNAP rescued MEFs showing its characteristic broad subcellular distribution reminiscent of endogenous NCT and PSEN1. Scale bar = 10 µm (D) FAC Sorting of NCT-SNAP-SiR/GFP-PSEN1 and NCT-SNAP-SiR/mEOS3.2-PSEN1 rescued tKO MEFs into four populations with varying combinations of expression: P5 (NCT low/PSEN1 low), P6 (NCT high/PSEN1 low), P7 (NCT low/PSEN1 high) and P8 (NCT high/PSEN1 high). Western blot analysis identifies populations with highest relative levels of mature vs immature NCT underscoring optimal rescue without overexpression artifacts (e.g. much higher immature NCT indicates insufficient mature complexes caused by too low levels of tagged-PSEN1). The NCT high/PSEN1high (population P8 in both cases) were used for subsequent experiments. (E) Western blot of the selected P8 population of (d) compared to tKO and single NCT-SNAP rescued tKO MEFs, demonstrating the lack of any mature glycosylation in the absence of PSEN1 expression, decreased PEN2 levels and increased APP-CTF, the direct substrate of γ-secretase. (F) Blue native PAGE and western blotting of DDM-extracts of single NCT-SNAP and NCT-SNAP/GFP-PSEN1 rescued tKO MEFs compared to WT. A 440 kDa band, denoting the full γ-secretase complex, is detected only in WT and NCT-SNAP/GFP-PSEN1 rescued tKO MEF extracts. Note the lower mobility of the full complex for NCT-SNAP/GFP-PSEN1 rescued tKO MEFs due to the introduced tags. The NCT-SNAP rescued tKO (lacking PSEN expression) extract shows only the dimeric NCT/APH1a subcomplex and monomeric NCT-SNAP. (G) Scheme of the procedure for PM isolation using aminolipid-SPIONs. Aminolipid-SPIONs adhere to the cell surface. After cell cracking, lysates are passed through a column held in a magnet to retain PM sheets. Withdrawal from the magnet allows to elute and concentrate the isolated PM fraction (see Material and methods for details). (H) Confocal microscopy ROIs of tKO NCT-SNAP GFP-PSEN1 (P8 fraction) rescued entire cells depicting LAMP1-positive vesicles containing co-localized NCT-SiR and GFP-PSEN1 signal (arrowheads).
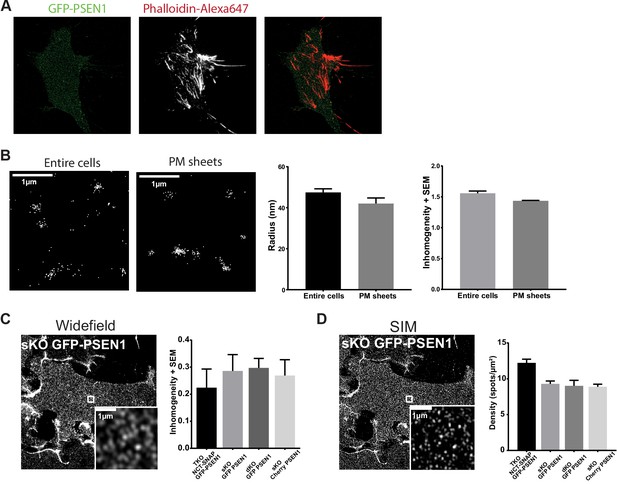
PM sheets controls.
(A) SIM on PM sheets derived from GFP-PSEN1 rescued sKO MEFs stained with Phalloidin-Alexa647. Some cytoskeletal elements are still detected on the isolated basal PM. (B) Comparison of PALM on PSEN1 sKO mEOS3.2-PSEN1 rescued sKO MEFs entire cells and PM sheets. Both the cluster radius and overall distribution of localizations at the PM are not affected by the PM sheet procedure (mean ± SEM, entire cells n = 2 cells; PM sheets n = 4 cells) Scale bar = 1 µm (C) PM sheets of the indicated rescued MEFs imaged in widefield were analyzed by QuASIMoDOH to determine the overall random distribution of PSEN1/γ-secretase at the cell surface. The degree of inhomogeneity of the spots remains constant across the different cell lines, regardless of tags or genetic background (mean ± SEM, NCT-SNAP/GFP-PSEN1 rescued tKO, n = 11 cells; GFP-PSEN1 rescued sKO, n = 18 cells; GFP-PSEN1 rescued dKO, n = 33 cells; cherry-PSEN1 rescued sKO, n = 25 cells). Scale bar = 1 µm (D) SIM of the same PM sheets to determine the density of spots (~10 spots/µm²) which was also unaltered across the different cell lines (mean ± SEM, NCT-SNAP/GFP-PSEN1 rescued tKO, n = 9 cells; GFP-PSEN1 rescued sKO, n = 13 cells; GFP-PSEN1 rescued dKO, n = 17 cells; cherry-PSEN1 rescued sKO, n = 20 cells). Scale bar = 1 µm. An example of GFP-PSEN1 rescued sKO MEFs is displayed for both (C) and (D).
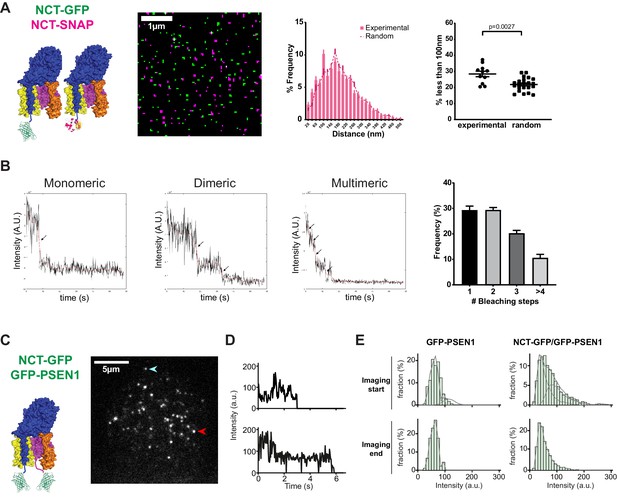
PSEN1/γ-secretase is monomeric and dimeric at the cell surface.
(A) SIM analysis of PM sheets derived from NCT KO MEFs rescued with both NCT-GFP and NCT-SNAP. Mask of SIM images shows no to little color overlap. Nearest-neighbor distances distribution and respective dot plot of distances (<100 nm) show little differences from a random distribution. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, n = 11 cells). (B) Photobleaching step examples of monomeric, dimeric and multimeric GFP-PSEN1 spots in PM sheets. Histogram shows the distribution of photobleaching steps per cell (mean ± SEM, n = 4 cells). (C) Live-cell single-molecule intensity measurements. NCT-GFP/GFP-PSEN1 rescued tKO or GFP-PSEN1 rescued sKO MEFs imaged in TIRF post-photogate, to diminish the molecular density of the ROI. After bleaching, unbleached molecules diffuse to the bleached area. Scale bar = 5 µm (D) Intensity measurement of individual spots marked by an arrow in (C). (E) Comparison of GFP-PSEN1 with NCT-GFP/GFP-PSEN1 intensity histograms. (Upper panels) Histograms of the first 100 frames of the movies show two populations, one with the unitary intensity, and one with twice the unitary intensity (dashed curves) for NCT-GFP/GFP-PSEN1; the second population is absent in GFP-PSEN1. (Lower panels) Histograms of the last 100 frames of the movies showed depletion of double intensity population in NCT-GFP/GFP-PSEN1 after photobleaching during the recording, leaving only single intensity molecules in both cases. See Figure 2—source data 1 and Figure 2—source data 2.
-
Figure 2—source data 1
Source Data for Nearest Neighbor Analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig2-data1-v1.zip
-
Figure 2—source data 2
Source Data for Photobleaching analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig2-data2-v1.zip
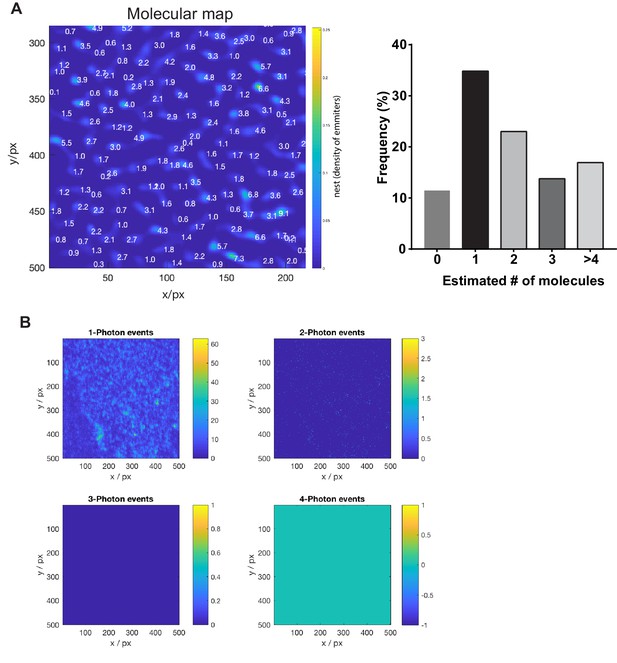
Molecular counting by photon statistics (CoPS) on PM sheets derived from NCT-YFP rescued NCT KO MEFs.
(A) Molecular map of NCT-YFP. As a molecule can only generate one photon at a time, photon coincidences were measured on a time-resolved confocal microscope to calculate the number of fluorescent emitters per spot. Molecular brightness and spatial density of fluorophores per image pixel were estimated. Finally, the emitter density image was segmented by a watershed algorithm. The histogram shows the distribution of the number of emitters per spot. (n = 1 cell) (B) Multi-photon detection events. Molecular brightness and emitter density were estimated from the relative frequencies of multi-photon detection events. By adding-up over selected pixels, the number of emitters per cluster was calculated.
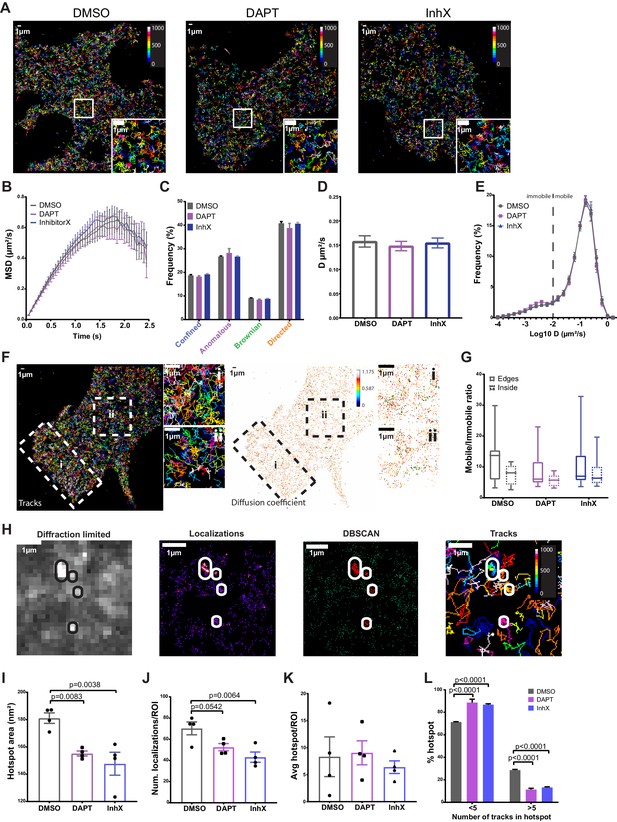
PSEN1/γ-secretase diffusion is fast at the cell surface and unaffected by GSI treatments, whereas hotspot areas display sensitivity to treatments.
Rescued MEFs were treated for 1 hr at 37°C with 1 µm of either DAPT or InhX (L-685,458) prior to imaging. DMSO (1:1000) was used as a control. (A) Representative cells showing similar distribution of mEOS3.2-PSEN1 tracks of cells treated with GSI or control (Bar code = frame of track initiation). Scale bar = 1 µm (B) Plot with the MSD over time shows an average anomalous movement (α = 0.844), which is unchanged upon GSI treatment (mean ± SEM, n = 14 cells). (C) Comparison of each motility frequency in the absence (control) or presence of GSIs (mean ± SEM, n = 14). (D) Average diffusion coefficients are not different between control and GSI treated cells (mean ± SEM, n = 14 cells). (E) Frequency distributions of diffusion coefficients (as Log10) identifies a major mobile and minor immobile pool. The dotted line marks the limit resolution of the microscope, rendering everything below it as immobile fraction. Only in the case of DAPT, the immobile pool is slightly increased (mean ± SEM, n = 14 cells). (F) Representative sptPALM image of mEOS3.2-PSEN1 tracks (left panel) and individual diffusion coefficient distribution (right panel, bar code = individual D) during ~1.5 min recording. Edge region marked by ‘i’ and boxed inside region by ‘ii’. Scale bar = 1 µm (G) Mobile/immobile ratio from tracks either in cell edges or in the inner region. There is a non-significant trend towards more mobile tracks in the cell periphery that is abolished after GSI treatments (mean, min-max, DMSO n = 12 cells, DAPT n = 6 cells, InhX n = 11 cells). (H) Diffraction-limited projection (left panel) of mEOS-PSEN1 hotspots (encircled). sptPALM localizations (left-middle panel) analyzed by DBSCAN revealed hotspots (right-middle panel) consisting of overlapping tracks (right panel). For cluster analysis, four to eight ROIs of the same size per cell were defined and wherein cell edges were avoided. Scale bar = 1 µm (I) Hotspot area is significantly reduced in GSI treated cells. Comparison analysis by one-way ANOVA (mean ± SEM, n = 4 cells, 30 hotspots/cell). (J) Number of localizations per hotspot shows a trend towards less localizations per hotspot upon treatment with GSI. Comparison analysis by one-way ANOVA (mean ± SEM, n = 4 cells, 30 hotspots/cell). (K) Mean number of hotspots per ROI is variable but not different in control and GSI treated cells (mean ± SEM, n = 4 cells, 30 hotspots/cell). (L) Number of tracks per hotspot is reduced upon GSI treatment. Multiple comparisons by two-way ANOVA (mean ± SEM, n = 4 cells, 30 hotspots/cell). See Figure 3—source data 1 and Figure 3—source data 2.
-
Figure 3—source data 1
Source Data for Hotspot analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig3-data1-v1.zip
-
Figure 3—source data 2
Source Data for SPT analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig3-data2-v1.zip
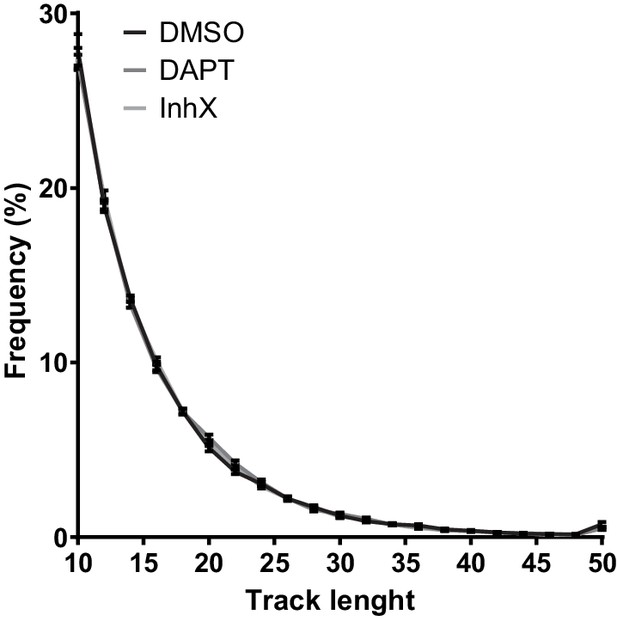
Track length information.
Distribution of track lengths of mEOS3.2-PSEN1 showing that short tracks are the most abundant in any condition.
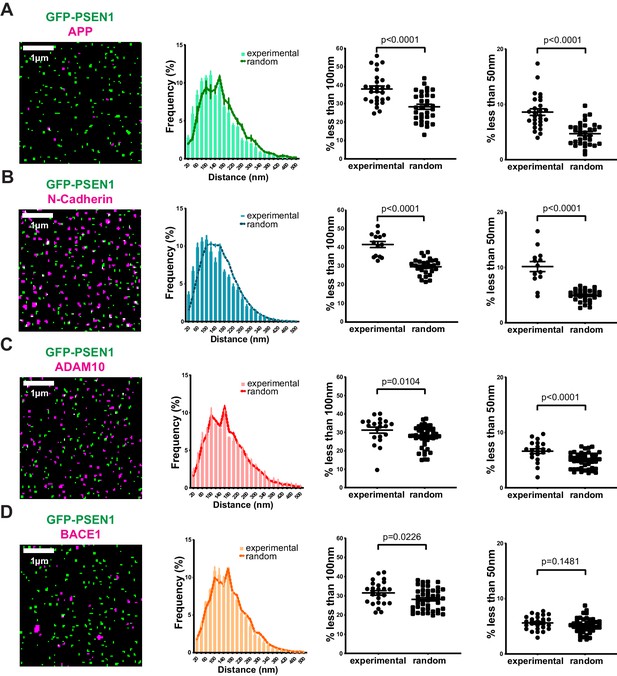
Correlation of PSEN1/γ-secretase complexes with substrates and sheddases at the cell surface.
(From left to right) SIM mask, nearest-neighbor distances distribution, percentage of spots present at a distance <100 nm and percentage of spots present below 50 nm. Each dot represents one cell. (A) Distance of endogenous APP and (B) N-cadherin to GFP-PSEN1 shows a clear association to PSEN1 (mean ± SEM, n = 27 cells and n = 14 cells, respectively). (C) ADAM10-SNAP distance to GFP-PSEN1 shows a limited association to PSEN1 (mean ± SEM, n = 20 cells) whereas (D) BACE1-SNAP distance to GFP-PSEN1 shows no association to PSEN1 (mean ± SEM, n = 25 cells). All comparison analysis by two-tail Mann-Whitney test. See Figure 4—source data 1.
-
Figure 4—source data 1
Source Data for Nearest neighbor analysis.
- https://cdn.elifesciences.org/articles/56679/elife-56679-fig4-data1-v1.zip
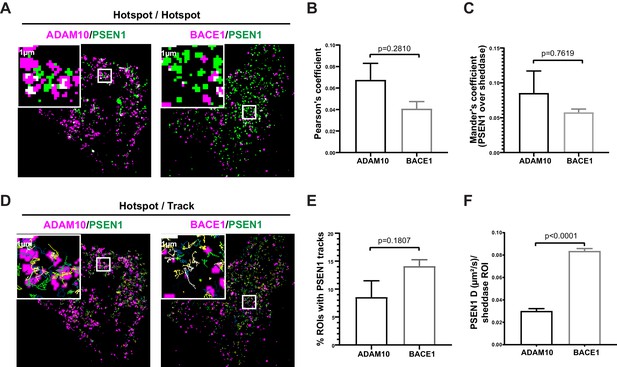
GSI-sensitive PSEN1/γ-secretase hotspots do not overlap with those of sheddases.
(A) Diffraction-limited hotspots of SNAP tagged-sheddases and mEOS3.2-PSEN1 show limited overlap (white). Scale bar = 1 µm. (B) Pearson’s coefficient of ADAM10 and BACE1 hotspots associated with PSEN1 hotspots show very low correlation. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, BACE1 n = 4 cells; ADAM10 n = 6 cells). (C) Mander’s coefficient of PSEN1 hotspots over ADAM10 or BACE1 hotspot showed limited overlap. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, BACE1 n = 4 cells; ADAM10 n = 6 cells). (D) Tracks of mEOS3.2-PSEN1 show some association with diffraction-limited hotspots of SNAP tagged-sheddases. Scale bar = 1 µm. (E) Percentage of sheddases hotspots associated with mEOS3.2-PSEN1 tracks show a tendency for higher association with BACE1 hotspots. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, BACE1 n = 7 cells; ADAM10 n = 6 cells) (F) Diffusion coefficient analysis of pooled tracks associated with a sheddase-hotspot shows a significantly decreased diffusion coefficient for tracks on ADAM10-SNAP, but not for tracks on BACE1 hotspots. Comparison analysis by two-tail Mann-Whitney test (mean ± SEM, BACE1 = 263 tracks, n = 7 cells; ADAM10 = 126 tracks, n = 6 cells each).
Videos
Molecular counting of GFP-tagged γ-secretase complexes.
An ROI was prebleached by PhotoGate and molecules were allowed to diffuse into the ROI prior to imaging. (left panel-control) GFP-TRCP4a transfected cell shows diffraction–limited spots containing 4x GFP intensities as these molecules are organized as tetramers. (middle panel) A tKO NCT-GFP GFP-PSEN1 cell shows diffraction-limited spots of 2x GFP intensity (one each on NCT and PSEN1) reflecting the 1:1 NCT-PSEN1 subunit stoichiometry. (right panel) A sKO GFP-PSEN1 cell shows diffraction-limited spots of 1x GFP intensity as γ-secretase consists of only one PSEN1 molecule per complex. Color table shows fluorescence intensities (a.u). Time stamp shows seconds of recording.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | XL10-Gold | Stratagene | 200314 | Competent cells |
| Cell line (M. musculus) | MEF NCT/PSEN1/PSEN2 TKO NCT-SNAP GFP-PSEN1 | This paper | Mouse embryonic fibroblast KO of NCT, PSEN1 and PSEN2, transduced to express mNCT-SNAP and GFP-hPSEN1 | |
| Cell line (M. musculus) | MEF PSEN1 KO mCherry-PSEN1 | Sannerud et al., 2016 | Mouse embryonic fibroblast KO of PSEN1, transduced to express mCherry-hPSEN1 | |
| Cell line (M. musculus) | MEF PSEN1 KO GFP-PSEN1 | Sannerud et al., 2016 | Mouse embryonic fibroblast KO of PSEN1, transduced to express GFP-hPSEN1 | |
| Cell line (M. musculus) | MEF PSEN1/PSEN2 KO GFP-PSEN1 | Sannerud et al., 2016 | Mouse embryonic fibroblast KO of PSEN1 and PSEN2, transduced to express GFP-hPSEN1 | |
| Cell line (M. musculus) | MEF NCT KO NCT-SNAP | This paper | Mouse embryonic fibroblast KO of NCT, transduced to express mNCT-SNAP | |
| Cell line (M. musculus) | MEF PSEN1 KO GFP-PSEN1 + BACE1-SNAP | This paper | Mouse embryonic fibroblast KO of PSEN1, transduced to express GFP-PSEN1 and hBACE1-SNAP | |
| Transfected construct (H. sapiens) | GFP-hPSEN1 | This paper | Construct to transduce MEF PSEN1 KO, tKO and dKO | |
| Transfected construct (H. sapiens) | mEOS3.2-hPSEN1 | This paper | Construct to transduce MEF PSEN1 KO, tKO and dKO | |
| Transfected construct (H. sapiens) | mCherry-hPSEN1 | This paper | Construct to transfect and express mADAM10-SNAP | |
| Transfected construct (H. sapiens) | mNCT-SNAP | This paper | Construct to transduce tKO and NCT KO | |
| Transfected construct (H. sapiens) | mNCT-GFP | This paper | Construct to transduce NCT KO | |
| Transfected construct (H. sapiens) | mNCT-YFP | This paper | Construct to transduce NCT KO | |
| Transfected construct (H. sapiens) | hBace1-SNAP | This paper | Construct to transduce PSEN1 KO GFP-PSEN1 and mEOS3.2-PSEN1 | |
| Transfected construct (H. sapiens) | mADAM10-SNAP | This paper | Construct to transfect and express mADAM10-SNAP | |
| Transfected construct (H. sapiens) | pX330-PSEN1 | This paper | CRISPR plasmid to KO PSEN1 | |
| Transfected construct (H. sapiens) | pX330-PSEN2 | This paper | CRISPR plasmid to KO PSEN2 | |
| Transfected construct (H. sapiens) | px459-NCT | This paper | CRISPR plasmid to KO NCT | |
| Antibody | anti-LAMP1 (rat monoclonal) | Santa Cruz | sc-19992; RRID:AB_2134495 | (1:200) |
| Antibody | anti-NCT (mouse monoclonal) | Esselens et al., 2004 | 9C3 | (1:7000) |
| Antibody | anti-PSEN1-NTF (rabbit polyclonal) | Abcam | ab71181; RRID:AB_1603935 | (1:2000) |
| Antibody | anti-PSEN1-NTF (rat polyclonal) | Millipore | MAB1563; RRID:AB_1671560 | (1:4000) |
| Antibody | anti-PSEN1-CTF (rabbit monoclonal) | Abcam | ab76083; RRID:AB_1310605 | (1:2000) |
| Antibody | anti-PEN2 (rabbit polyclonal) | Abcam | ab18189; RRID:AB_444310 | (1:1000) |
| Antibody | anti-APP-CTF (rabbit polyclonal) | Esselens et al., 2004 | B63 | (1:10,000) |
| Antibody | anti-transferrin receptor (mouse monoclonal) | Invitrogen | 136800; RRID:AB_2533029 | (1:4000) |
| Antibody | anti-rabbit (HRP-goat) | Bio-Rad | 1706515; RRID:AB_11125142 | (1:10000) |
| Antibody | anti-mouse (HRP-goat) | Bio-Rad | 1706516; RRID:AB_11125547 | (1:10000) |
| Antibody | anti-GFP (rabbit polyclonal) | Bio-Rad | A11122; RRID:AB_221569 | (1:10000) |
| Antibody | anti-PSEN1-CTF (mouse monoclonal) | Millipore | MAB5232; RRID:AB_95175 | (1:1000) |
| Antibody | anti-mouse Alexa Fluor790 (goat) | Invitrogen | A11375; RRID:AB_2534146 | (1:15,000) |
| Recombinant DNA reagent | pSNAPf (plasmid) | New England Biolabs | N9183S | SNAPtag sequence |
| Peptide, recombinant protein | γ-secretase | Szaruga et al., 2017 | ||
| Commercial assay or kit | NEBuilder HiFI DNA assembly mix | New England Biolabs | E5520 | Gibson assembly |
| Commercial assay or kit | Q5 site-directed mutagenesis | New England Biolabs | E0554S | Mutagenesis |
| Commercial assay or kit | NativePAGE | ThermoFisher | Native protein electrophoresis | |
| Commercial assay or kit | MycoAlert | Lonza | LT07-118 | Mycoplasma detection kit |
| Commercial assay or kit | DAPT | Tocris Bioscience | 2634/10 | γ-secretase inhibitor |
| Commercial assay or kit | Inhibitor X | Calbiochem | 565771 | γ-secretase inhibitor |
| Commercial assay or kit | DMSO | VWR | A3672 0250 | Treatment vector |
| Software, algorithm | BD FACS | Stall and AC Technologies, 2008 BD Biosciences | RRID:SCR_005400 | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| Software, algorithm | QuASIMoDOH | Paparelli et al., 2016 | Software to analyze spots distribution | |
| Software, algorithm | Nearest Neighbour analysis | MATLAB | RRID:SCR_001622 | findNearestNeighbors(ptCloud,point,K) |
| Software, algorithm | B-unwarpJ | Schindelin et al., 2012 ImageJ | https://imagej.net/BUnwarpJ | |
| Software, algorithm | H-watershed | Schindelin et al., 2012 ImageJ | https://imagej.net/Interactive_Watershed | |
| Software, algorithm | Spot intensity analysis | Schindelin et al., 2012 ImageJ | https://imagej.net/Spot_Intensity_Analysis | |
| Software, algorithm | Molecular counting by photon statistics | Grußmayer and Herten, 2017 PicoQuant | ||
| Software, algorithm | qSR | Andrews, 2017 | https://github.com/cisselab/qSR | |
| Software, algorithm | PALMtracer | University of Bordeaux | https://www.iins.u-bordeaux.fr/team-sibarita-PALMTracer | |
| Software, algorithm | Thunderstorm | Ovesný et al., 2014 | RRID:SCR_016897 | https://zitmen.github.io/thunderstorm/ |
| Software, algorithm | SR Tesseler | Levet et al., 2015 | https://www.iins.u-bordeaux.fr/team-sibarita-sr-tesseler | |
| Other | SNAP-Cell 647-SiR substrate | New England Biolabs | S9102S | Cell permeable substrate to label SNAPtag |
| Other | Tetraspeck beads | Invitrogen | S9102S | Fiducial Markers (1:1000) |





