Dithranol targets keratinocytes, their crosstalk with neutrophils and inhibits the IL-36 inflammatory loop in psoriasis
Figures
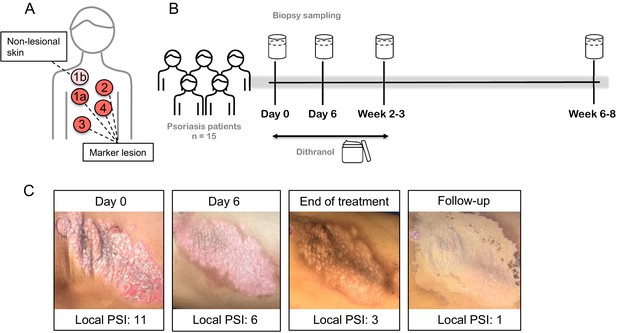
Dithranol leads to a fast reduction of psoriatic skin lesions as determined by local psoriasis severity index (PSI) of marker lesions.
(A,B) 15 psoriasis patients were treated with dithranol. Skin biopsies were taken from marker lesions at multiple timepoints: 1a = lesional skin at baseline, 2 = lesional skin at day 6, 3 = lesional skin at end of treatment, week 2–3, 4 = lesional skin at follow-up, 4–6 weeks after end of treatment. In addition, non-lesional skin at baseline (1b) was sampled. (C) Representative images of lesional psoriatic skin and local psoriasis severity index (PSI; sum of erythema (0–4), induration (0–4) and scaling (0–4); 0 = none, 1 = mild, 2 = moderate, 3 = severe, 4 = very severe) at different time points.
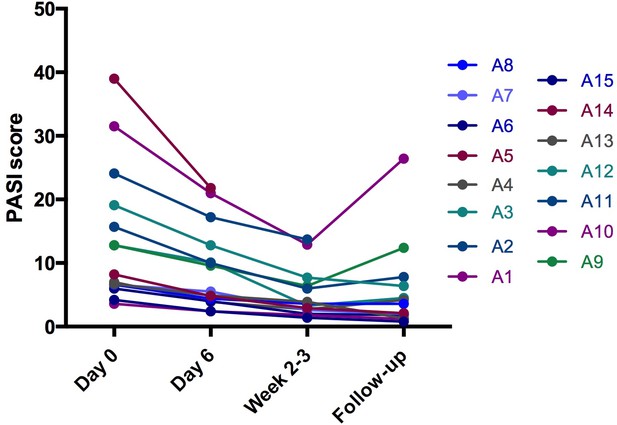
Psoriasis area and severity (PASI) score at baseline (day 0), early during treatment (day 6), at end of treatment (week 2–3) and at follow-up (4–6 weeks after end of treatment) for all 15 patients (A1–A15) treated with dithranol.
-
Figure 1—figure supplement 1—source data 1
Values displayed in graph shown in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/56991/elife-56991-fig1-figsupp1-data1-v1.xlsx
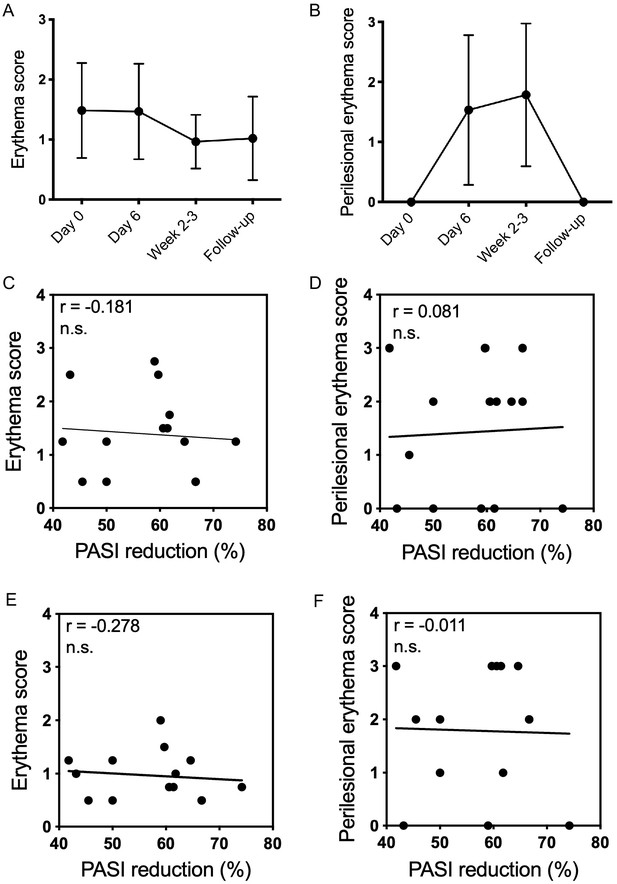
Lesional and perilesional erythema does not correlate with reduction in PASI score.
(A) Mean erythema score (0 = none, 1 = mild, 2 = moderate, 3 = severe 4 = very severe erythema on lesional skin) and B) Mean perilesional erythema score (0 = none, 1 = mild, 2 = moderate, 3 = severe erythema on perilesional skin) at baseline (day 0), early during dithranol treatment (day 6), at end of treatment (week 2–3) and at follow-up (4–6 weeks after end of treatment). Mean ± SD (n = 15) is shown. (C–D) Erythema score (C) and perilesional erythema score (D) at day six during dithranol treatment was compared with reduction in PASI score at end of treatment using Spearman correlation. (E–F) Erythema score (E) and perilesional erythema score (F) at end of treatment (week 2–3) was compared with reduction in PASI score at end of treatment using Spearman correlation. n.s. = not significant.
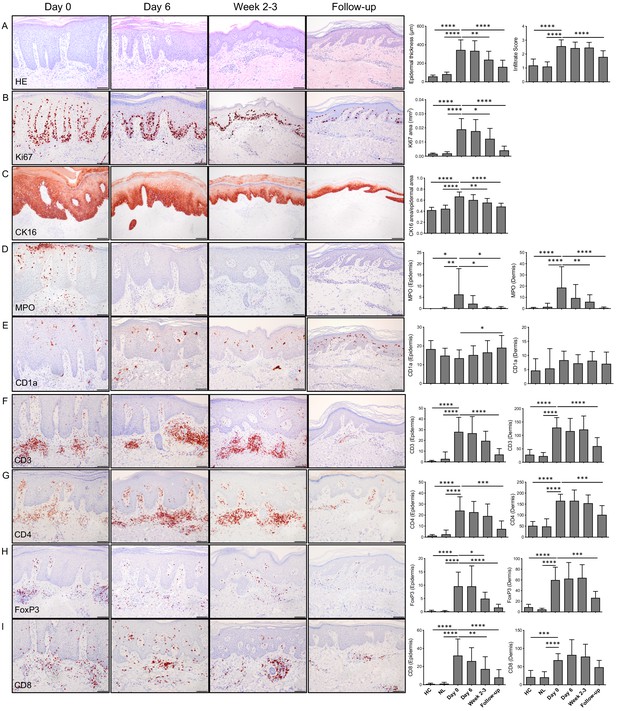
Histological and immunohistochemical analysis of lesional skin before (day 0), during (day 6), at end of treatment (week 2–3) and 4–6 weeks after ending treatment (follow-up).
(A) Representative H and E images, epidermal thickness and cellular infiltrate scoring. In treated psoriatic lesions, epidermal thickness was significantly decreased at week 2–3 and at follow-up. Infiltrate score (0 = none, 0.5 = none/low, 1 = low, 1.5 = low/moderate, 2 = moderate, 2.5 = moderate/high, 3 = high infiltration of immune cells) was significantly higher in untreated psoriatic lesions compared to non-lesional skin (NL) and healthy controls (HC). In treated psoriatic lesions, infiltrate score was significantly decreased at follow-up. (B) Ki67 staining in epidermis was significantly reduced at week 2–3 and follow-up. (C) CK16 staining in epidermis was significantly reduced at week 2–3 and follow-up. (D–I) Representative IHC images and mean cell counts for epidermis and dermis. Neutrophil cell counts were significantly reduced at week 2–3 and follow-up in epidermis and dermis (D). Langerhans cell (CD1a+) numbers in epidermis were significantly increased at follow-up (E). Epidermal FoxP3 (H) and CD8 (I) cell counts were significantly reduced at week 2–3. Dermal CD3 (F), CD4 (G), and FoxP3 (H) positive cell counts display significant reduction at follow-up. One-way ANOVA with Dunnet’s multiple comparisons test was used for statistics. Bars represent mean ± SD; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001; scale bar = 100 µm.
-
Figure 2—source data 1
Values displayed in bar plots shown in Figure 2.
- https://cdn.elifesciences.org/articles/56991/elife-56991-fig2-data1-v1.xlsx
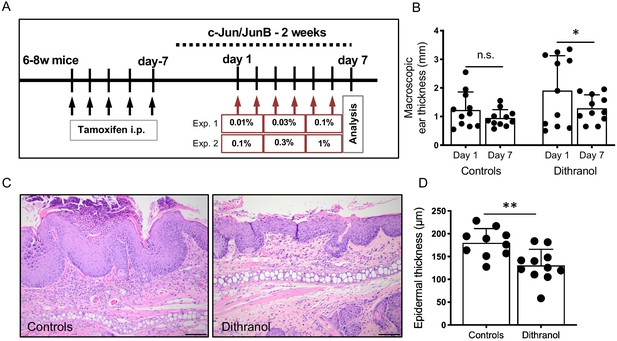
Topical application of dithranol ameliorates psoriasis-like skin lesions in c-Jun/JunB knockout mice.
(A) Schematic representation of experimental set-up. Red arrows indicate dithranol application in series of increasing concentrations. (Exp. 1 and 2 = experiment 1 and 2) (B) Macroscopic ear thickness on day 1 compared to day 7. Dithranol treatment led to a significant reduction in ear thickness. Controls (n = 11), dithranol group (n = 11); Paired t-test was used for statistics. (C) Representative H and E images of untreated and dithranol-treated ears. (D) Dithranol treatment led to a significant reduction in epidermis thickness. Controls (n = 10), dithranol group (n = 11); unpaired t-test was used for statistics. Data from the two experiments was pooled (D). Bars represent mean ± SD; n.s. = not significant; *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001; scale bar = 100 µm.
-
Figure 3—source data 1
Values displayed in scatter plots shown in Figure 3.
- https://cdn.elifesciences.org/articles/56991/elife-56991-fig3-data1-v1.xlsx
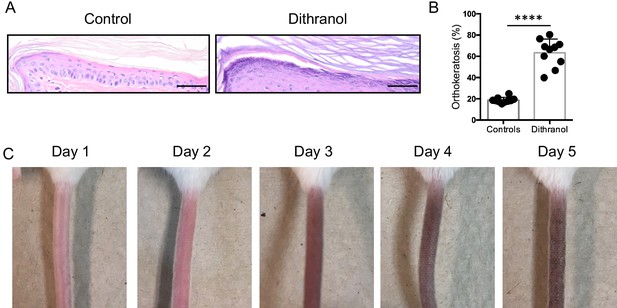
Strong inducing effect of dithranol in the mouse-tail test.
(A) Representative H and E images of vehicle-treated and dithranol-treated mouse tail skin. (B) Dithranol treatment significantly increased orthokeratosis. (C) Representative images of erythema and typical brownish discoloration of skin caused by dithranol. Bars represent mean ± SD (n = 10). Mann-Whitney test was used for statistics. *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001; Scale bar = 50 µm.
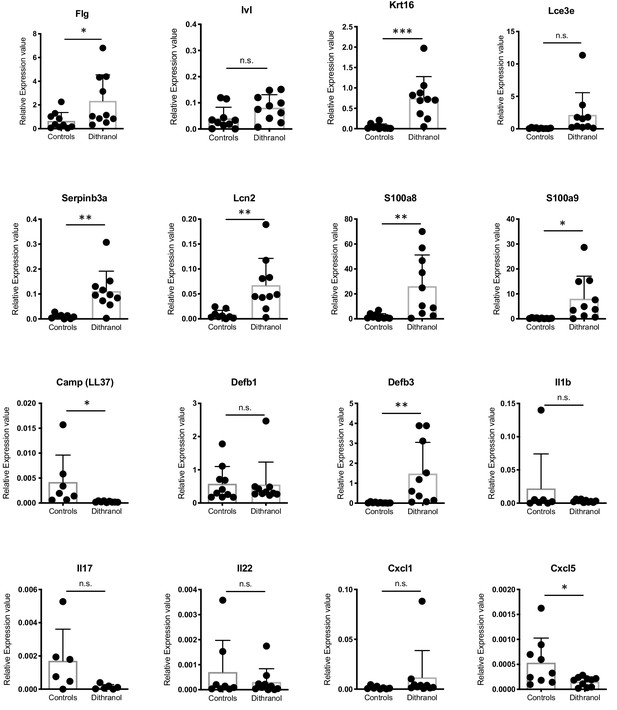
Gene expression analysis by RT-PCR of keratinization markers (Flg, Ivl, Krt16, Lce3e, Serpinb3a), AMPs (Lcn2, S100a8, S100a9, Camp, Defb1, Defb3) and inflammatory markers (Il1b, Il17, Il22, Cxcl1, Cxcl5) of dithranol- and vehicle-treated murine tail skin.
Bars represent mean ± SD (n = 10) of relative expression values (deltaCT) normalized to Ubc. Unpaired t-test was used for statistics (n.s. = not significant) Flg, filaggrin; Ivl, involucrin, Krt16, keratin16; Lce3e, Late Cornified Envelope 3e; Camp, Cathelicidin Antimicrobial Peptide; Defb1, Beta-Defensin 1; Defb3, Beta-Defensin 3; Il1b, Interleukin one beta; Il17, Interleukin 17; Il22, Interleukin 22; Cxcl1, C-X-C Motif Chemokine Ligand 1; Cxcl5, C-X-C Motif Chemokine Ligand 5.
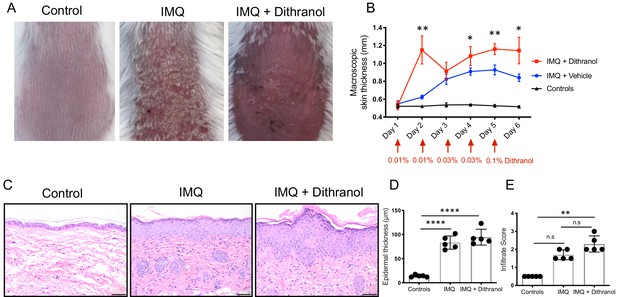
Dithranol causes exacerbation of psoriasis-like skin lesions in imiquimod mouse model.
(A) Representative images of untreated, IMQ-treated and IMQ and dithranol-treated dorsal skin of mice. (B) Macroscopic skin thickness (red arrows indicate concentration of dithranol), (C–D) epidermal thickness measurements with corresponding H and E images and E) cellular infiltrate score show exacerbation of psoriasis-like dermatitis in IMQ and Dithranol-treated mice. Infiltrate score (0 = none, 0.5 = none/low, 1 = low, 1.5 = low/moderate, 2 = moderate, 2.5 = moderate/high, 3 = high infiltration of immune cells); Tukey’s multiple comparisons test (B), One-way ANOVA with Tukey’s multiple comparisons test (D) and Kruskal Wallis test with Dunn’s multiple comparisons test (E) was used for statistics. Bars represent mean ± SD (n = 5); *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001; Scale bar = 50 µm.
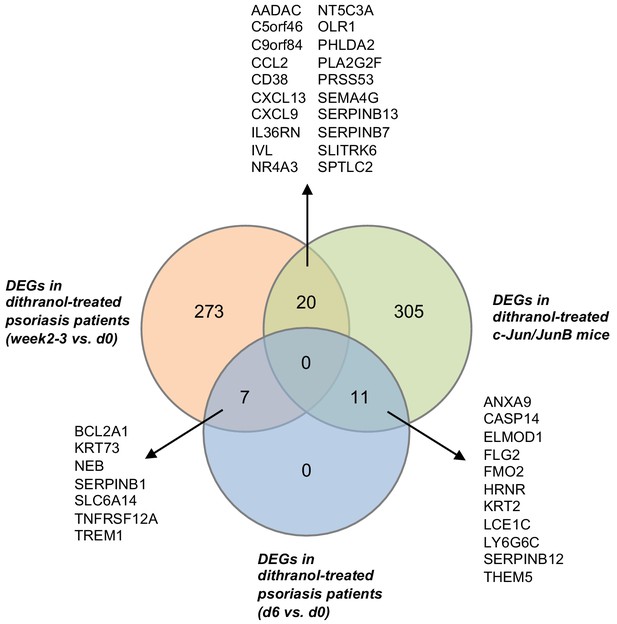
Venn diagram (created using InteractiVenn Heberle et al., 2015) showing comparison and overlap between differentially expressed genes (DEGs) in dithranol-treated human psoriatic skin at week 2–3 vs. day 0, DEGs in dithranol-treated human psoriatic skin at day 6 vs. day 0 and DEGs in dithranol-treated c-Jun/JunB psoriatic skin.
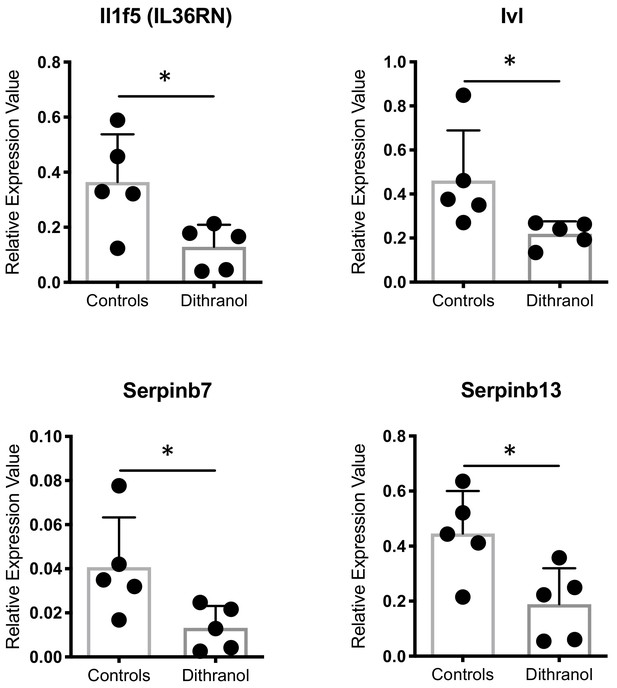
Verification of microarray gene expression analysis: Gene expression analysis by RT-PCR of selected genes based on microarray data in dithranol-treated c-Jun/JunB knockout mice vs. vehicle-treated controls.
Bars represent mean ± SD (n = 5) of relative expression values (deltaCT) normalized to Ywhaz. Unpaired t-test was used for statistics. Il1f5 (IL36RN), interleukin 1 family member 5 (IL-36 receptor antagonist); Ivl, Involucrin; Serpinb7, Serine peptidase inhibitor, clade B member 7; Serpinb13, Serine peptidase inhibitor, clade B member 13.
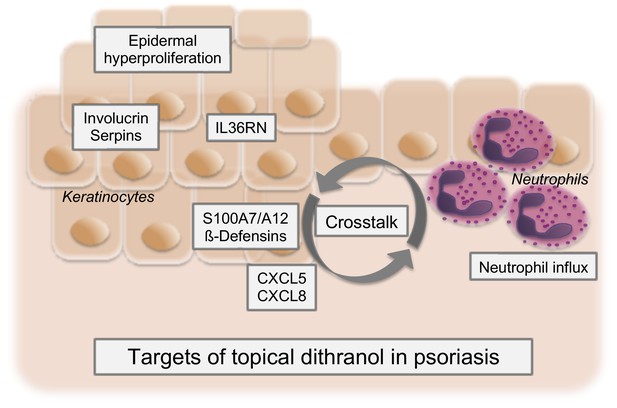
Proposed model of dithranol’s mechanism of action in psoriasis.
Dithranol decreases expression of keratinocyte differentiation regulators (involucrin and serpins), IL-36-related genes, keratinocyte-derived antimicrobial peptides (AMPs) (S100A7/A12 and ß-defensins) and chemotactic factors for neutrophils (CXCL5, CXCL8). This disrupts the crosstalk between keratinocytes and neutrophils and leads to diminished neutrophilic infiltration, thereby halting the inflammatory feedback loop in psoriasis. Together, this results in clearance of psoriatic lesions.
Tables
Psoriasis area and severity index (PASI) and psoriasis severity index (PSI) from 15 psoriasis patients.
| Parameter | Time point | ||||
|---|---|---|---|---|---|
| Baseline | Day 6 | End of treatment | Follow-up | ||
| PASI | |||||
| mean ± SD | 13.6 ± 10.3 | 9.0 ± 6.3 (p<0.0001)* | 5.1 ± 3.8 (p=0.0001)* | 5.7 ± 6.7 (p=0.0002)* | |
| %reduction, mean ± SD (range) | - | 32.9 ± 8.0 (16.7–44.3) | 57.5 ± 9.5 (41.8–74.2) | 56.1 ± 23.3 (3.1–81.0) | |
| Local PSI | |||||
| mean ± SD | 6.7 ± 1.1 | 3.3 ± 1.6 (p<0.0001)* | 2.0 ± 1.5 (p<0.0001)* | 1.6 ± 1.3 (p<0.0001)* | |
| %reduction, mean ± SD (range) | - | 52.6 ± 21.9 (14.3–100) | 69.0 ± 23.9 (16.7–100) | 76.3 ± 18.3 (37.5–100) | |
-
*P value was determined using Wilcoxon test comparing indicated value to baseline.
-
Table 1—source data 1
Values displayed in Table 1.
- https://cdn.elifesciences.org/articles/56991/elife-56991-table1-data1-v1.xlsx
Top 45 differentially regulated genes (p-value<0.05, fold change >1.5) in dithranol-treated lesional skin after 6 days compared to baseline from 15 patients with psoriasis.
| Probe set ID | Gene symbol | Gene title | Day 6 vs. baseline (fold change) |
|---|---|---|---|
| 16693318 | FLG2 | Filaggrin family member 2 | 2.89 |
| 16693303 | HRNR | Hornerin | 2.64 |
| 16903552 | NEB | Nebulin | 2.18 |
| 16765017 | KRT2 | Keratin 2 | 2.04 |
| 16693308 | FLG | Filaggrin | 1.97 |
| 16761221 | CLEC2A | C-type lectin domain family 2, member A | 1.92 |
| 16730782 | ELMOD1 | ELMO/CED-12 domain containing 1 | 1.91 |
| 16735288 | OVCH2 | Ovochymase 2 | 1.83 |
| 16990787 | SPINK7 | Serine peptidase inhibitor, Kazal type 7 (putative) | 1.78 |
| 16852824 | SERPINB12 | Serpin peptidase inhibitor, clade B (ovalbumin), member 12 | 1.74 |
| 16693341 | LCE1C | Late cornified envelope 1C | 1.71 |
| 16693249 | THEM5 | Thioesterase superfamily member 5 | 1.70 |
| 16921644 | MIRLET7C | MicroRNA let-7c | 1.69 |
| 16670681 | ANXA9 | Annexin A9 | 1.68 |
| 16821186 | CLEC3A | C-type lectin domain family 3, member A | 1.68 |
| 16859090 | CASP14 | Caspase 14 | 1.68 |
| 16765005 | KRT73 | Keratin 73 | 1.66 |
| 16945497 | COL6A5 | Collagen, type VI, alpha 5 | 1.66 |
| 16976615 | SULT1E1 | Sulfotransferase family 1E, estrogen-preferring, member 1 | 1.65 |
| 16898858 | CD207 | CD207 molecule, langerin | 1.63 |
| 16861126 | UPK1A | Uroplakin 1A | 1.60 |
| 16704475 | FAM35DP | Family with sequence similarity 35, member A pseudogene | 1.60 |
| 17085015 | FRMPD1 | FERM and PDZ domain containing 1 | 1.59 |
| 16817034 | CHP2 | Calcineurin-like EF-hand protein 2 | 1.56 |
| 16671082 | LCE1A | Late cornified envelope 1A | 1.55 |
| 16671037 | LCE2D | Late cornified envelope 2D | 1.55 |
| 16748835 | PIK3C2G | Phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit | 1.54 |
| 17039517 | LY6G6C | Lymphocyte antigen six complex, locus G6C | 1.53 |
| 16673713 | FMO2 | Flavin containing monooxygenase 2 (non-functional) | 1.52 |
| 16812344 | BCL2A1 | BCL2-related protein A1 | −1.52 |
| 16968213 | ANXA3 | Annexin A3 | −1.54 |
| 17104519 | RNY4P23 | RNA, Ro-associated Y4 pseudogene 23 | −1.55 |
| 16948835 | MIR1224 | MicroRNA 1224 | −1.55 |
| 16815310 | TNFRSF12A | Tumor necrosis factor receptor superfamily, member 12A | −1.60 |
| 17015084 | SERPINB1 | Serpin peptidase inhibitor, clade B (ovalbumin), member 1 | −1.67 |
| 17106398 | SLC6A14 | Solute carrier family 6 (amino acid transporter), member 14 | −1.69 |
| 16693375 | SPRR2F | Small proline-rich protein 2F | −1.69 |
| 17065458 | DEFB4A | Defensin, beta 4A | −1.75 |
| 17074361 | DEFB4B | Defensin, beta 4B | −1.77 |
| 16698947 | RNU5A-8P | RNA, U5A small nuclear 8, pseudogene | −1.78 |
| 16976827 | CXCL5 | Chemokine (C-X-C motif) ligand 5 | −1.81 |
| 16976821 | PPBP | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | −1.82 |
| 17019056 | TREM1 | Triggering receptor expressed on myeloid cells 1 | −1.99 |
| 17050251 | SLC26A4 | Solute carrier family 26 (anion exchanger), member 4 | −2.19 |
| 16967771 | CXCL8 | Chemokine (C-X-C motif) ligand 8 | −3.36 |
Top 45 differentially regulated genes in dithranol-treated lesional skin at end of treatment compared to baseline from 15 patients with psoriasis (p<0.05, fold change >1.5).
| Probe set ID | Gene symbol | Gene title | End of treatment vs. baseline (fold change) |
|---|---|---|---|
| 16947045 | AADAC | Arylacetamide deacetylase | 3.07 |
| 16765005 | KRT73 | Keratin 73 | 2.31 |
| 16976868 | BTC | Betacellulin | 2.27 |
| 16924792 | CLDN8 | Claudin 8 | 2.11 |
| 17125092 | CNTNAP3 | Contactin associated protein-like 3 | 2.09 |
| 17097358 | SLC46A2 | Solute carrier family 46, member 2 | 2.07 |
| 16780133 | SLITRK6 | SLIT and NTRK-like family, member 6 | 2.00 |
| 16834436 | RAMP2 | Receptor (G protein-coupled) activity modifying protein 2 | 1.96 |
| 17123970 | ZDHHC11B | Zinc finger, DHHC-type containing 11B | 1.88 |
| 17104313 | AR | Androgen receptor | 1.85 |
| 16951140 | MIR548I2 | MicroRNA 548i-2 | 1.85 |
| 17063366 | ATP6V0A4 | ATPase, H+ transporting, lysosomal V0 subunit a4 | 1.84 |
| 16974224 | MIR548I2 | MicroRNA 548i-2 | 1.83 |
| 16687914 | CYP2J2 | Cytochrome P450, family 2, subfamily J, polypeptide 2 | 1.82 |
| 17125106 | CNTNAP3B | Contactin associated protein-like 3B | 1.80 |
| 16903552 | NEB | Nebulin | 1.80 |
| 17125094 | CNTNAP3B | Contactin associated protein-like 3B | 1.80 |
| 16770284 | TMEM116 | Transmembrane protein 116 | 1.79 |
| 16765041 | KRT77 | Keratin 77 | 1.79 |
| 17123972 | ZDHHC11B | Zinc finger, DHHC-type containing 11B | 1.78 |
| 16688210 | MIR3671 | MicroRNA 3671 | 1.78 |
| 17125218 | CNTNAP3 | Contactin associated protein-like 3 | 1.74 |
| 16764923 | KRT6A | Keratin 6A | −3.69 |
| 16767261 | IL22 | Interleukin 22 | −3.87 |
| 17065453 | DEFB103A | Defensin, beta 103A | −3.94 |
| 17074366 | DEFB103A | Defensin, beta 103A | −3.94 |
| 16671027 | LCE3C | Late cornified envelope 3C | −4.14 |
| 16743751 | MMP12 | Matrix metallopeptidase 12 (macrophage elastase) | −4.42 |
| 16979444 | TNIP3 | TNFAIP3 interacting protein 3 | −4.48 |
| 17106398 | SLC6A14 | Solute carrier family 6 (amino acid transporter), member 14 | −4.60 |
| 16686734 | CYP4Z2P | Cytochrome P450, family 4, subfamily Z, polypeptide 2 | −4.89 |
| 16671144 | S100A7A | S100 calcium binding protein A7A | −5.00 |
| 16764907 | KRT6C | Keratin 6C | −5.01 |
| 16730157 | HEPHL1 | Hephaestin-like 1 | −5.15 |
| 16967831 | EPGN | Epithelial mitogen | −5.31 |
| 16842517 | NOS2 | Nitric oxide synthase 2, inducible | −5.38 |
| 16693409 | S100A12 | S100 calcium binding protein A12 | −6.12 |
| 16813112 | RHCG | Rh family, C glycoprotein | −6.18 |
| 16738803 | TCN1 | Transcobalamin I (vitamin B12 binding protein, R binder family) | −6.45 |
| 16924785 | CLDN17 | Claudin 17 | −8.09 |
| 16693365 | SPRR2C | Small proline-rich protein 2C (pseudogene) | −8.72 |
| 16967771 | CXCL8 | Chemokine (C-X-C motif) ligand 8 | −9.14 |
| 17074361 | DEFB4B | Defensin, beta 4B | −9.39 |
| 16693375 | SPRR2F | Small proline-rich protein 2F | −10.12 |
| 16884602 | IL36A | Interleukin 36, alpha | −10.50 |
Top 20 significantly enriched pathways as determined by Gene Ontology (GO) enrichment analysis in dithranol-treated lesional skin after 6 days compared to baseline from 15 patients with psoriasis.
(GO was done using Cytoscape software [Bindea et al., 2009; Shannon et al., 2003]; a.o. = among others).
| GOID | GO term | P-Value | % Associated Genes | Associated genes found |
|---|---|---|---|---|
| GO:0001533 | Cornified envelope | 3.8E-12 | 10.45 | FLG, HRNR, KRT2, LCE1A, LCE1C, LCE2D, SPRR2F |
| GO:0031424 | Keratinization | 7.7E-12 | 3.93 | CASP14, FLG, HRNR, KRT2, KRT73, LCE1A, LCE1C, LCE2D, SPRR2F |
| GO:0030216 | Keratinocyte differentiation | 79.0E-12 | 3.02 | CASP14, FLG, HRNR, KRT2, KRT73, LCE1A, LCE1C, LCE2D, SPRR2F |
| GO:0018149 | Peptide cross-linking | 300.0E-12 | 9.84 | FLG, KRT2, LCE1A, LCE1C, LCE2D, SPRR2F |
| GO:0070268 | Cornification | 12.0E-9 | 5.31 | CASP14, FLG, KRT2, KRT73, LCE1A, SPRR2F |
| GO:0004168 | Receptor CXCR2 binds ligands CXCL1 to 7 | 1.0E-6 | 33.33 | CXCL5, CXCL8, PPBP |
| GO:0042379 | Chemokine receptor binding | 5.4E-6 | 6.25 | CXCL5, CXCL8, DEFB4A, PPBP |
| GO:0045236 | CXCR chemokine receptor binding | 6.0E-6 | 18.75 | CXCL5, CXCL8, PPBP |
| GO:0061436 | Establishment of skin barrier | 18.0E-6 | 13.04 | FLG, FLG2, HRNR |
| GO:0033561 | Regulation of water loss via skin | 21.0E-6 | 12.00 | FLG, FLG2, HRNR |
| GO:0004657 | IL-17 signaling pathway | 21.0E-6 | 4.30 | CXCL5, CXCL8, DEFB4A, DEFB4B |
| GO:0007874 | Keratin filament formation | 21.0E-6 | 4.17 | FLG, KRT2, KRT73, SPRR2F |
| GO:0030593 | Neutrophil chemotaxis | 21.0E-6 | 4.17 | CXCL5, CXCL8, PPBP, TREM1 |
| GO:1990266 | Neutrophil migration | 27.0E-6 | 3.85 | CXCL5, CXCL8, PPBP, TREM1 |
| GO:0071621 | Granulocyte chemotaxis | 38.0E-6 | 3.31 | CXCL5, CXCL8, PPBP, TREM1 |
| GO:1902622 | Regulation of neutrophil migration | 45.0E-6 | 7.50 | CXCL5, CXCL8, PPBP |
| GO:0071622 | Regulation of granulocyte chemotaxis | 71.0E-6 | 5.66 | CXCL5, CXCL8, PPBP |
| GO:0030104 | Water homeostasis | 130.0E-6 | 4.00 | FLG, FLG2, HRNR |
| GO:0002690 | Positive regulation of leukocyte chemotaxis | 120.0E-6 | 3.30 | CXCL5, CXCL8, PPBP |
| GO:0004867 | Serine-type endopeptidase inhibitor activity | 79.0E-6 | 3.00 | SERPINB1, SERPINB12, SPINK7 |
Top 20 significantly enriched pathways as determined by Gene Ontology (GO) enrichment analysis in dithranol-treated lesional skin at end of treatment compared to baseline from 15 patients with psoriasis.
(GO was done using Cytoscape software; Bindea et al., 2009; Shannon et al., 2003 a.o. = among others).
| GOID | GO term | P-Value | % Associated Genes | Associated genes found |
|---|---|---|---|---|
| GO:0006954 | Inflammatory response | 17.0E-18 | 6.93 | IL17A, IL1B, IL20, IL22, IL36A, IL36G, IL36RN, a.o. |
| GO:0006952 | Defense response | 52.0E-18 | 4.60 | CXCL8, CXCL9, DEFB103A, DEFB4A, IFNG, IL17A, IL1B, IL20, IL22, IL36A, IL36G, IL36RN, IL4R, IRAK2, IRF1, KRT16, a.o. |
| GO:0051707 | Response to other organism | 56.0E-18 | 6.02 | CCL2, CCL20, CCL22, CD24, CD80, COTL1, CXCL13, CXCL16, CXCL8, CXCL9, DDX21, DEFB103A, DEFB4A, a.o. |
| GO:0050663 | Cytokine secretion | 480.0E-18 | 13.78 | IFNG, IL17A, IL1B, IL26, IL36RN, IL4R, LYN, MMP12, NOS2, PAEP, PANX1, PNP, S100A12, S100A8, S100A9, a.o. |
| GO:0001816 | Cytokine production | 680.0E-18 | 6.76 | IDO1, IFNG, IL17A, IL1B, IL26, IL36A, IL36RN, IL4R, IRF1, LTF, LYN, MB21D1, MMP12, NOS2, a.o. |
| GO:0012501 | Programmed cell death | 1.0E-15 | 4.05 | CASP5, CASP7, CCL2, CCL21, CD24, CD274, CD38, IFNG, IL17A, IL1B, IRF1, IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, KRT74, KRT77, a.o. |
| GO:0051240 | Positive regulation of multicellular organismal process | 1.5E-15 | 4.57 | CXCL17, CXCL8, FBN2, FERMT1, GBP5, GPR68, HPSE, HRH2, IDO1, IFNG, IL17A, IL1B, IL20, IL26, IL36A, S100A8, S100A9, SERPINB3, SERPINB7, a.o. |
| GO:0001817 | Regulation of cytokine production | 2.0E-15 | 7.04 | CCL2, CCL20, CD24, CD274, CD80, CD83, CXCL17, IFNG, IL17A, IL1B, IL26, IL36A, IL36RN, IL4R, IRF1, TNFRSF9, WNT5A, a.o. |
| GO:0002237 | Response to molecule of bacterial origin | 15.0E-15 | 9.22 | S100A8, S100A9, SELE, SOD2, TIMP4, TNFRSF9, TNIP3, WNT5A, ZC3H12A, a.o. |
| GO:0070268 | Cornification | 100.0E-15 | 17.70 | DSC2, DSG3, IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, KRT74, KRT77, PI3, SPRR2A, SPRR2B, SPRR2D, a.o. |
| GO:0043588 | Skin development | 190.0E-15 | 8.17 | IL20, IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, KRT74, KRT77, LCE3A, LCE3C, LCE3E, a.o. |
| GO:0008544 | Epidermis development | 220.0E-15 | 7.63 | DSC2, DSG3, EPHA2, FERMT1, FOXE1, FURIN, HPSE, IL20, IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, KRT74, KRT77, a.o. |
| GO:0009617 | Response to bacterium | 240.0E-15 | 6.63 | CCL2, CCL20, CD24, CD80, CXCL13, CXCL16, CXCL8, CXCL9, DEFB103A, DEFB4A, S100A12, S100A8, S100A9, TREM1, a.o. |
| GO:0001819 | Positive regulation of cytokine production | 1.4E-12 | 7.89 | CCL2, CCL20, CD274, CD80, CD83, CXCL17, FERMT1, GBP5, HPSE, IDO1, IFNG, IL17A, IL1B, IL26, IL36A, a.o. |
| GO:0050707 | Regulation of cytokine secretion | 2.4E-12 | 13.10 | AIM2, CASP5, CD274, FERMT1, GBP1, IFNG, IL17A, IL1B, IL26, IL36RN, IL4R, LYN, MMP12, PAEP, PANX1, a.o. |
| GO:0005125 | Cytokine activity | 4.4E-12 | 10.64 | CCL20, CCL21, CCL22, CCL4L2, CXCL13, CXCL16, CXCL8, CXCL9, FAM3D, IFNG, IL17A, IL1B, IL20, IL22, IL26, a.o. |
| GO:0031424 | Keratinization | 21.0E-12 | 10.48 | IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, KRT74, KRT77, LCE3A, LCE3C, LCE3E, PI3, SPRR2A, SPRR2B, a.o. |
| GO:0002790 | Peptide secretion | 97.0E-12 | 6.25 | CD274, CD38, DOC2B, FAM3D, FERMT1, GBP1, GBP5, GLUL, GPR68, IFNG, IL17A, IL1B, IL26, IL36RN, IL4R, LYN, MMP12, NOS2, TREM1, WNT5A, a.o. |
| GO:0032940 | Secretion by cell | 140.0E-12 | 4.04 | AIM2, AMPD3, CASP5, IL36RN, IL4R, LCN2, LRG1, LTF, LYN, MMP12, NOS2, NR1D1, NR4A3, OLR1, PAEP, PANX1, PLA2G3, a.o. |
| GO:0009913 | Epidermal cell differentiation | 190.0E-12 | 7.91 | DSC2, DSG3, EPHA2, FURIN, IL20, IVL, KLK13, KRT16, KRT17, KRT31, KRT6A, KRT6C, KRT73, LCE3E, PI3, SLITRK6, SPRR2A, SPRR2B, SPRR2D, SPRR2E, a.o. |
Top 45 differentially regulated genes in dithranol-treated psoriatic skin of c-Jun/JunB knockout mice compared to that of vehicle-treated controls (p<0.05, fold change (FC) >1.5).
| Probe set ID | Gene symbol | Gene title | Fold change |
|---|---|---|---|
| TC0Y00000006.mm.2 | Eif2s3y | Eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | 10,37 |
| TC0Y00000233.mm.2 | Uty | Ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome | 6,38 |
| TC0Y00000235.mm.2 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 5,40 |
| TC0900000047.mm.2 | Mmp3 | Matrix metallopeptidase 3 | 3,64 |
| TC0Y00000079.mm.2 | Ssty2 | Spermiogenesis specific transcript on the Y 2 | 3,31 |
| TC0100001632.mm.2 | Ifi209 | Interferon activated gene 209 | 3,23 |
| TC0500002755.mm.2 | Cxcl9 | Chemokine (C-X-C motif) ligand 9 | 2,85 |
| TC0300003133.mm.2 | Ifi44 | Interferon-induced protein 44 | 2,69 |
| TC0100003591.mm.2 | Ifi213 | Interferon activated gene 213 | 2,53 |
| TC0100001634.mm.2 | Ifi208 | Interferon activated gene 208 | 2,49 |
| TC1900000217.mm.2 | Ms4a4c | Membrane-spanning 4-domains, subfamily A, member 4C | 2,45 |
| TC0300002684.mm.2 | Chil3 | Chitinase-like 3 | 2,39 |
| TC0100003550.mm.2 | Slamf7 | SLAM family member 7 | 2,33 |
| TC0500000922.mm.2 | Cxcl13 | Chemokine (C-X-C motif) ligand 13 | 2,26 |
| TC1900000500.mm.2 | Ifit2 | Interferon-induced protein with tetratricopeptide repeats 2 | 2,20 |
| TC0500002840.mm.2 | Plac8 | Placenta-specific 8 | 2,20 |
| TC0700001630.mm.2 | Adm | Adrenomedullin | 2,15 |
| TC1900000501.mm.2 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 2,13 |
| TC1800000610.mm.2 | Iigp1 | Interferon inducible GTPase 1 | 2,07 |
| TC0300001446.mm.2 | Gbp3 | Guanylate binding protein 3 | 2,07 |
| TC0300003134.mm.2 | Ifi44l | Interferon-induced protein 44 like | 2,05 |
| TC1900000502.mm.2 | Ifit3b | Interferon-induced protein with tetratricopeptide repeats 3B | 2,02 |
| TC0400003296.mm.2 | Skint5 | Selection and upkeep of intraepithelial T cells 5 | −2,37 |
| TC0300000482.mm.2 | Aadac | Arylacetamide deacetylase | −2,37 |
| TC1100000469.mm.2 | Fndc9 | Fibronectin type III domain containing 9 | −2,39 |
| TC0300002399.mm.2 | Lce6a | Late cornified envelope 6A | −2,42 |
| TC0300002409.mm.2 | Lce1i | Late cornified envelope 1I | −2,44 |
| TC0300002412.mm.2 | Kprp | Keratinocyte expressed, proline-rich | −2,44 |
| TC0300000846.mm.2 | Hrnr | Hornerin | −2,48 |
| TC0300002407.mm.2 | Lce1g | Late cornified envelope 1G | −2,49 |
| TC1600000324.mm.2 | Fetub | Fetuin beta | −2,53 |
| TC1300001377.mm.2 | Akr1c18 | Aldo-keto reductase family 1, member C18 | −2,56 |
| TC0300002406.mm.2 | Lce1f | Late cornified envelope 1F | −2,57 |
| TC1500001714.mm.2 | Slurp1 | Secreted Ly6/Plaur domain containing 1 | −2,57 |
| TC1500002344.mm.2 | Gsdmc2 | Gasdermin C2 | −2,59 |
| TC1000000145.mm.2 | Il20ra | Interleukin 20 receptor, alpha | −2,66 |
| TC0700000443.mm.2 | Cyp2b19 | Cytochrome P450, family 2, subfamily b, polypeptide 19 | −2,70 |
| TC0100000103.mm.2 | Ly96 | Lymphocyte antigen 96 | −2,89 |
| TC0700000778.mm.2 | Klk5 | Kallikrein related-peptidase 5 | −3,15 |
| TC0300003252.mm.2 | Clca3a2 | Chloride channel accessory 3A2 | −3,20 |
| TC0900002312.mm.2 | Elmod1 | ELMO/CED-12 domain containing 1 | −3,20 |
| TC0300002401.mm.2 | Lce1a1 | Late cornified envelope 1A1 | −3,26 |
| TC0700000484.mm.2 | Fcgbp | Fc fragment of IgG binding protein | −3,83 |
| TC0300002405.mm.2 | Lce1e | Late cornified envelope 1E | −4,16 |
| TC1500002274.mm.2 | Krt2 | Keratin 2 | −5,95 |
Top 20 significantly enriched pathways as determined by Gene Ontology (GO) enrichment analysis in dithranol-treated psoriatic skin of c-Jun/JunB knockout mice compared to that of vehicle-treated controls.
(GO was done using Cytoscape software Bindea et al., 2009; Shannon et al., 2003; a.o. = among others).
| Go id | GO term | P-Value | % Associated Genes | Associated genes found |
|---|---|---|---|---|
| GO:0043588 | Skin development | 14,0E-18 | 11,11 | Casp14, Cldn1, Flg2, Gjb3, Hrnr, Ivl, Krt2, Lce1a1, Lce1a2, Lce1l, Lce1m, Lor, Pou2f3, Ptgs1, Scel, Tfap2b, a.o. |
| GO:0008544 | Epidermis development | 73,0E-15 | 9,18 | Acer1, Alox8, Casp14, Cnfn, Cst6, Dnase1l2, Hrnr, Ivl, Krt2, Lce1c, Lce1d, Lce1e, Lce1f, Lce1g, Lce1h, Lce1i, Lce1j, a.o. |
| GO:0071345 | Cellular response to cytokine stimulus | 1,7E-12 | 6,26 | Ccdc3, Ccl2, Ccl5, Ccr9, Cxcl13, Cxcl9, Edn1, Il18, Il1f5, Il1f8, Il1rl1, Il20ra, Stat2, a.o. |
| GO:0034097 | Response to cytokine | 7,1E-12 | 5,62 | Cd38, Chad, Cldn1, Csf3, Edn1, Gbp2, Gbp3, Gbp4, Gbp7, Gbp8, Gbp9, Gm4951, Ifi203, Ifi204, Ifi209, Ifit1, Ifit2, Ifit3, Ifit3b, Xaf1, a.o. |
| GO:0035456 | Response to interferon-beta | 12,0E-12 | 25,00 | Gbp2, Gbp3, Gbp4, Gm4951, Ifi203, Ifi204, Ifi209, Ifit1, Ifit3, Iigp1, Irgm1, Xaf1 |
| GO:0006952 | Defense response | 63,0E-12 | 4,11 | Ccl2, Ccl5, Cd180, Cd59a, Cxcl13, Cxcl9, Cybb, Defb6, Drd1, Herc6, Hp, Il18, Il1f5, Il1f8, Il1rl1, Irgm1, Kalrn, Klk5, a.o. |
| GO:0030855 | Epithelial cell differentiation | 430,0E-12 | 5,51 | Casp14, Cdkn1a, Cldn1, Cnfn, Dlx3, Dnase1l2, Gsdmc2, Gstk1, Hrnr, Ivl, Klf15, Krt2, Lce1a1, Lor, Pou2f3, a.o. |
| GO:0020005 | Symbiont-containing vacuole membrane | 1,4E-9 | 66,67 | Gbp2, Gbp3, Gbp4, Gbp7, Gbp9, Iigp1 |
| GO:0044216 | Other organism cell | 5,6E-9 | 30,77 | C4b, Gbp2, Gbp3, Gbp4, Gbp7, Gbp9, Iigp1, Tap1 |
| GO:0044406 | Adhesion of symbiont to host | 120,0E-9 | 37,50 | Ace2, Gbp2, Gbp3, Gbp4, Gbp7, Gbp9 |
| GO:0045087 | Innate immune response | 390,0E-9 | 4,35 | Ccl2, Ccl5, Cd180, Cfb, Cldn1, Ifit3, Iigp1, Il1f5, Il1f8, Irgm1, Lbp, Ly96, Mx1, Parp9, Sla, Slamf6, Slamf7, Stat2, Tlr7, Trim62, a.o. |
| GO:0034341 | Response to interferon-gamma | 1,0E-6 | 10,58 | Ccl2, Ccl5, Cldn1, Edn1, Gbp2, Gbp3, Gbp4, Gbp7, Gbp8, Gbp9, Parp9 |
| GO:0006954 | Inflammatory response | 5,9E-6 | 4,29 | C3, C4b, Ccl2, Ccl5, Cd180, Cd59a, Chil3, Crip2, Ctla2a, Cxcl13, Cxcl9, Cybb, Hp, Il18, Il1f5, Il1f8, Il1rl1, Lbp, Ly96, a.o. |
| GO:0042832 | Defense response to protozoan | 9,4E-6 | 19,35 | Gbp2, Gbp3, Gbp4, Gbp7, Gbp9, Iigp1 |
| GO:0035457 | Cellular response to interferon-alpha | 21,0E-6 | 36,36 | Ifit1, Ifit2, Ifit3, Ifit3b |
| GO:0030414 | Peptidase inhibitor activity | 22,0E-6 | 6,19 | C3, C4b, Cst6, Ctla2b, Fetub, R3hdml, Serpinb12, Serpinb13, Serpinb2, Serpinb7, Spink14, Tfap2b, Wfdc12, Wfdc5 |
| GO:0098542 | Defense response to other organism | 34,0E-6 | 4,05 | Adm, Ccl5, Cxcl13, Cxcl9, Defb6, Gbp2, Gbp3, Hp, Ifit1, Ifit2, Ifit3, Ifit3b, Iigp1, Il1f5, Klk5, Lbp, Mx1, Oas1f, Plac8, Stat2, Tlr7, Wfdc12, a.o. |
| GO:0031424 | Keratinization | 43,0E-6 | 15,00 | Casp14, Cnfn, Hrnr, Ivl, Krt2, Lor |
| GO:0044403 | Symbiosis, encompassing mutualism through parasitism | 55,0E-6 | 4,11 | Ace2, Acta2, Atg16l2, Ccl5, Cxcl9, Gbp2, Gbp3, Gbp4, Gbp7, Gbp9, Ifit1, Ifit2, Ifit3, Ifit3b, Lbp, Mx1, Oas1f, Pou2f3, Rab9, Stat2, Tap1, Tlr7, Trim62 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-CD1a; mouse monoclonal | Immunotech, Beckman Coulter | Clone: O10 RRID:AB_10547704 | (undiluted) |
| Antibody | anti-CD3; mouse monoclonal | Novocastra, Leica Biosystems | Clone: PS1 RRID:AB_2847892 | (1:100) |
| Antibody | anti-CD4; mouse monoclonal | Novocastra, Leica Biosystems | Clone: 1F6 RRID:AB_563559 | (1:30) |
| Antibody | anti-CD8; mouse monoclonal | Dako, Agilent | Clone: C8/144b RRID:AB_2075537 | (1:25) |
| Antibody | anti-CK16; rabbit monoclonal | Abcam | Clone: EPR13504 RRID:AB_2847885 | (1:1000) |
| Antibody | anti-FoxP3; mouse monoclonal | Bio-Rad | Clone: 236A/E7 RRID:AB_2262813 | (1:100) |
| Antibody | anti-Ki-67; mouse monoclonal | Dako, Agilent | Clone: MIB-1 RRID:AB_2631211 | (1:50) |
| Antibody | anti-MPO; mouse monoclonal | Dako, Agilent | Clone: MPO-7 RRID:AB_578599 | (1:100) |
| Strain, strain background Mus musculus | BALB/c, wild-type | Charles River Laboratories | RRID:IMSR_CRL:028 Charles River Strain code#: 028 | |
| Strain, strain background Mus musculus | JunBf/f c-Junf/f K5-Cre-ERT | PMID:16163348 | Obtained from the laboratory of Maria Sibilia (Medical University of Vienna) | |
| Commercial assay or kit | miRNeasy Mini Kit | Qiagen | Cat #: 217004 | |
| Commercial assay or kit | iScript Reverse Transcription Supermix | Bio-Rad | Cat #: 1708841 | |
| Commercial assay or kit | GoTag qPCR Master Mix | Promega | Cat #: A6001 | |
| Commercial assay or kit | Human GeneChip2.0 ST arrays | Affymetrix, ThermoFisher Scientific | Cat #:902113 | |
| Commercial assay or kit | mouse Clariom S Assay | Affymetrix, ThermoFisher Scientific | Cat #:902919 | |
| Commercial assay or kit | GeneChip WT PLUS Reagent Kit | ThermoFisher Scientific | Cat #: 902280 | |
| Commercial assay or kit | GeneChip WT Terminal Labeling Kit | ThermoFisher Scientific | Cat #: 900671 | |
| Commercial assay or kit | GeneChip Hybridization, Wash and Stain Kit | ThermoFisher Scientific | Cat #: 900720 | |
| Commercial assay or kit | nCounter GX Custom codeset | NanoString Technologies | Custom codeset (80 target genes, four reference genes | |
| Chemical compound, drug | Aldara (Imiquimod)5% cream | MEDA Pharmaceuticals | Cat #: 111981 | |
| Chemical compound, drug | Tamoxifen | Sigma-Aldrich | Cat #: T5648 | |
| Chemical compound, drug | Dithranol (1,8-Dihydroxy-9(10H)-anthracenone) | Gatt-Koller GmbH Pharmaceuticals | Cat #: 8069994 | Dissolved in vaseline and provided by the pharmacy of the Medical University of Graz, Austria |
| Software, algorithm | GraphPad Prism version 8 | GraphPad | RRID:SCR_002798 https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | Interacti Venn | PMID:25994840 | http://www.interactivenn.net/ | |
| Software, algorithm | Cytoscape | PMID:19237447 | RRID:SCR_003032 https://cytoscape.org/ | |
| Software, algorithm | Transcriptome Analysis Console (TAC) 4.0.2 | ThermoFisher Scientific | https://www.thermofisher.com/at/en/home/life-science/microarray-analysis/microarray-analysis-instruments-software-services/microarray-analysis-software/affymetrix-transcriptome-analysis-console-software.html | |
| Software, algorithm | Partek Genomics Suite version 6.6 | Partek Inc | RRID:SCR_011860 https://www.partek.com/partek-genomics-suite/ | |
| Software, algorithm | R Version 3.5.1 | The R Project for Statistical Computing | RRID:SCR_001905 https://www.r-project.org/ | |
| Software, algorithm | nSolver 2.5 Software | NanoString Technologies | https://www.nanostring.com/products/analysis-software/nsolver |
Additional files
-
Supplementary file 1
Top 45 upregulated genes in lesional psoriatic skin compared to non-lesional skin at baseline from 15 patients with psoriasis (p<0.05, fold change >1.5).
- https://cdn.elifesciences.org/articles/56991/elife-56991-supp1-v1.docx
-
Supplementary file 2
Verification of microarray results of dithranol-treated skin samples of psoriasis patients with Nanostring analysis (nCounter GX Custom codeset) of 80 target genes and four reference genes.
FC, fold change
- https://cdn.elifesciences.org/articles/56991/elife-56991-supp2-v1.docx
-
Supplementary file 3
Top 20 significantly enriched pathways as determined by Gene Ontology (GO) enrichment analysis in histological responders compared to non-responders in the psoriasis patient cohort (GO was done using Cytoscape software; [Bindea et al., 2009; Shannon et al., 2003] a.o. = among others).
- https://cdn.elifesciences.org/articles/56991/elife-56991-supp3-v1.docx
-
Supplementary file 4
qPCR Primer sequences and corresponding annealing temperatures.
- https://cdn.elifesciences.org/articles/56991/elife-56991-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56991/elife-56991-transrepform-v1.pdf





