The novel ciliogenesis regulator DYRK2 governs Hedgehog signaling during mouse embryogenesis
Figures
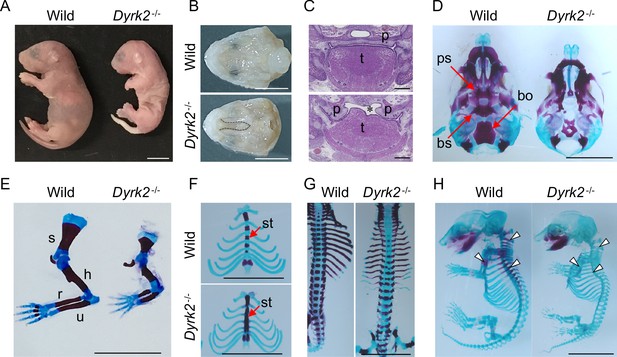
Deletion of DYRK2 shows skeletal defects in mouse development.
(A) Whole embryo gross images of wild-type and homozygous Dyrk2-/- embryos at birth. (B, C) Palatal and tongue abnormalities in Dyrk2-/- embryos. Gross images of the palate with mandible removed from wild-type and Dyrk2-/- embryos at E18.5 (B), and HE staining from the coronal plane at E13.5 (C). Dotted lines in (B) and an asterisk in (C) indicate cleft of the secondary palate. (D–H) Arizarin red and alcian blue staining of the craniofacial skeleton (D), forelimbs (E), sternum (F), and vertebra (G) from wild-type and Dyrk2-/- embryos at E18.5, and whole skeleton staining at E16.5 (H). Arrowheads in (H) indicate regions that decreasing bone mineralization. bo, basioccipital bone; bs, basisphenoid; h, humerus; r, radius; p, palatal shelves; ps, presphenoid; s, scapula; st, sternebrae; t, tongue; u, ulna. Scale bars, 5 mm.
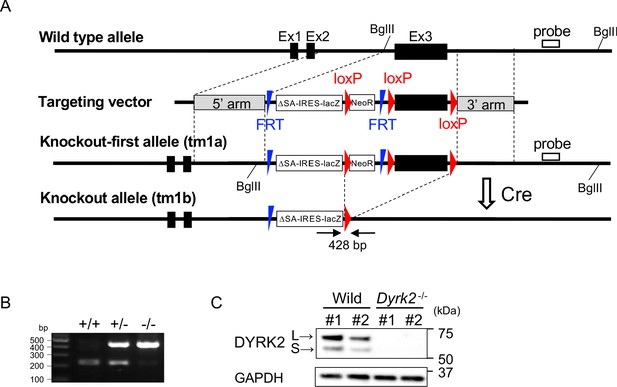
Generation of Dyrk2-/- mice schematic representation of the Dyrk2-/- allele (Dyrk2tm1b).
(A) Cre-mediated recombination was used to generate the Dyrk2-/- allele (Dyrk2tm1b) from the floxed allele (Dyrk2tm1a). The black boxes, red arrowheads, and blue arrowheads indicate exons, loxP sites, and FRT sites, respectively. (B) PCR-confirmed mutagenesis. (C) Immunoblotting of DYRK2 in extracts from each wild-type and Dyrk2-/- embryo at E13.5. L and S indicate long and short transcriptional isoforms of DYRK2, respectively.
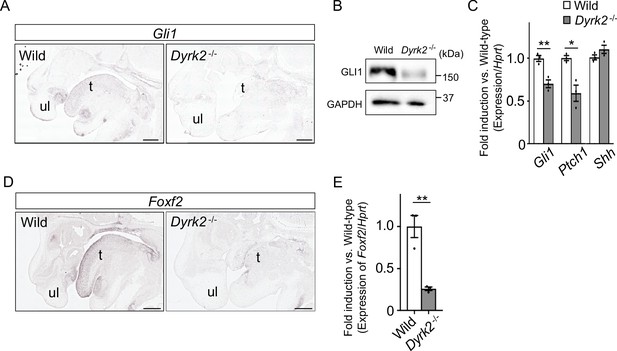
Deletion of DYRK2 affects activation of Hh signaling in mouse development.
(A) In situ hybridization of Gli1 in the craniofacial region in wild-type and Dyrk2-/- embryos from the sagittal plane at E14.5. (B) Immunoblotting of GLI1 in extracts from the limbs of wild-type and Dyrk2-/- embryos at E13.5. GAPDH serves as a loading control. (C) qPCR of Gli1, Ptch1, and Shh in the limbs from wild-type and Dyrk2-/- embryos at E13.5. (D, E) Repression of Foxf2-expression in the craniofacial region of Dyrk2-/- mice. (D) In situ hybridization of Foxf2 in the craniofacial region in wild-type and Dyrk2-/- embryos from the sagittal plane at E14.5. (E) qPCR of Foxf2 in the mandibular arch from wild-type and Dyrk2-/- embryos at E10.5. Hypoxanthine phosphoribosyltransferase (Hprt) in (C and E) was used as an internal standard, and fold change was calculated by comparing expression levels relative to those of wild-type. Data are presented as the means ± SEM (n = 3 biological replicates). The statistical significance between wild-type and Dyrk2-/- was determined by the Student’s t-test. (*) p<0.05, (**) p<0.01. t, tongue; ul, upper lip. Scale bars, 500 µm.
-
Figure 2—source data 1
Source data for Figure 2C and E.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig2-data1-v1.xlsx
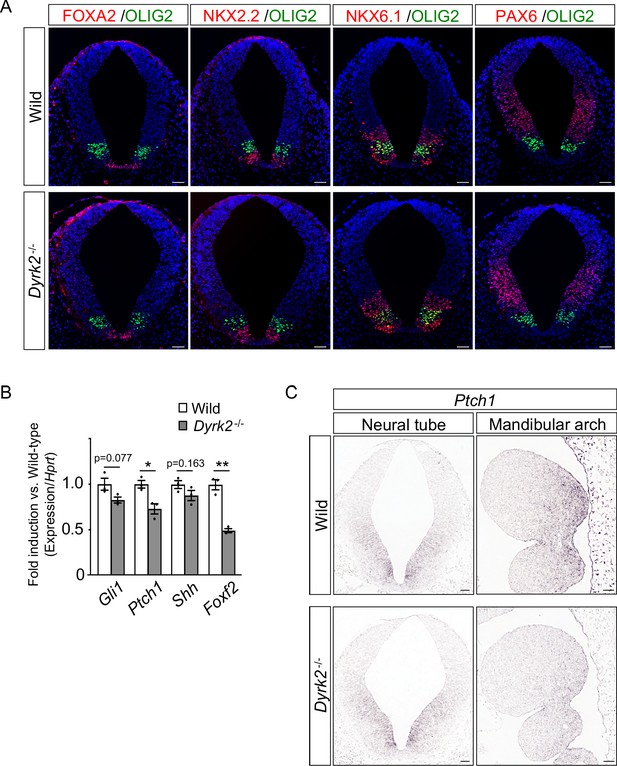
Dorsal-ventral patterning of the neural tube in Dyrk2-/- mice.
(A) Transverse sections of wild-type and Dyrk2-/- embryos at E10.5 (at the branchial level) were stained for markers of ventral (FOXA2, NKX2.2, OLIG2, and NKX6.1) and dorsal (PAX6) regions. Nuclei were stained with DAPI (blue). (B) qPCR of Gli1, Ptch1, Shh, and Foxf2 in the whole embryos from wild-type and Dyrk2-/- embryos at E9.5. Data are presented as the means ± SEM (n = 3 biological replicates). The statistical significance between wild-type and Dyrk2-/- was determined by the Student’s t-test. (*) p<0.05, (**) p<0.01. (C) In situ hybridization of Ptch1 in the neural tube (left panels) and mandibular arch (right panels) in wild-type and Dyrk2-/- embryos at E10.5 from the transverse and sagittal plane, respectively. Scale bars, 50 µm.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig2-figsupp1-data1-v1.xlsx
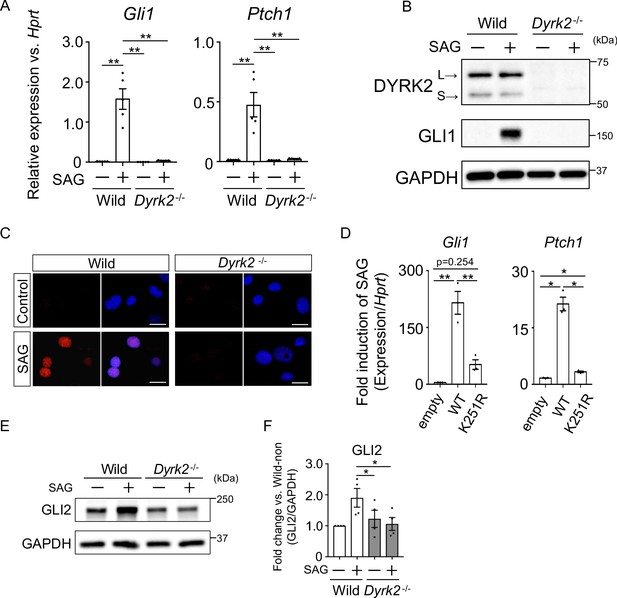
Deletion of Dyrk2 suppresses activation of Hh signaling in vitro.
(A) Expression of the Hh target genes Gli1 and Ptch1 in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG was measured by qPCR. Data are shown as relative expression to Hprt. (B) Protein levels of GLI1 and DYRK2 in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG were measured by immuno-blotting. L and S indicate long and short transcriptional isoforms of DYRK2, respectively. (C) Wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG were immune-cytostained for GLI1 (red). Nuclei were stained with DAPI (blue). Scale bars, 5 µm. (D) Expression of Gli1 and Ptch1 in Dyrk2-/- MEFs overexpressing human DYRK2 or DYRK2-K251R (kinase dead) constructs via adenovirus infection was measured by qPCR. Data indicates fold induction of 100 nM SAG against vehicle after normalization to Hprt. (E, F) Immunoblotting for GLI2 in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG. Protein level as fold changes of GLI2 (E) was calculated by comparing protein levels relative to those of wild-type MEFs in the absence of SAG after normalization to the GAPDH loading control in (F). Data are presented as the means ± SEM (n = 5, 3, and 4 biological replicates per condition in A, D, and F, respectively). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (*) p<0.05, (**) p<0.01.
-
Figure 3—source data 1
Source data for Figure 3A and D.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Source data for Figure 3F.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig3-data2-v1.xlsx
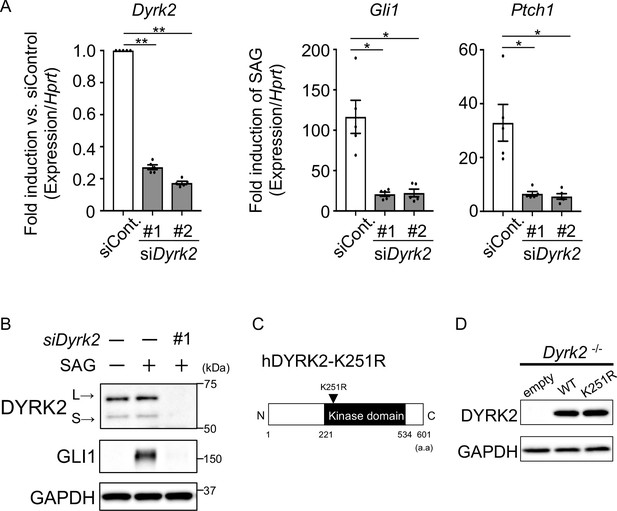
A transient knockdown of Dyrk2 suppresses activation of Hh signaling.
(A) Expression of Gli1 and Ptch1 in wild-type MEFs treated with two independent siDyrk2 for 48 hr was measured by qPCR. Hprt was used as an internal standard, and fold change of Dyrk2 was calculated by comparing expression levels relative to those of siControl. Data for Gli1 and Ptch1 indicate fold induction of 100 nM SAG against vehicle after normalization to Hprt. Data are presented as the means ± SEM (n = 5 biological replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (*) p<0.05, (**) p<0.01. (B) Protein levels of GLI1 and DYRK2 in wild-type MEFs treated with siDyrk2 for 48 hr in the absence or presence of 100 nM SAG were measured by immune-blotting. L and S indicate long and short transcriptional isoforms of DYRK2, respectively. (C) Schematic representation of a kinase dead human DYRK2 protein. (D) Immunoblotting for over-expressed short form of hDYRK2 or DYRK2-K251R (kinase dead) via adenovirus infection in Dyrk2-/- MEFs. GAPDH serves as a loading control.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig3-figsupp1-data1-v1.xlsx
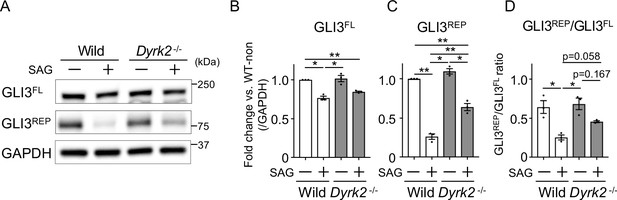
Deletion of Dyrk2 affects the stabilities of GLI3 Immuno-blotting for GLI3 in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG.
Protein levels as fold changes of GLI3FL, and GLI3REP (A) were calculated by comparing protein levels relative to those of wild-type MEFs in the absence of SAG after normalization to the GAPDH loading control in (B and C), respectively. The ratio of GLI3REP/GLI3FL was calculated directly according to each band intensity value (D). Data are presented as the means ± SEM (n = 3 biological replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (*) p<0.05, (**) p<0.01.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2B–D.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig3-figsupp2-data1-v1.xlsx
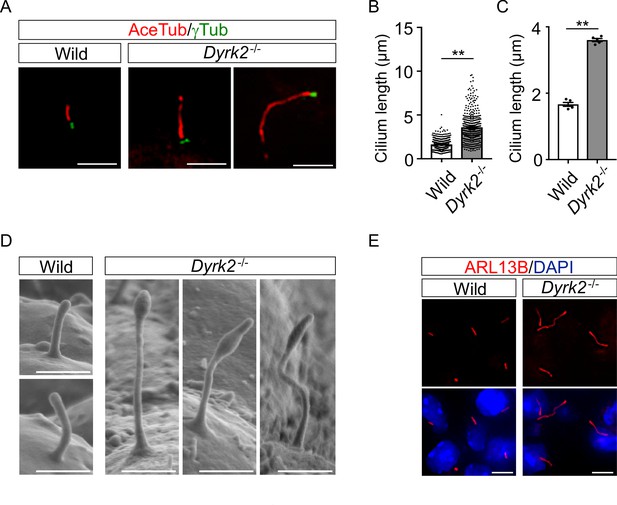
DYRK2 constrains the length of primary cilia.
(A–C) Elongation of primary cilia in Dyrk2-/- MEFs. Primary cilia of wild-type and Dyrk2-/- MEFs were immunostained with acetylated-tubulin and gamma-tubulin antibodies. (B, C) Measurements of cilia length in wild-type and Dyrk2-/- MEFs using acetylated-tubulin as a cilia axoneme marker. Cilia lengths are presented as pooled from five MEFs derived from independent embryos of each genotype (B) and the average of each MEF (C). Data are presented as the means ± SEM (n = 5 biological replicates per condition). The statistical significance between wild-type and Dyrk2-/- was determined by the Student’s t-test. (**) p<0.01. (D) Scanning electron microscopy showing wild-type and Dyrk2-/- embryos in the frontonasal prominence at E10.5. (E) Immunohistochemistry of primary cilia in wild-type and Dyrk2-/- embryos. ARL13B was immuno-stained in wild-type and Dyrk2-/- mesenchymal cells at the craniofacial region at E13.5. Nuclei were stained with DAPI. Scale bars, 5 µm (A and E) and 1 µm (D).
-
Figure 4—source data 1
Source data for Figure 4B–C.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig4-data1-v1.xlsx
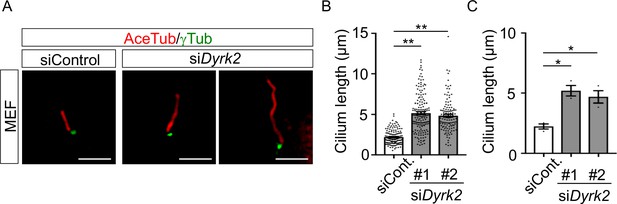
Elongation of primary cilia in wild-type MEFs treated with siDyrk2.
(A) Primary cilia of wild-type and Dyrk2-/- MEFs treated with siControl or two independent siDyrk2 were immunostained with acetylated-tubulin and gamma-tubulin antibodies. Scale bars, 5 µm. (B, C) Measurements of cilia length in wild-type MEFs treated with siControl or siDyrk2 using acetylated-tubulin as a cilia axoneme marker. Cilia lengths are presented as pooled from three MEFs derived from independent wild-type embryos (B) and represent the average of each MEF (C). Data are presented as the means ± SEM (n = 3 biological replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (*) p<0.05, (**) p<0.01.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1B–C.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig4-figsupp1-data1-v1.xlsx
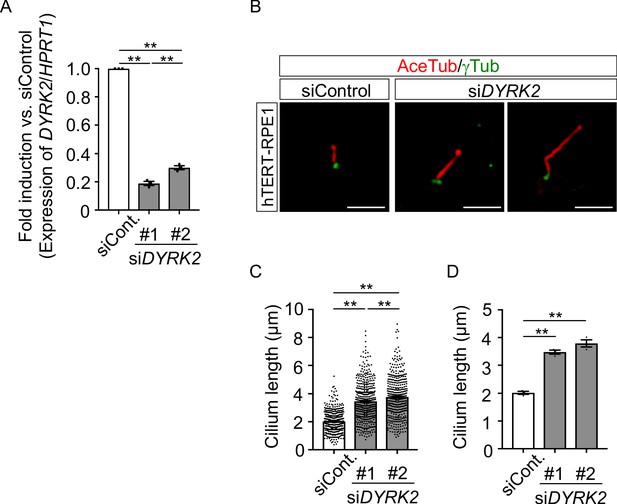
Elongation of primary cilia in hTERT-RPE1 cells treated with siDYRK2.
(A) Knockdown efficiency of DYRK2-expression in hTERT-RPE1 cells treated with two independent siDYRK2 for 48 hr was measured by qPCR. HPRT1 was used as an internal standard, and fold change was calculated by comparing expression levels relative to those of siControl. (B) Primary cilia of hTERT-RPE1 cells treated with siControl or siDYRK2 were immunostained with acetylated-tubulin and gamma-tubulin antibodies. (C, D) Measurements of cilia length in hTERT-RPE1 cells treated with siControl or two independent siDYRK2 using acetylated-tubulin as a cilia axoneme marker. Scale bars, 5 µm. Cilia lengths are presented as pooled from three independent experiments (C) and represent the average of each condition (D). Data are presented as the means ± SEM (n = 3 replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (**) p<0.01.
-
Figure 4—figure supplement 2—source data 1
Source data for Figure 4—figure supplement 2A and C–D.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig4-figsupp2-data1-v1.xlsx
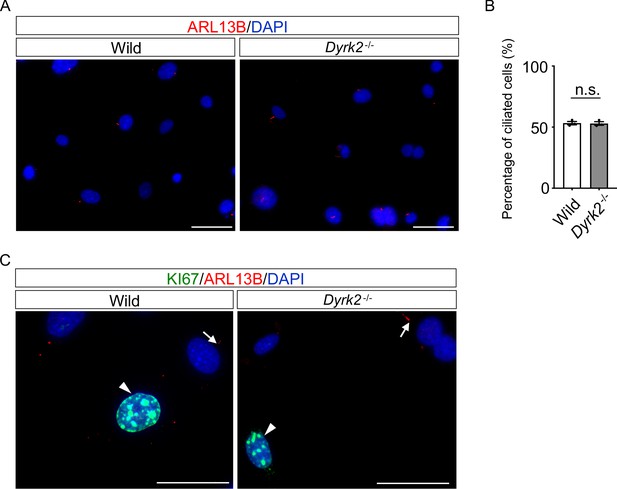
Quantification of the proportion of ciliated cells in wild-type and Dyrk2-/- MEFs.
(A, B) Proportion of ciliated cells in wild-type and Dyrk2-/- MEFs. Primary cilia of wild-type and Dyrk2-/- MEFs were immunostained with ARL13B (A). Measurements of proportion of ciliated cells in wild-type and Dyrk2-/- MEFs using ARL13B as a cilia axoneme marker (B). Data are presented as the means ± SEM (n = 3 biological replicates per condition;>150 cells were scored for each experiment). The statistical significance between wild-type and Dyrk2-/- was determined by the Student’s t-test. (C) Proportion of ciliated cells in cell-cycling wild-type and Dyrk2-/- MEFs. Wild-type and Dyrk2-/- MEFs cultured under 10% FBS containing medium at low density were immunostained with KI67 and ARL13B. Nuclei were stained with DAPI. Arrowheads and arrows indicate non-ciliated/KI67-positive cycling cells and ciliated/KI67-negative ones, respectively. Scale bars, 50 µm.
-
Figure 4—figure supplement 3—source data 1
Source data for Figure 4—figure supplement 3B.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig4-figsupp3-data1-v1.xlsx
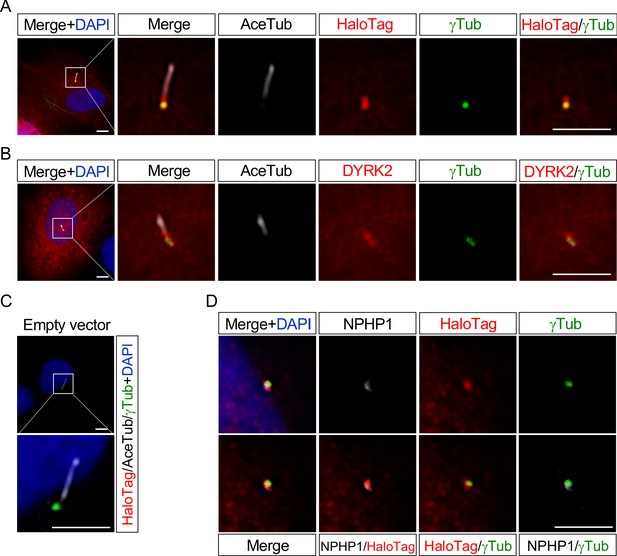
DYRK2 localizes at basal bodies and transition zone (TZ) in primary cilia.
Cultured hTERT-RPE1 cells were transfected with a mouse DYRK2-HaloTag overexpression construct and immunostained using anti-HaloTag (A) or anti-DYRK2 (B) with acetylated-tubulin (white) and gamma-tubulin antibodies. (C) Cultured hTERT-RPE1 cells transfected with an empty vector (pFN22K-Halo Tag-CMVd1-Flexi-vector) and immunostained using anti-HaloTag with acetylated-tubulin (white) and gamma-tubulin antibodies. (D) Co-localization of DYRK2 and a TZ marker, NPHP1. Cultured hTERT-RPE1 cells overexpressed with a mouse DYRK2-HaloTag were immunostained using anti-HaloTag, NPHP1 (white), and gamma-tubulin antibodies. Nuclei were stained with DAPI. Scale bars, 5 µm.
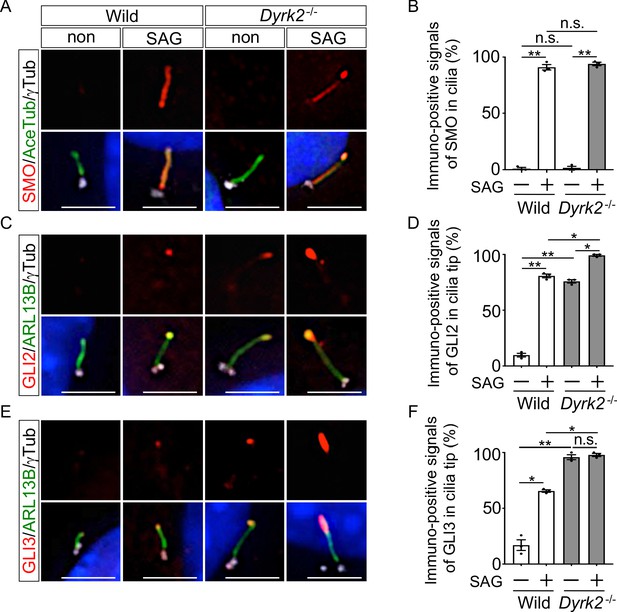
Depletion of Dyrk2 induces abnormal ciliary trafficking of endogenous Hh components.
Ciliary localization of endogenous SMO, GLI2, and GLI3 in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG. Primary cilia were immuno-stained for SMO (A), GLI2 (C), or GLI3 (E) with ARL13B and gamma-tubulin (white) antibodies. Nuclei were stained with DAPI (blue). The percentage of cells with SMO (B) at the cilia or foci of GLI2 (D) or GLI3 (F) at the cilia tips was determined. Data are presented as the means ± SEM (n = 3 biological replicates for each condition;>110 cells were scored for each experiment). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (*) p<0.05, (**) p<0.01. Scale bars, 5 µm.
-
Figure 6—source data 1
Source data for Figure 6B,D and F.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig6-data1-v1.xlsx
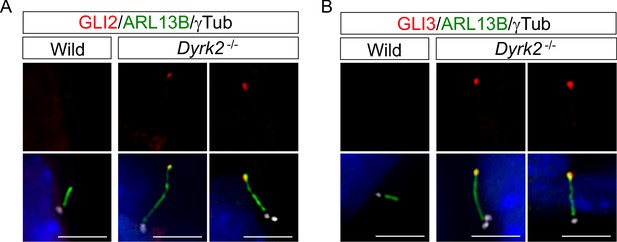
Depletion of Dyrk2 induces abnormal ciliary trafficking of endogenous GLI2 and GLI3 in vivo.
Immunohistochemistry for GLI2 and GLI3 in wild-type and Dyrk2-/- mesenchymal cells in the craniofacial region at E10.5 tissues. Primary cilia were immuno-stained for GLI2 (A) or GLI3 (B) with ARL13B and gamma-tubulin (white) antibodies. Nuclei were stained with DAPI (blue). Scale bars, 5 µm.
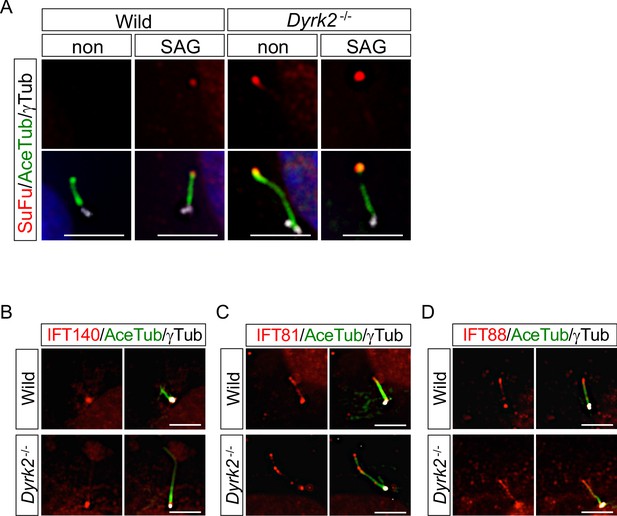
Immunocytochemistry of endogenous SuFu and IFTs.
(A) Ciliary localization of endogenous SuFu in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG. Primary cilia were immunostained for SuFu with ARL13B and gamma-tubulin (white) antibodies. (B–D) Ciliary localization of endogenous IFTs in wild-type and Dyrk2-/- MEFs. Primary cilia were immuno-stained for IFT140 (B), IFT81 (C), or IFT88 (D) with acetylated-tubulin and gamma-tubulin (white) antibodies. Nuclei were stained with DAPI (blue). Scale bars, 5 µm.
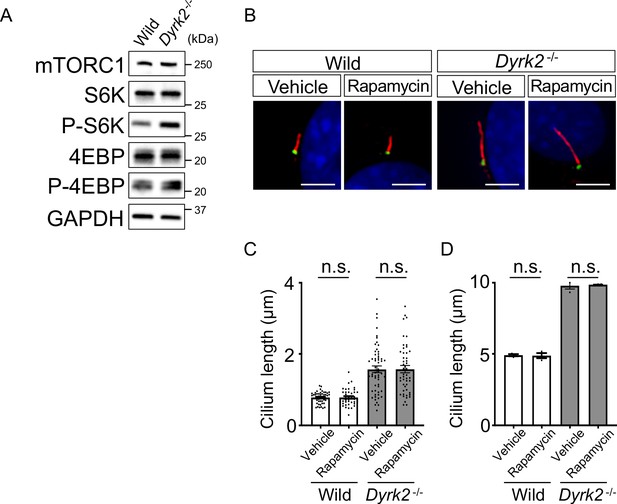
Effects of rapamycin treatment on cilia.
(A) Phosphorylated protein levels of S6K and 4EBP in wild-type and Dyrk2-/- MEFs were measured by immunoblotting. GAPDH serves as a loading control. (B) Primary cilia in wild-type and Dyrk2-/- MEFs treated with vehicle (DMSO) or 0.5 µM rapamycin for 24 hr were immunostained with acetylated-tubulin (red) and gamma-tubulin (green) antibodies. Nuclei were stained with DAPI (blue). Scale bars, 5 µm. (C, D) Measurements of cilia length in wild-type and Dyrk2-/- MEFs treated with vehicle (DMSO) or 0.5 µM rapamycin using acetylated-tubulin as a cilia axoneme marker. Cilia lengths are presented as pooled from three MEFs derived from independent embryos of each genotype (C) and the average of each MEF (D). Data are presented as the means ± SEM (n = 3 biological replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test.
-
Figure 6—figure supplement 3—source data 1
Source data for Figure 6—figure supplement 3C–D.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig6-figsupp3-data1-v1.xlsx
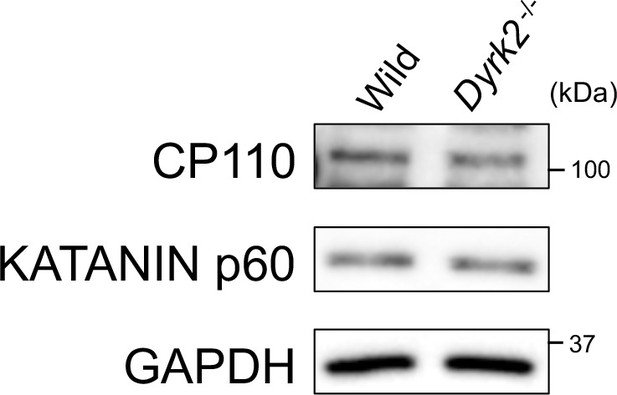
Protein levels of CP110 and KATANIN p60 in Dyrk2-/- MEFs.
Protein levels of CP110 and KATANIN p60 in wild-type and Dyrk2-/- were measured by immune-blotting. GAPDH serves as a loading control.
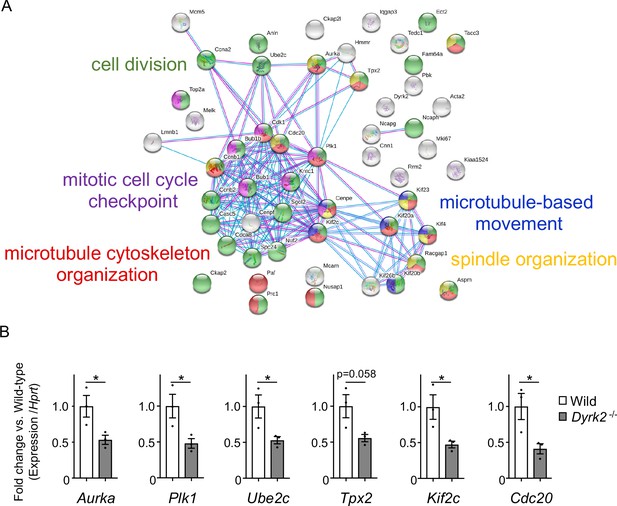
Changes in mRNA expression of genes in Dyrk2-/- MEFs.
(A) STRING GO analyses of the 53 differentially downregulated genes in Dyrk2-/- MEFs reveals protein-protein interaction networks. Robust networks for cell division (green, GO: 0051301), microtubule cytoskeleton organization (red, GO:0000226), spindle organization (yellow, GO:0007051), mitotic cell cycle checkpoint function (purple, GO:0007093), and microtubule-based movement (blue, GO:0007018) were extracted. (B) Confirmation of downregulation of genes related to ciliary resorption mechanisms in Dyrk2-/- MEFs by qPCR. Hprt was used as an internal standard, and fold change was calculated by comparing expression levels relative to those of wild-type. Data are presented as the means ± SEM (n = 3 biological replicates per condition). The statistical significance between wild-type and Dyrk2-/- MEFs was determined using the Student’s t-test. (*) p<0.05.
-
Figure 7—source data 1
Source data for Figure 7B.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig7-data1-v1.xlsx
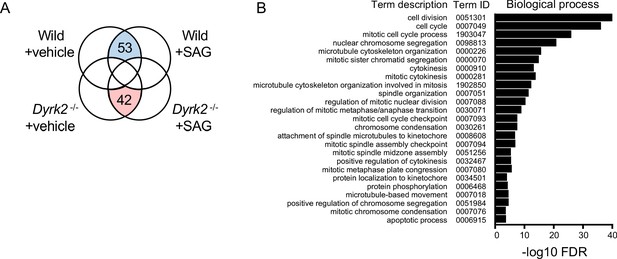
Transcriptome analysis in Dyrk2-/- MEFs.
(A) Venn diagrams revealing the similarities and differences among genes that were differentially expressed more than 1.5-fold from RNA-seq experiments in wild-type and Dyrk2-/- MEFs in the absence or presence of 100 nM SAG. (B) Significantly enriched gene ontology terms belonging to ‘biological process’ (false discovery rate: FDR < 0.005) among 53 differentially downregulated genes in Dyrk2-/- MEFs.
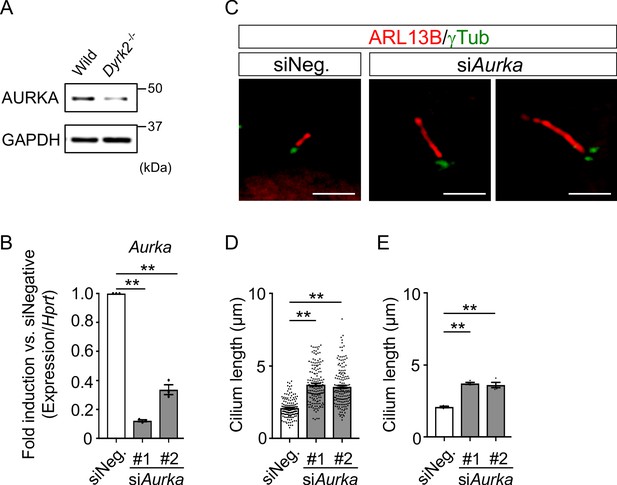
Elongation of primary cilia in wild-type MEFs treated with siAurka.
(A) Immunoblotting of AURKA in wild-type and Dyrk2-/- MEFs. GAPDH serves as a loading control. (B) Knockdown efficiency of Aurka-expression in wild-type MEFs treated with two independent siAurka for 48 hr was measured by qPCR. Hprt was used as an internal standard, and fold change was calculated by comparing expression levels relative to those of siNegative (siNeg.). Data are presented as the means ± SEM (n = 3 biological replicates per condition). (C) Primary cilia in wild-type cells treated with siNegative (siNeg.) or two independent siAurka were immuno-stained with ARL13B and gamma-tubulin antibodies. Scale bars, 5 µm. (D, E) Measurements of cilia length in wild-type MEFs treated with siNeg. or two independent siAurka using ARL13B and acetylated-tubulin as a cilia axoneme marker. Cilia lengths are presented as pooled from four MEFs derived from independent wild-type embryos (D) and represent an average of each MEF (E). Data are presented as the means ± SEM (n = 4 biological replicates per condition). The statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test. (**) p<0.01.
-
Figure 8—source data 1
Source data for Figure 8B and D–E.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig8-data1-v1.xlsx
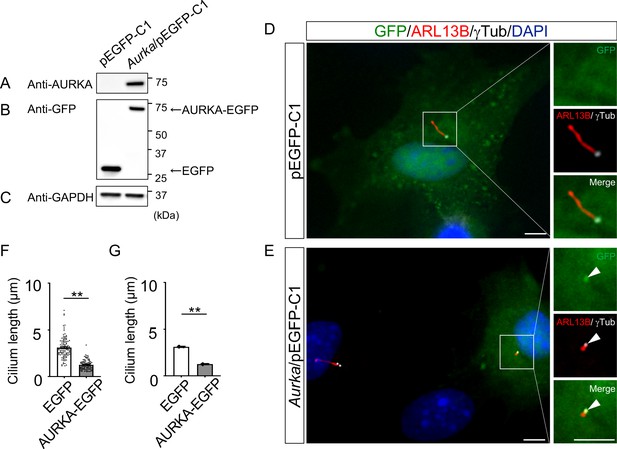
Reduction of the length of primary cilia in Dyrk2-/- MEFs by over-expression of AURKA.
(A–C) Immunoblotting by anti-AURKA (A), anti-GFP (B), and anti-GAPDH (C) in cells transfected with pEGFP-C1 or mouse Aurka/pEGFP-C1. GAPDH serves as a loading control. (D, E) Primary cilia in Dyrk2-/- MEFs over-expressed with EGFP (D) or AURKA-EGFP (E) were immunostained with GFP, ARL13B, and gamma-tubulin (white) antibodies. Arrowheads in (E) indicate signals for AURKA-EGFP in gamma-tubulin-positive basal body. Scale bars, 5 µm. (F, G) Measurements of cilia length in EGFP- or AURKA-EGFP-over-expressed Dyrk2-/- MEFs using ARL13B as a cilia axoneme marker. Cilia lengths in EGFP- or AURKA-EGFP-positive cells are presented as pooled from three MEFs derived from independent Dyrk2-/- embryos (F) and represent an average of each MEF (G). Data are presented as the means ± SEM (n = 3 biological replicates per condition). The statistical significance between EGFP- and AURKA-EGFP-positive cells was determined by the Student’s t-test. (**) p<0.01.
-
Figure 9—source data 1
Source data for Figure 9F and G.
- https://cdn.elifesciences.org/articles/57381/elife-57381-fig9-data1-v1.xlsx
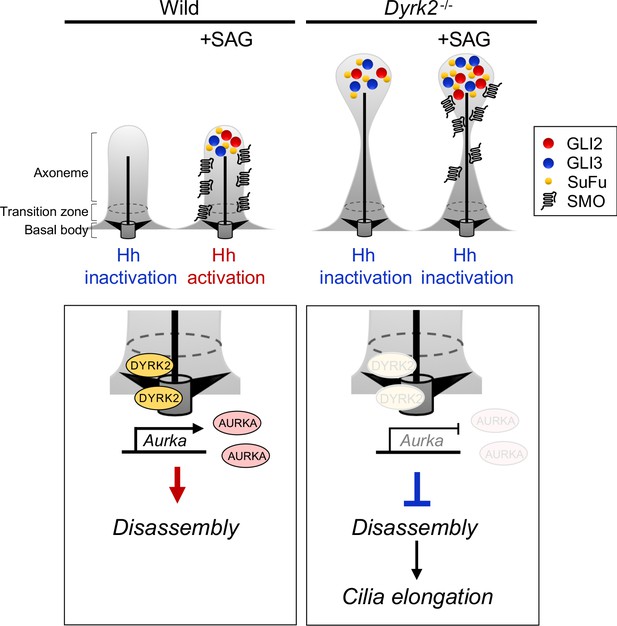
Schematic representation of DYRK2 in ciliogenesis and Hh signaling.
(Left panel) A schematic model of normal ciliogenesis and response to stimulation with Hh ligand. (Right panel) A schematic model ciliogenesis and response to stimulation with Hh ligand in Dyrk2-deletion. The morphology of primary cilia in Dyrk2-/- MEFs was elongated and often bulged at the tips. In Dyrk2-/- cells, downregulation of Aurka and other ciliary disassembly genes caused suppression of disassembly and elongation of primary cilia. Furthermore, abnormal ciliary trafficking caused accumulation of GLI2, GLI3, and SuFu in Dyrk2-/- cells. Consequently, the induction of Hh signaling is drastically suppressed by deletion of Dyrk2.
Tables
A list of downregulated or upregulated genes in Dyrk2-/- MEFs
| Down-regulated genes in Dyrk2-/- | ||||
|---|---|---|---|---|
| ID | GeneSymbol | Description | Ratio of Dyrk2-/-per wild-type in the presence of SAG | Ratio of Dyrk2-/-per wild-type in the absence of SAG |
| ENSMUSG00000028630 | Dyrk2 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | 0.02 | 0.03 |
| ENSMUSG00000035683 | Melk | Maternal embryonic leucine zipper kinase | 0.23 | 0.22 |
| ENSMUSG00000074476 | Spc24 | NDC80 kinetochore complex component%2C homolog (S. cerevisiae) | 0.25 | 0.21 |
| ENSMUSG00000020808 | Pimreg | PICALM interacting mitotic regulator | 0.28 | 0.28 |
| ENSMUSG00000033952 | Aspm | Abnormal spindle microtubule assembly | 0.31 | 0.25 |
| ENSMUSG00000026683 | Nuf2 | NDC80 kinetochore complex component | 0.31 | 0.30 |
| ENSMUSG00000037466 | Tedc1 | Tubulin epsilon and delta complex 1 | 0.31 | 0.26 |
| ENSMUSG00000030867 | Plk1 | Polo-like kinase 1 | 0.31 | 0.17 |
| ENSMUSG00000022033 | Pbk | PDZ binding kinase | 0.33 | 0.29 |
| ENSMUSG00000027326 | Knl1 | Kinetochore scaffold 1 | 0.33 | 0.20 |
| ENSMUSG00000041431 | Ccnb1 | Cyclin B1 | 0.33 | 0.26 |
| ENSMUSG00000036777 | Anln | Anillin actin binding protein | 0.33 | 0.26 |
| ENSMUSG00000001403 | Ube2c | Ubiquitin-conjugating enzyme E2C | 0.33 | 0.25 |
| ENSMUSG00000027496 | Aurka | Aurora kinase A | 0.34 | 0.26 |
| ENSMUSG00000001349 | Cnn1 | Calponin 1 | 0.34 | 0.31 |
| ENSMUSG00000032218 | Ccnb2 | Cyclin B2 | 0.34 | 0.28 |
| ENSMUSG00000026039 | Sgo2a | Shugoshin 2A | 0.34 | 0.25 |
| ENSMUSG00000015880 | Ncapg | Non-SMC condensin I complex subunit G | 0.34 | 0.34 |
| ENSMUSG00000027379 | Bub1 | BUB1 mitotic checkpoint serine/threonine kinase | 0.36 | 0.23 |
| ENSMUSG00000040084 | Bub1b | BUB1B mitotic checkpoint serine/threonine kinase | 0.36 | 0.29 |
| ENSMUSG00000045328 | Cenpe | Centromere protein E | 0.36 | 0.22 |
| ENSMUSG00000032254 | Kif23 | Kinesin family member 23 | 0.37 | 0.25 |
| ENSMUSG00000028873 | Cdca8 | Cell division cycle associated 8 | 0.37 | 0.30 |
| ENSMUSG00000032135 | Mcam | Melanoma cell adhesion molecule | 0.37 | 0.29 |
| ENSMUSG00000027469 | Tpx2 | TPX2microtubule-associated | 0.37 | 0.33 |
| ENSMUSG00000028678 | Kif2c | Kinesin family member 2C | 0.37 | 0.24 |
| ENSMUSG00000027715 | Ccna2 | Cyclin A2 | 0.38 | 0.23 |
| ENSMUSG00000048327 | Ckap2l | Cytoskeleton associated protein 2-like | 0.39 | 0.23 |
| ENSMUSG00000040204 | Pclaf | PCNA clamp associated factor | 0.40 | 0.19 |
| ENSMUSG00000029414 | Kntc1 | Kinetochore associated 1 | 0.42 | 0.24 |
| ENSMUSG00000034311 | Kif4 | Kinesin family member 4 | 0.42 | 0.24 |
| ENSMUSG00000031004 | Mki67 | Antigen identified by monoclonal antibody Ki 67 | 0.42 | 0.21 |
| ENSMUSG00000020914 | Top2a | Topoisomerase (DNA) II alpha | 0.42 | 0.21 |
| ENSMUSG00000033031 | Cip2a | Cell proliferation regulating inhibitor of protein phosphatase 2A | 0.42 | 0.32 |
| ENSMUSG00000035783 | Acta2 | Actin alpha two smooth muscle aorta | 0.43 | 0.48 |
| ENSMUSG00000024795 | Kif20b | Kinesin family member 20B | 0.43 | 0.30 |
| ENSMUSG00000038943 | Prc1 | Protein regulator of cytokinesis 1 | 0.43 | 0.26 |
| ENSMUSG00000026494 | Kif26b | Kinesin family member 26B | 0.43 | 0.25 |
| ENSMUSG00000023015 | Racgap1 | Rac GTPase-activating protein 1 | 0.43 | 0.26 |
| ENSMUSG00000026605 | Cenpf | Centromere protein F | 0.44 | 0.25 |
| ENSMUSG00000027306 | Nusap1 | Nucleolar and spindle associated protein 1 | 0.45 | 0.28 |
| ENSMUSG00000028068 | Iqgap3 | IQ motif containing GTPase activating protein 3 | 0.46 | 0.21 |
| ENSMUSG00000003779 | Kif20a | Kinesin family member 20A | 0.47 | 0.25 |
| ENSMUSG00000005410 | Mcm5 | Minichromosome maintenance complex component 5 | 0.47 | 0.26 |
| ENSMUSG00000034906 | Ncaph | Non-SMC condensin I complex subunit H | 0.47 | 0.27 |
| ENSMUSG00000006398 | Cdc20 | Cell division cycle 20 | 0.48 | 0.29 |
| ENSMUSG00000037313 | Tacc3 | Transforming acidic coiled-coil containing protein 3 | 0.48 | 0.36 |
| ENSMUSG00000027699 | Ect2 | ect2 oncogene | 0.48 | 0.26 |
| ENSMUSG00000020330 | Hmmr | Hyaluronan-mediated motility receptor (RHAMM) | 0.50 | 0.28 |
| ENSMUSG00000020649 | Rrm2 | Ribonucleotide reductase M2 | 0.50 | 0.26 |
| ENSMUSG00000019942 | Cdk1 | Cyclin-dependent kinase 1 | 0.50 | 0.34 |
| ENSMUSG00000024590 | Lmnb1 | Lamin B1 | 0.51 | 0.33 |
| ENSMUSG00000037725 | Ckap2 | Cytoskeleton associated protein 2 | 0.55 | 0.42 |
| Upregulated genes in Dyrk2-/- | ||||
| ID | GeneSymbol | Description | Ratio of Dyrk2-/-per wild-type in the presence of SAG | Ratio of Dyrk2-/-per wild-type in the absence of SAG |
| ENSMUSG00000056673 | Kdm5d | Lysine (K)-specific demethylase 5D | Inf | Inf |
| ENSMUSG00000068457 | Uty | Ubiquitously transcribed tetratricopeptide repeat gene Y chromosome | Inf | Inf |
| ENSMUSG00000069049 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3 Y-linked | Inf | 8278 |
| ENSMUSG00000069045 | Eif2s3y | Eukaryotic translation initiation factor 2 subunit three structural gene Y-linked | Inf | Inf |
| ENSMUSG00000112616 | Gm47434 | Predicted gene 47434 | 719 | Inf |
| ENSMUSG00000025582 | Nptx1 | Neuronal pentraxin 1 | 4.74 | 11.91 |
| ENSMUSG00000024164 | C3 | Complement component 3 | 4.47 | 11.59 |
| ENSMUSG00000039457 | Ppl | Periplakin | 4.30 | 11.11 |
| ENSMUSG00000025784 | Clec3b | C-type lectin domain family three member b | 3.99 | 8.60 |
| ENSMUSG00000002944 | Cd36 | CD36 molecule | 3.20 | 3.45 |
| ENSMUSG00000035385 | Ccl2 | Chemokine (C-C motif) ligand 2 | 2.86 | 2.84 |
| ENSMUSG00000095478 | Gm9824 | Predicted pseudogene 9824 | 2.60 | 4.14 |
| ENSMUSG00000038642 | Ctss | Cathepsin S | 2.58 | 3.19 |
| ENSMUSG00000043719 | Col6a6 | Collagen type VI alpha 6 | 2.44 | 4.64 |
| ENSMUSG00000033327 | Tnxb | Tenascin XB | 2.37 | 3.61 |
| ENSMUSG00000069516 | Lyz2 | Lysozyme 2 | 2.30 | 3.08 |
| ENSMUSG00000016494 | Cd34 | CD34 antigen | 2.29 | 2.26 |
| ENSMUSG00000042129 | Rassf4 | Ras association (RalGDS/AF-6) domain family member 4 | 2.29 | 3.43 |
| ENSMUSG00000004730 | Adgre1 | Adhesion G-protein-coupled receptor E1 | 2.27 | 2.49 |
| ENSMUSG00000030144 | Clec4d | C-type lectin domain family member d | 2.26 | 3.74 |
| ENSMUSG00000029816 | Gpnmb | Glycoprotein (transmembrane) nmb | 2.22 | 2.66 |
| ENSMUSG00000042286 | Stab1 | Stabilin 1 | 2.18 | 2.70 |
| ENSMUSG00000020120 | Plek | Pleckstrin | 2.18 | 2.99 |
| ENSMUSG00000040254 | Sema3d | Sema domain immunoglobulin domain (Ig) short basic domain secreted (semaphorin) 3D | 2.17 | 2.89 |
| ENSMUSG00000005268 | Prlr | Prolactin receptor | 2.17 | 4.44 |
| ENSMUSG00000024621 | Csf1r | Colony-stimulating factor one receptor | 2.10 | 2.74 |
| ENSMUSG00000074896 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | 2.04 | 3.96 |
| ENSMUSG00000002985 | Apoe | Apolipoprotein E | 2.03 | 2.51 |
| ENSMUSG00000057137 | Tmem140 | Transmembrane protein 140 | 2.02 | 3.18 |
| ENSMUSG00000002289 | Angptl4 | Angiopoietin-like 4 | 2.02 | 5.94 |
| ENSMUSG00000050335 | Lgals3 | Lectin galactose binding soluble 3 | 1.99 | 2.66 |
| ENSMUSG00000090877 | Hspa1b | Heat-shock protein 1B | 1.98 | 2.13 |
| ENSMUSG00000054404 | Slfn5 | Schlafen 5 | 1.96 | 3.77 |
| ENSMUSG00000031209 | Heph | Hephaestin | 1.92 | 2.48 |
| ENSMUSG00000027996 | Sfrp2 | Secreted frizzled-related protein 2 | 1.91 | 5.68 |
| ENSMUSG00000050953 | Gja1 | Gap junction protein alpha 1 | 1.90 | 2.45 |
| ENSMUSG00000005413 | Hmox1 | Heme oxygenase 1 | 1.90 | 1.97 |
| ENSMUSG00000046805 | Mpeg1 | Macrophage expressed gene 1 | 1.85 | 2.57 |
| ENSMUSG00000022037 | Clu | Clusterin | 1.83 | 3.06 |
| ENSMUSG00000026389 | Steap3 | STEAP family member 3 | 1.81 | 2.24 |
| ENSMUSG00000041577 | Prelp | Proline arginine-rich end leucine-rich repeat | 1.81 | 2.01 |
| ENSMUSG00000027339 | Rassf2 | Ras association (RalGDS/AF-6) domain family member 2 | 1.80 | 2.72 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Dyrk2-/- mouse | This paper | N/A | Maintained in K. Yoshida lab. |
| Cell line (M. musculus) | Wild-type and Dyrk2-/- MEFs | This paper | N/A | Maintained in K. Yoshida lab. |
| Cell line (H. sapiens) | hTERT-RPE1 | ATCC | Cat# CRL-4000 RRID:CVCL_4388 | |
| Transfected construct (M. musculus) | mouse Aurka/pEGFP-C1 | This paper | N/A | See Materials and methods subsection ‘Plasmid constructs’ |
| Transfected construct (M. musculus) | mouse Dyrk2/FN22K-Halo Tag-CMVd1-Flexi-vector | This paper | N/A | See Materials and methods subsection ‘Plasmid constructs’ |
| Transfected construct (M. musculus) | Dyrk2 targeting vector | Knockout Mouse Project Repository | PG00105_X_1_G09, PG00105_X_1_E04 | See Materials and methods subsection ‘Plasmid constructs’ |
| Recombinant DNA regent | Plasmid pEGFP-C1 (empty vector) | TaKaRa Bio | Cat# 6084–1 | |
| Recombinant DNA regent | Plasmid pFN22K-Halo Tag-CMVd1-Flexi-vector (empty vector) | Promega | Cat# G2851 | |
| Transfected construct (M. musculus) | Dyrk2 targeting vector | Knockout Mouse Project Repository | PG00105_X_1_G09, PG00105_X_1_E04 | |
| Biological sample (Adenovirus) | Adenovirus-Cre | Yokoyama-Mashima et al., 2019 doi: 10.1016/j.canlet.2019.02.046. | N/A | |
| Biological sample (Adenovirus) | Adenovirus-human DYRK2 | Yokoyama-Mashima et al., 2019 doi: 10.1016/j.canlet.2019.02.046. | N/A | |
| Biological sample (Adenovirus) | Adenovirus-human DYRK2-K251R | Yokoyama-Mashima et al., 2019 doi: 10.1016/j.canlet.2019.02.046. | N/A | |
| Biological sample (Adenovirus) | Adenovirus-GFP | Yokoyama-Mashima et al., 2019 doi: 10.1016/j.canlet.2019.02.046. | N/A | |
| Antibody | Anti-Acetylated-tubulin (Mouse monoclonal) | Sigma-Aldrich | Cat# T7451, RRID:AB_609894 | ICC (1:2000) |
| Antibody | Anti-ARL13B (Mouse monoclonal) | Abcam | Cat# ab136648,N/A | ICC (1:300) |
| Antibody | Anti-ARL13B (Rabbit polyclonal) | Proteintech | Cat# 17711–1-AP, RRID:AB_2060867 | ICC (1:400) |
| IHC (1:400) | ||||
| Antibody | Anti-AURKA (Mouse monoclonal) | BD Transduction | Cat# 610938, RRID:AB_398251 | WB (1:1000) |
| Antibody | Anti-DYRK2 (Rabbit polyclonal) | Sigma-Aldrich | Cat# HPA027230, RRID:AB_1847925 | WB (1:1000) |
| ICC (1:400) | ||||
| Antibody | Anti-FOXA2 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 4C7, RRID:AB_528207 | IHC (1:8) |
| Antibody | Anti-CP110 (Rabbit polyclonal) | Proteintech | Cat# 12780–1-AP, RRID:AB_10638480 | WB (1:1000) |
| Antibody | Anti-GAPDH (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-32233, RRID:AB_627679 | WB (1:3000) |
| Antibody | Anti-GFP (Chicken polyclonal IgY) | Aves Labs | Cat# GFP-1020, RRID:AB_10000240 | ICC (1:500) |
| Antibody | Anti-GFP (Rabbit monoclonal) | Abcam | Cat# ab183734, RRID:AB_2732027 | WB (1:30000) |
| Antibody | Anti-GLI1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 2534, RRID:AB_2294745 | WB (1:500) |
| ICC (1:100) | ||||
| Antibody | Anti-GLI2 (Goat polyclonal) | R and D systems | Cat# AF3635, RRID:AB_2111902 | WB (1:500) |
| ICC (1:50) | ||||
| IHC (1:50) | ||||
| Antibody | Anti-GLI3 (Goat polyclonal) | R and D systems | Cat# AF3690, RRID:AB_2232499 | WB (1:200) |
| ICC (1:100) | ||||
| IHC (1:150) | ||||
| Antibody | Anti-gamma-tubulin (Goat polyclonal) | Santa Cruz Biotechnology | Cat# sc-7396, RRID:AB_2211262 | ICC (1:3500) |
| Antibody | Anti-gamma-tubulin (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-17787, RRID:AB_628417 | ICC (1:400) |
| IHC (1:400) | ||||
| Antibody | Anti-HaloTag (Rabbit polyclonal) | Promega | Cat# G9281, RRID:AB_713650 | ICC (1:700) |
| Antibody | Anti-IFT140 (Rabbit polyclonal) | Proteintech | Cat# 17460–1-AP, RRID:AB_2295648 | ICC (1:100) |
| Antibody | Anti-IFT81 (Rabbit polyclonal) | Proteintech | Cat# 11744–1-AP, RRID:AB_2121966 | ICC (1:50) |
| Antibody | Anti-IFT88 (Rabbit polyclonal) | Proteintech | Cat# 13967–1-AP, RRID:AB_2121979 | ICC (1:100) |
| Antibody | Anti-KATANIN p60 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-373814, RRID:AB_11014191 | WB (1:1000) |
| Antibody | Anti-KI67 (Rabbit monoclonal) | Abcam | Cat# ab16667, RRID:AB_302459 | ICC (1:500) |
| Antibody | Anti-NPHP1 (Mouse monoclonal) | SIGMA-Aldrich | Cat# MABS2185,N/A | ICC (1:100) |
| Antibody | Anti-mTORC1 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2972, RRID:AB_330978 | WB (1:1000) |
| Antibody | Anti-NKX2.2 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 74.5A5, RRID:AB_531794 | IHC (1:10) |
| Antibody | Anti-NKX6.1 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# F55A10, RRID:AB_532378 | IHC (1:100) |
| Antibody | Anti-OLIG2 (Rabbit monoclonal) | abcam | Cat# ab109186, RRID:AB_10861310 | IHC (1:500) |
| Antibody | Anti-PAX6 (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-81649, RRID:AB_1127044 | IHC (1:400) |
| Antibody | Anti-Phosho-S6 (Ser 235/236) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2211, RRID:AB_331679 | WB (1:2000) |
| Antibody | Anti-P-4EBP1(Thr 37/46) (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2855, RRID:AB_560835 | WB (1:1500) |
| Antibody | Anti-SMO (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-166685, RRID:AB_2239686 | ICC (1:100) |
| Antibody | Anti-SuFu (Mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-137014, RRID:AB_2197315 | ICC (1:100) |
| Antibody | Anti-S6 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 2217, RRID:AB_331355 | WB (1:2000) |
| Antibody | Anti-4EBP1 (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9644, RRID:AB_2097841 | WB (1:3000) |
| Sequence-based reagent | Human DYRK2 siRNA#1 | BEX | 608481 | |
| Sequence-based reagent | Human DYRK2 siRNA#2 | ThermoFisher Scientific | HSS112284 | |
| Sequence-based reagent | Mouse Dyrk2 siRNA#1 | ThermoFisher Scientific | 4390771 (s87545) | |
| Sequence-based reagent | Mouse Dyrk2 siRNA#2 | ThermoFisher Scientific | 4390771 (s87546) | |
| Sequence-based reagent | Mouse Aurka siRNA#1 | Integrated DNA Technologies | mm.Ri.Aurka.13.1 | |
| Sequence-based reagent | Mouse Aurka siRNA#2 | Integrated DNA Technologies | mm.Ri.Aurka.13.4 | |
| Sequence-based reagent | Mouse Cdc20 siRNA | Integrated DNA Technologies | mm.Ri.Cdc20.13.2 | |
| Sequence-based reagent | Mouse Kif2c siRNA | Integrated DNA Technologies | mm.Ri.Kif2c.13.3 | |
| Sequence-based reagent | Mouse Plk1 siRNA | Integrated DNA Technologies | mm.Ri.Plk1.13.1 | |
| Sequence-based reagent | Mouse Tpx2 siRNA | Integrated DNA Technologies | mm.Ri.Tpx2.13.1 | |
| Sequence-based reagent | Mouse Ube2c siRNA | Integrated DNA Technologies | mm.Ri.Ube2c.13.1 | |
| Sequence-based reagent | Negative Control DsiRNA (siNegative) | Integrated DNA Technologies | 51-01-14 | |
| Sequence-based reagent | Silencer Select Negative Control (siControl) | ThermoFisher Scientific | 4390843 | |
| Chemical compound, drug | InSolution SAG | Merck | 566660 | |
| Chemical compound, drug | Rapamycin | LC Laboratories | R-5000 | |
| Software, algorithm | BZ-X800 Analyzer | Keyence | BZ-X800 Analyzer | |
| Software, algorithm | Excel | Microsoft | Mac2019 | |
| Software, algorithm | Fusion | M and S Instruments | Fusion | |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software Inc | Mac OS X | |
| Software, algorithm | PikoReal Software 2.1 | ThermoFisher Scientific | PikoReal Software 2.1 |
List of primer sets.
| For genotyping | |||
|---|---|---|---|
| Gene | Sequence (5'→3') | Accession number | |
| Dyrk2 tm1b-WT | Forward | TGGGTCCAAATGCAAAGAAACGCCA | NC_000076.6 |
| Reverse | GCTTCTCGTTCCGCACCATCTTCAG | ||
| Dyrk2 tm1b-KO | Forward | CCTTCTCCCTCCTCCACTCTGACCCA | NC_000076.6 |
| Reverse | CCACACCTCCCCCTGAACCTGAAAC | ||
| For amplification of the probes for in situ hybridization or Southern blotting | |||
| Gene | Sequence (5'→3') | Accession number | |
| Mouse Foxf2 | Forward | GAGATTAACCCTCACTAAAGGGAGGTTATGGTGGCCTCGACAT | NM_010225.2 |
| Reverse | GAGTAATACGACTCACTATAGGGACACACACACCTCCCTTTTCA | ||
| Mouse Gli1 | Forward | GAGTATTTAGGTGACACTATAGAAGCAGGGAAGAGAGCAGACTG | NM_010296.2 |
| Reverse | GAGTAATACGACTCACTATAGGGGCTGAGTGTTGTCCAGGTC | ||
| Mouse Ptch1 | Forward | GAGATTAACCCTCACTAAAGGGACATGGCCTCGGCTGGTAAC | NM_008957.3 |
| Reverse | GAGTAATACGACTCACTATAGGGTGTACCCATGGCCAACTTCG | ||
| Southern for Dyrk2 | Forward | CTTCGAATCCTTTTATCCTTCAGGC | NC_000076.6 |
| Reverse | ACATCATGTTCATTGGTTTTGCTCT | ||
| For cloning | |||
| Gene | Sequence (5'→3') | Accession number | |
| Mouse Aurka CDS | Forward | GGACTCAGATCTCGAGACATGGCTGTTGAGGGCG | NM_011497.4 |
| Reverse | GTCGACTGCAGAATTCCTAAGATGATTTGCTGGTTG | ||
| Mouse Dyrk2 CDS | Forward | GTGCGCGATCGCCATGTTAACCAGGAAACCTTCGGC | NM_001014390.2 |
| Reverse | CTCCGTTTAAACGCTAACGAGTTTCGGCAACAC | ||
| For real-time PCR | |||
| Gene | Sequence (5'→3') | Accession number | |
| Human DYRK2 | Forward | GGGGAGAAAACGTCAGTGAA | NM_006482.3 |
| Reverse | TCTGCGCCAAATTAGTCCTC | ||
| Human HPRT1 | Forward | GGACTAATTATGGACAGGACTG | NM_000194.3 |
| Reverse | GCTCTTCAGTCTGATAAAATCTAC | ||
| Mouse Aurka | Forward | CACACGTACCAGGAGACTTACAGA | NM_011497.4 |
| Reverse | AGTCTTGAAATGAGGTCCCTGGCT | ||
| Mouse Cdc20 | Forward | GAGCTCAAAGGACACACAGC | NM_023223.2 |
| Reverse | GCCACAACCGTAGAGTCTCA | ||
| Mouse Dyrk2 | Forward | CTACCACTACAGCCCACACG | NM_001014390.2 |
| Reverse | TCTGTCCGTGGCTGTTGA | ||
| Mouse Foxf2 | Forward | AGCATGTCTTCCTACTCGTTG | NM_010225.2 |
| Reverse | TCTTTCCTGTCGCACACT | ||
| Mouse Gli1 | Forward | GCACCACATCAACAGTGAGC | NM_010296.2 |
| Reverse | GCGTCTTGAGGTTTTCAAGG | ||
| Mouse Hprt | Forward | CTCATGGACTGATTATGGACAGGAC | NM_013556.2 |
| Reverse | GCAGGTCAGCAAAGAACTTATAGCC | ||
| Mouse Kif2c | Forward | GAGAGCAAGCTGACCCAGG | NM_134471.4 |
| Reverse | CCTGGTGAGATCATGGCGATC | ||
| Mouse Plk1 | Forward | CCAAGCACATCAACCCAGTG | NM_011121.4 |
| Reverse | TGAGGCAGGTAATAGGGAGACG | ||
| Mouse Ptch1 | Forward | CTCTGGAGCAGATTTCCAAGG | NM_008957.3 |
| Reverse | TGCCGCAGTTCTTTTGAATG | ||
| Mouse Shh | Forward | GTGAAGCTGCGAGTGACCG | NM_009170.3 |
| Reverse | CCTGGTCGTCAGCCGCCAGCACGC | ||
| Mouse Tpx2 | Forward | GCGAGGTTGTCAGGTGTGTA | NM_001141977.1 |
| Reverse | TTGATAAAGTCGGTGGGGGC | ||
| Mouse Ube2c | Forward | CTGCTAGGAGAACCCAACATC | NM_026785.2 |
| Reverse | GCTGGAGACCTGCTTTGAATA | ||





