Multiplexed measurement of variant abundance and activity reveals VKOR topology, active site and human variant impact
Figures
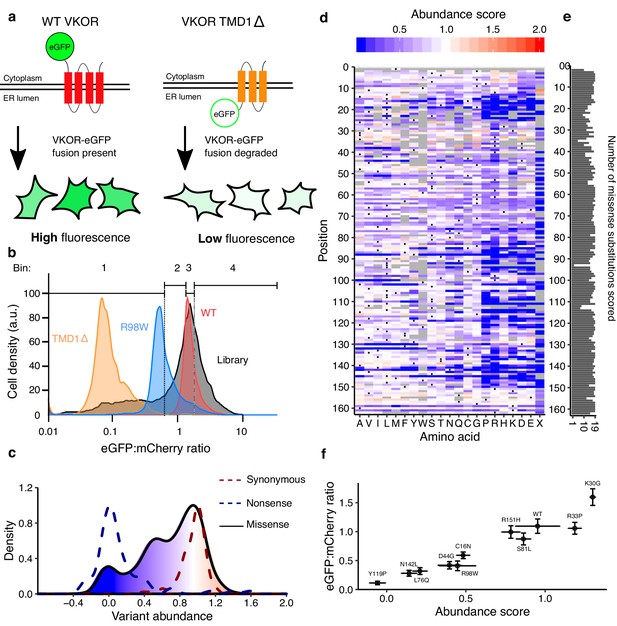
Multiplexed measurement of VKOR variant abundance using VAMP-seq.
(a) To measure abundance, an eGFP reporter is fused to VKOR. eGFP-tagged WT VKOR is folded correctly, leading to high eGFP fluorescence. However, a destabilized variant is degraded by protein quality control machinery, leading to low eGFP fluorescence. (b) Flow cytometry is used to bin cells based on their eGFP:mCherry fluorescence intensity. Density plots of VKOR library expressing cells (grey, n = 12,109) relative to three controls: WT VKOR (red, n = 4,756), VKOR 98W (blue, n = 2,453), and VKOR TMD1Δ (orange, n = 2,204) are shown. Quartile bins for FACS of the library are marked. (c) Abundance score density plots of nonsense variants (dashed blue line, n = 88), synonymous variants (dashed red line, n = 127), and missense variants (filled, solid line, n = 2,695). The missense variant density is colored as a gradient between the lowest 10% of abundance scores (blue), the WT abundance score (white) and abundance scores above WT (red). (d) Heatmap showing abundance scores for each substitution at every position within VKOR. Heatmap color indicates abundance scores scaled as a gradient between the lowest 10% of abundance scores (blue), the WT abundance score (white), and abundance scores above WT (red). Grey bars indicate missing variants. Black dots indicate WT amino acids. (e) Number of substitutions scored at each position for abundance. (f) Scatterplot comparing VAMP-seq derived abundance scores to mean eGFP:mCherry (n = 1 replicate) ratios measured individually by flow cytometry. Variants were selected at random to span the abundance score range. Error bars show standard error for abundance scores and standard error for eGFP:mCherry ratio.
-
Figure 1—source data 1
VKOR variant abundance and activity scores.
VKOR variant abundance and activity scores.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig1-data1-v1.csv
-
Figure 1—source data 2
Flow cytometry for monoclonal validation of variants.
Flow cytometry for monoclonal validation of variants. 11 variants were run individually, values show mean and error for VAMP-seq score and eGFP:mcherry intensity.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig1-data2-v1.csv
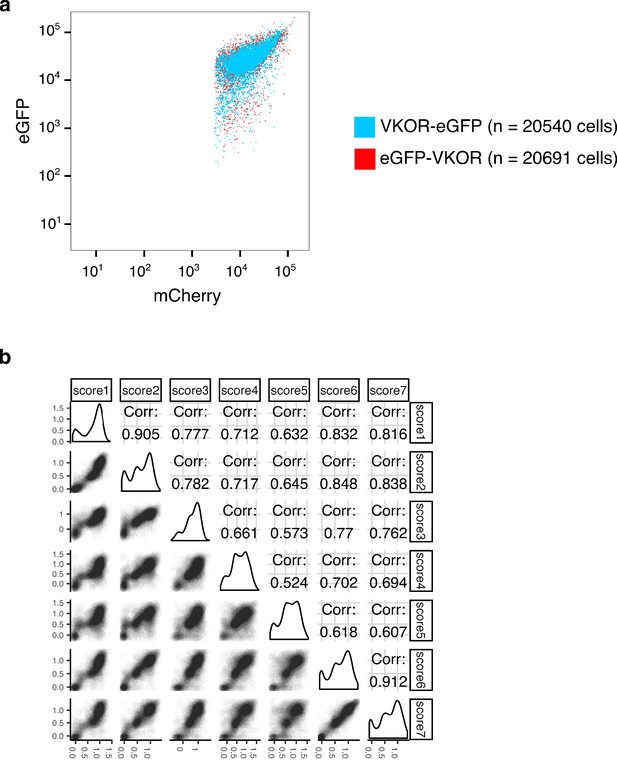
VKOR abundance assay pilot experiment and replicate correlations.
(a) Scatterplot of eGFP vs. mCherry fluorescence for cells expressing either C-terminally eGFP-tagged VKOR (VKOR-eGFP, blue) or N-terminally eGFP-tagged VKOR (eGFP-VKOR, red). (b) Pairwise abundance score correlations between replicate sorting experiments. Seven VAMP-seq replicates were performed. Pearson’s correlation coefficients are shown. Score numbers in this figure correspond to replicate numbers shown in Supplementary file 1.
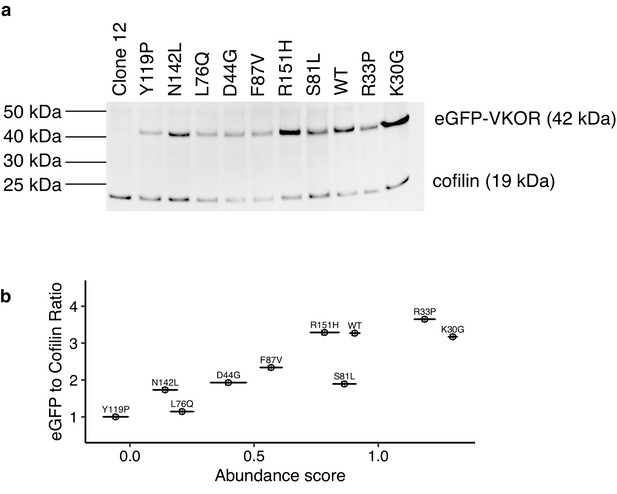
Western blot validation of VKOR abundance scores.
(a) Ten variants that spanned the range of abundance scores were assayed individually via Western blot. Protein abundance was measured using a GFP-specific antibody, resulting in a band at ~42 kDa, the predicted size of an eGFP-VKOR fusion. A cofilin-specific antibody was used as the loading control. (b) Ratios of eGFP band intensity to cofilin band intensity from the Western blot were plotted versus the variant’s abundance score (Pearson’s R = 0.87).
-
Figure 1—figure supplement 2—source data 1
Western blot intensity values derived from Figure 1—figure supplement 2a using Image Lab 6.0.1 (Bio-Rad).
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig1-figsupp2-data1-v1.csv
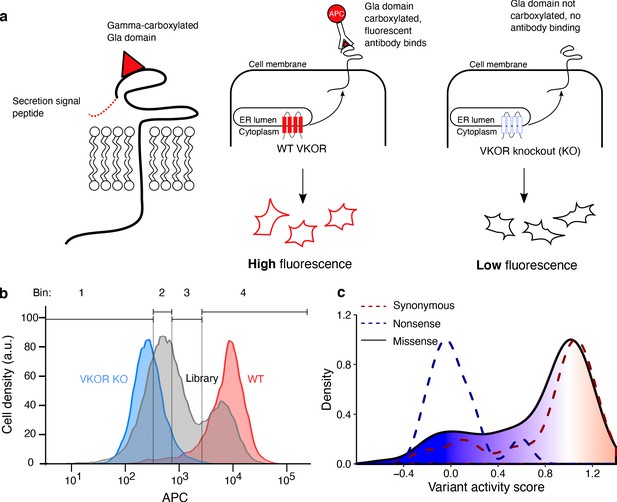
Multiplexed measurement of VKOR variant activity using a gamma-glutamyl carboxylation reporter.
(a) Left panel, A Factor IX Gla domain reporter is expressed inHEK293 cells and consists of a prothrombin pre-pro-peptide which allows for processing and secretion, a Factor IX Gla domain, and Proline rich Gla protein 2 (PRGP2) transmembrane and cytoplasmic domains. Middle panel, Cells expressing WT VKOR carboxylate the reporter Gla domain, which, upon trafficking to the cell surface, can be stained using a carboxylation-specific antibody conjugated to the fluorophore APC. Right panel, VKOR knockout cells do not carboxylate the reporter, so the fluorescent antibody does not bind. (b) Density plots of HEK293 activity reporter cells stained with APC-labeled carboxylation-specific antibody expressing no VKOR (blue, n = 7,188), WT VKOR (red, n = 4,107), or the VKOR variant library (grey, n = 41,418). Quartile bins for FACS of the library are marked. (c) Activity score density plots of nonsense variants (dashed blue line, n = 14), synonymous variants (dashed red line, n = 35), and missense variants (filled, solid line, n = 697). The missense variant density is colored as a gradient between the lowest 10% of activity scores (blue), the WT activity score (white) and activity scores above WT (red).
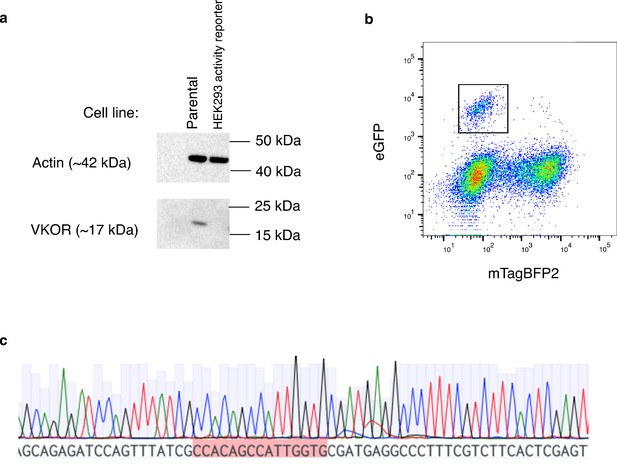
HEK293 VKOR activity reporter cell line characterization.
(a) Western blot of parental cell line vs. HEK293 activity reporter cell line. Loading control is actin (42 kDa). VKOR was probed using an antibody generated against a peptide from the C-terminal of VKOR (FRKVQEPQGKAKRH) (Hallgren et al., 2006). The band for VKOR at 17 kDA is visible in the parental cell line but is not present in the HEK293 activity reporter cell line. (b) Scatterplot showing mTagBFP2 vs. eGFP mean fluorescence intensities for HEK293 activity reporter cells recombined with a construct encoding WT VKOR followed by internal ribosomal entry sequence and eGFP. The emergence of a distinct recombined population that is eGFP positive and mTagBFP2 negative (black outline, n = 768 cells) supports the presence of a single landing pad into the cell genome, and not multiple insertions. (c) A chromatogram showing the barcode sequence of the landing pad inserted at the AAVS1 locus in the HEK293 activity reporter cell line. The presence of a single barcode, highlighted in red, instead of mixed peaks, supports insertion of one landing pad rather than multiple landing pads.
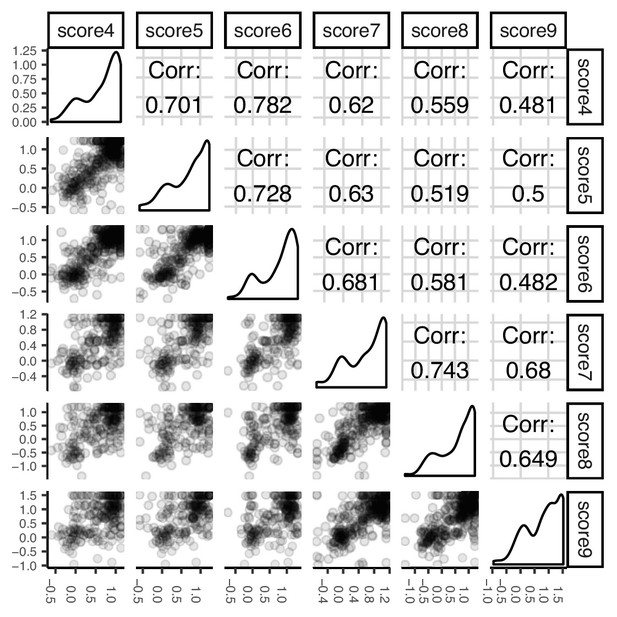
Correlations of activity assay replicates.
Pairwise score correlations between replicate sorting experiments of VKOR activity. Six replicates of the activity assay were performed. Pearson’s correlation coefficients are shown. Score numbers in this panel correspond to replicate numbers shown in Supplementary file 2.
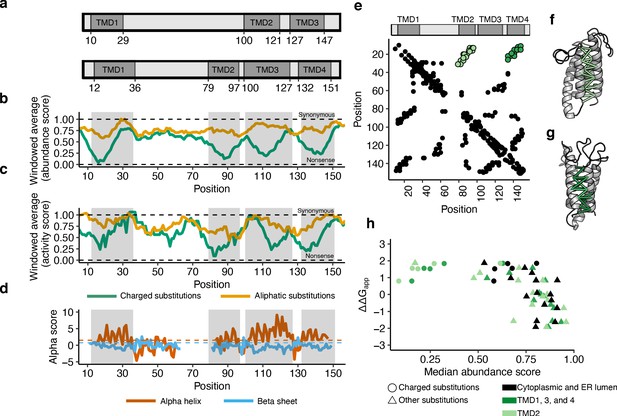
Abundance, activity, and evolutionary data support four transmembrane domains.
(a) Three and four transmembrane domain (TMD) models of VKOR, with TMDs in dark grey (Li et al., 2010; Tie et al., 2012). (b) Windowed abundance score means (width = 10 positions) for charged substitutions (green) and aliphatic substitutions (gold). Dark grey boxes correspond to TMDs proposed in the four-domain model. Dashed lines show median synonymous and the nonsense abundance scores. (c) Windowed activity score means (width = 10 positions) for charged substitutions (green) and aliphatic substitutions (gold). Boxes and dashed lines as described in b. (d) Secondary structure classification from local evolutionary couplings shown as alpha scores calculated for alpha helices (red) and beta sheets (blue). Dashed lines show significance cut-offs for alpha helices (1.5, red) and beta sheets (0.75, blue) (Toth-Petroczy et al., 2016). (e) A contact map derived from evolutionary couplings. Black points show pairs of positions with significant coupling. Light green points show predicted contacts between TMD1 and TMD2. Dark green points show predicted contacts between TMD1 and TMD4. (f) Predicted tertiary contacts between TMD1-TMD2 (shown in light green in e) and g, TMD1-TMD4 (shown in dark green in e) shown on the evolutionary couplings-derived hVKOR structural model. (h) Scatterplot comparing change in free energy for membrane insertion (Elazar et al., 2016a) (∆∆Gapp) to median abundance score for each amino acid substitution. Cytoplasmic and lumenal positions shown in black, TMD2 in light green, and TMDs 1, 3, and four in dark green. Charged substitutions shown as circles, all other substitutions as triangles.
-
Figure 3—source data 1
Evolutionary couplings secondary structure predictions.
Evolutionary couplings secondary structure predictions. Rows show position, with columns showing alpha helix or beta sheet values and predictions.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig3-data1-v1.csv
-
Figure 3—source data 2
Evolutionary couplings 3D contact predictions.
Evolutionary couplings 3D contact predictions. Rows show pairs of residues with contact probabilities.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig3-data2-v1.csv
-
Figure 3—source data 3
Insertion energies from Elazar et al., 2016b.
Insertion energies from Elazar et al., 2016b. Amino acids with calculated insertion energy.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig3-data3-v1.csv
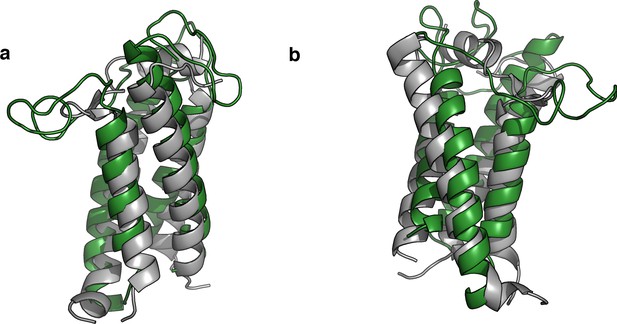
Bacterial VKOR structure and EV-couplings folded model are highly similar.
(a) Pymol graphic showing overlap between EVcouplings-folded model of VKOR (shown as a cartoon in green) compared to the bacterial structure (PDB: 4NV5, shown as a cartoon in grey). RMSD is 3.915903 Å over 120 residues. (b) Shows the same two structures, rotated 120°C.
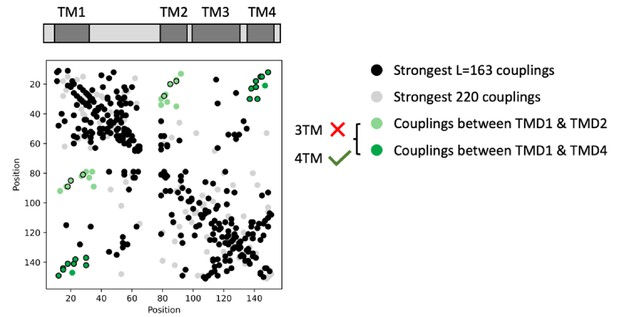
Four transmembrane domain and not three transmembrane domain topology is supported by couplings from eukaryote sequences alone.
Evolutionary couplings (black) from alignment of 1118 eukaryote sequences show an overall topology consistent with the bacterial 4-TM VKOR, and include contacts (green and dark-green) between helices in contact in 4-TM but not in a 3-TM topology. Based on fewer sequences, contact predictions are noisier than those from the full alignment. To best show the topology signal, we plot couplings beyond the top L strongest (gray).
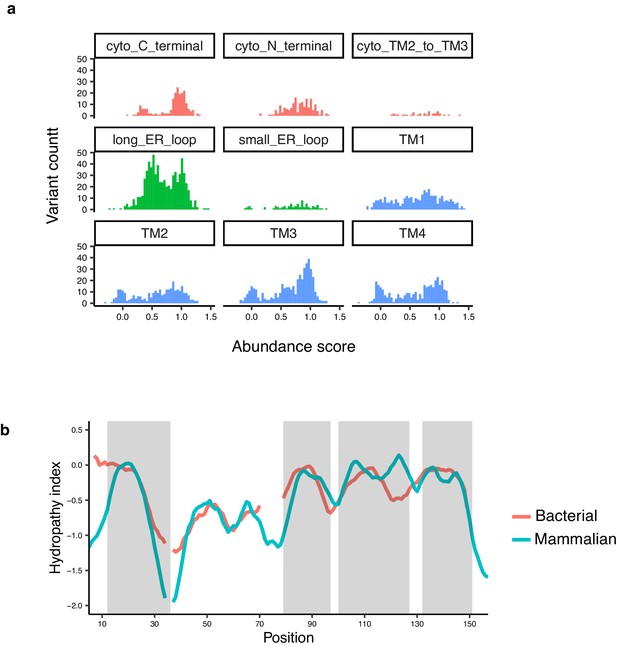
Specific domain abundance scores and hydrophobicity of bacterial and mammalian multiple sequence alignments (MSAs).
(a) Histograms of abundance scores for missense variants, grouped by domain and colored by cytoplasmic, ER lumenal, or transmembrane localization. b, Hydrophobicity index of a bacterial MSA (red) and a mammalian MSA (blue), calculated using Hessa et al. scale (Hessa et al., 2005) and AlignMe server (Stamm et al., 2014).
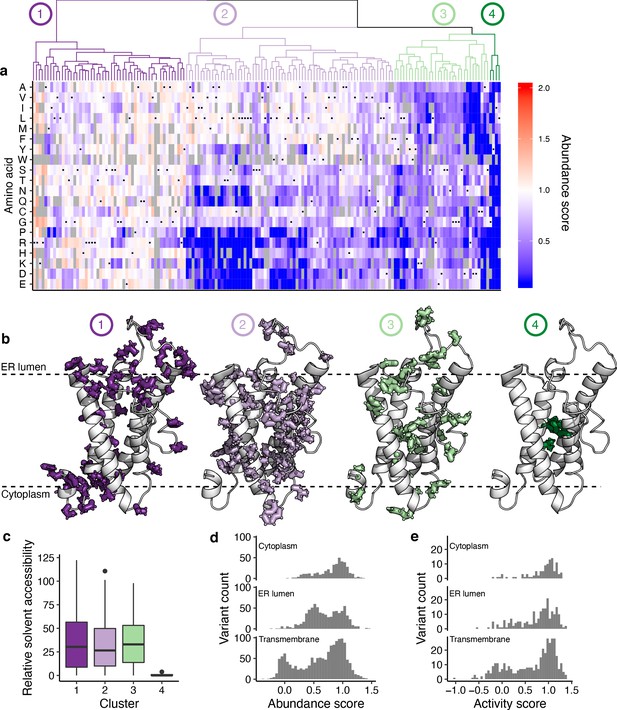
Hierarchical clustering of abundance scores and distributions of abundance and activity scores by domain.
(a) A heatmap showing hierarchical clustering of positions based on abundance score vectors, with the dendrogram above. Groups of positions, chosen based on the dendrogram, are numbered and colored. Heatmap color indicates abundance scores scaled as a gradient between the lowest 10% of abundance scores (blue), the WT abundance score (white) and abundance scores above WT (red). Grey bars indicate missing variants. Black dots indicate WT amino acids. (b) Positions in groups 1–4 shown on the VKOR homology model, with numbers and colors corresponding to panel a. (c) Boxplot showing relative solvent accessibility of positions in each cluster determined using DSSP (Kabsch and Sander, 1983; Touw et al., 2015) and colored as in b. Bold black line shows median, box shows 25th and 75th percentile. Line shows 1.5 interquartile range above and below percentiles, and outliers are shown as black points. (d) Histograms of abundance scores for missense variants in the cytoplasmic, ER lumenal, or transmembrane domains. (e) Histograms of activity scores for missense variants in the cytoplasmic, ER lumenal, or transmembrane domains.
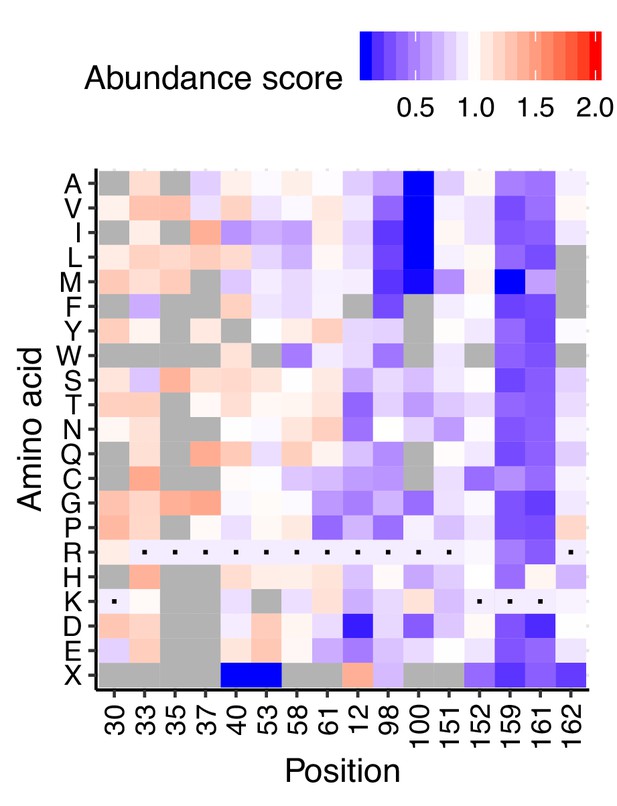
TMD1-adjacent positive residues show pattern of increased abundance.
Heatmap of abundance scores for all arginines and lysines in VKOR. First four positions (K30, K33, K35, K37) are in or proximal to transmembrane domain 1. Heatmap color indicates abundance scores scaled as a gradient between the lowest 10% of abundance scores (blue), the WT abundance score (white) and abundance scores above WT (red). Grey bars indicate missing variants. Black dots indicate WT amino acids.
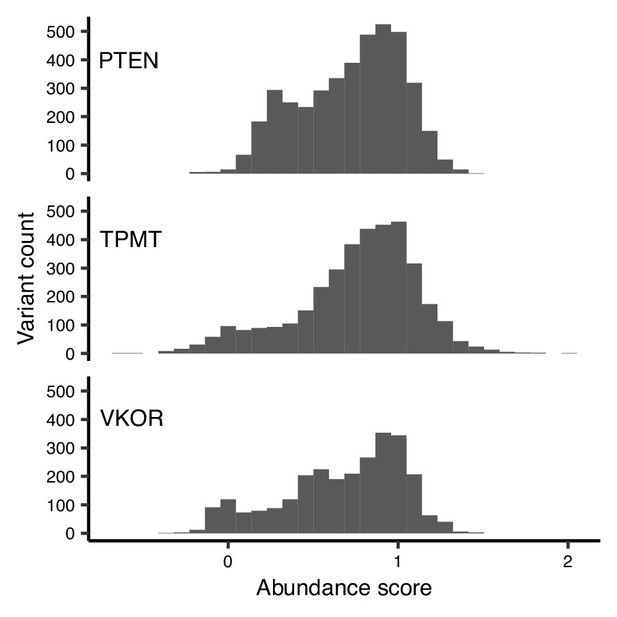
Trimodality of missense variant abundance scores is unique to VKOR.
Histograms of abundance scores for missense variants for three proteins: PTEN, TPMT, and VKOR.
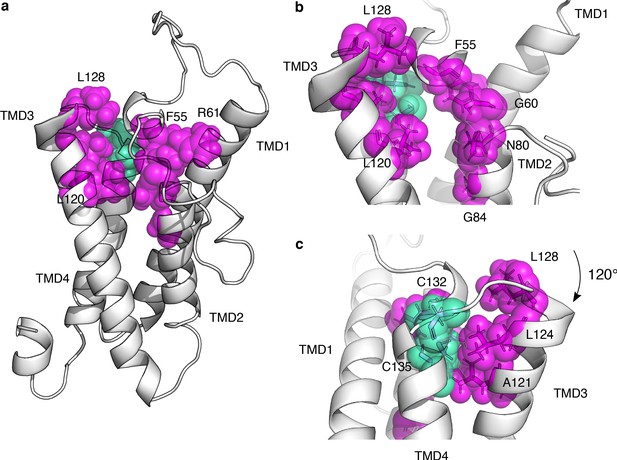
Functionally constrained positions reveal VKOR active site and critical cysteines.
(a) Positions with the lowest 12.5% of median specific activity scores and at least four variants scored for activity are shown as magenta spheres on the VKOR homology model. Cysteines C132, and C135, also in the bottom 12.5% of median specific activity scores, are shown in green spheres. (b) Magnified view of the redox center cysteines (positions 132, and 135, green spheres) and surrounding residues that define the active site (magenta spheres). Residues shown in transparent spheres, with side chains also shown in sticks. (c) Panel b rotated 120°.
-
Figure 5—source data 1
VKOR positional abundance and activity scores.
VKOR positional abundance and activity scores. Rows show positions, with columns showing median abundance score, median activity score, rescaled scores, and specific activity score.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig5-data1-v1.csv
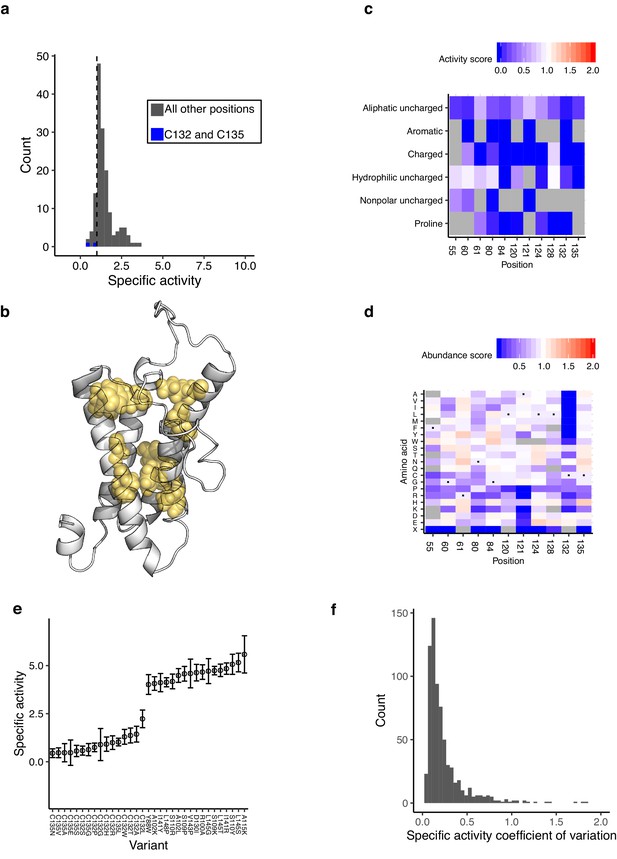
VKOR active site analysis.
(a) Histogram of specific activity, with catalytic cysteines C132 and C135 labeled in blue. Dashed line demarcates bottom 12.5%. (b) Active site positions as defined by computational docking, shown on the homology model as yellow spheres (Czogalla et al., 2017). (c) Heatmap of activity scores for residues with lowest 12.5% of specific activity scores, collapsed by amino acid class. Color indicates activity scores scaled as a gradient between the lowest 10% of activity scores (blue), the WT activity score (white) and activity scores above WT (red). Grey indicates missing data. (d) Heatmap of abundance scores for residues with lowest 12.5% of specific activity scores. Color legend same as described in c, applied to abundance scores. (e) Specific activity scores of a subset of variants, with error bars showing standard deviation. (f) Histogram of coefficient of variation for the specific activity value.
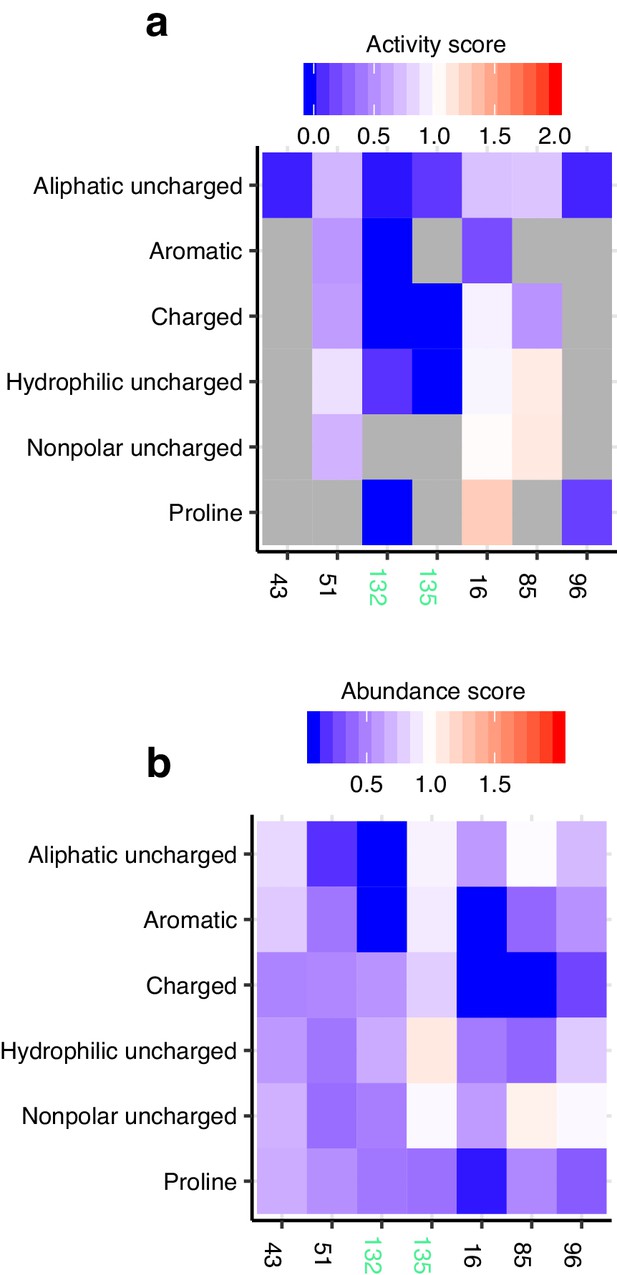
Conserved cysteine analysis.
(a) Heatmap of activity scores for cysteines. Catalytic cysteines C132 and C135 labeled in green. Color indicates activity scores scaled as a gradient between the lowest 10% of activity scores (blue), the WT activity score (white) and activity scores above WT (red). Grey indicates missing data. (b) Heatmap of abundance scores for cysteines. Catalytic cysteines C132 and C135 labeled in green. Color legend same as described in a, applied to abundance scores.

VKOR localization motif analysis.
(a) Heatmap of abundance scores for diarginine ER retention motif. X-axis shows residues and position. Color indicates abundance scores scaled as a gradient between the lowest 10% of abundance scores (blue), the WT abundance score (white) and abundance scores above WT (red). Grey indicates missing data. (b) Heatmap of abundance score for dilysine ER retention motif. X-axis shows residues and position.
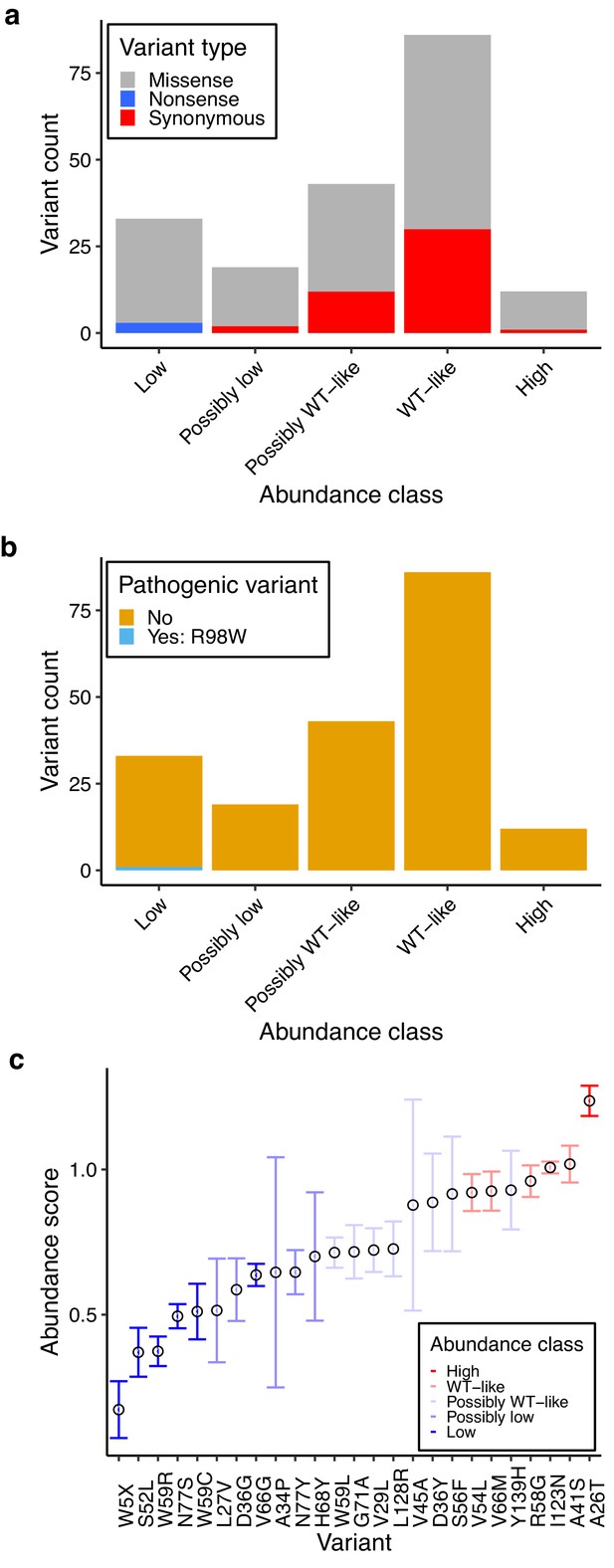
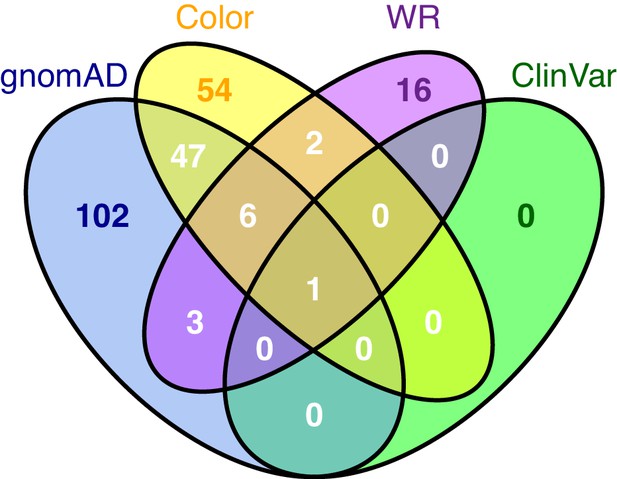
Characterization of human variants using abundance and activity data.
(a) Histogram of abundance classifications for variants from gnomAD, ClinVar, and Color Genomics. Nonsense variants colored in blue, synonymous in red, and missense in grey. (b) Histogram of abundance classifications for same variants in a, colored by pathogenicity. The only variant known to cause disease, R98W, is colored in blue. All other variants shown in yellow. (c) Scatterplot showing abundance scores for literature-curated warfarin resistance variants. Bars show standard error and are colored by abundance class. Variants are arranged in order of abundance score.
-
Figure 6—source data 1
Abundance and activity data for human variants found in ClinVar, gnomAD v2 and v3, and Color Genomics dataset.
- https://cdn.elifesciences.org/articles/58026/elife-58026-fig6-data1-v1.csv
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Murine anti-Factor IX carboxylated Gla domain (mouse monoclonal) | Green Mountain Antibodies | Cat#GMA-001 | (1:100) |
| Antibody | HRP Anti-beta-actin antibody (mouse monoclonal) | Abcam | Cat#ab20272; RRID:AB_445482 | (1:10,000) |
| Antibody | Amersham ECL Mouse IgG, HRP-linked whole Ab (sheep polyclonal) | GE Healthcare | Cat#NA931; RRID:AB_772210 | (1:10,000) |
| Antibody | Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (goat polyclonal) | ThermoFisher | Cat#:A32723; RRID:AB_2633275 | (1:10,000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (goat polyclonal) | ThermoFisher | Cat#:A-21244; RRID:AB_2535812 | (1:10,000) |
| Antibody | anti-Cofilin (D3F9) XP Rabbit mAb (rabbit monoclonal) | Cell Signaling Technology | Cat#:5175; RRID:AB_10622000 | (1:1000) |
| Antibody | Anti-GFP from mouse IgG1κ (clones 7.1 and 13.1) (mouse monoclonal) | Roche | Cat#:11814460001; RRID:AB_390913 | (1:1000) |
| Antibody | Anti-VKOR (rabbit monoclonal) | PMID:16634640 | (1:1000) | |
| Strain, strain background (Escherichia coli) | E. coli electrocompetent | NEB | Cat#C3020K | |
| Commercial assay or kit | LYNX Rapid APC conjugation kit | BioRad | Cat#LNK033APC | |
| Commercial assay or kit | SuperSignal West Dura Extended Duration Substrate | ThermoFisher | Cat#34076 | |
| Commercial assay or kit | Library Quantification Kit (Illumina) | KAPA Biosystems | Cat#KK4854 | |
| Commercial assay or kit | MiSeq Reagent Kit v3 (600 cycles) | Illumina | Cat#MS-102–3003; RRID:SCR_016379 | |
| Commercial assay or kit | NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | Cat#TG-160–2005; RRID:SCR_016381 | |
| Commercial assay or kit | Qiagen miniprep kit | Qiagen | Cat#27106 | |
| Commercial assay or kit | GenElute HP Plasmid midiprep kit | Sigma | Cat#NA0200 | |
| Commercial assay or kit | DNA clean and concentrator kit | Zymo | Cat#D4031 | |
| Software, algortihm | PyMOL | Schrodinger | https://pymol.org | |
| Software, algortihm | Enrich2 | PMID:28784151 | https://github.com/FowlerLab/Enrich2 | |
| Software, algortihm | R | R | https://cran.r-project.org/ | |
| Other | Fugene 6 | Promega | Cat#E2691 | |
| Other | Lipofectamine 3000 | ThermoFisher | L3000015 | |
| Other | Atomic coordinates, bacterial VKOR structure | Protein Data Bank | PDB: 4NV5 | |
| Other | Raw and analyzed data | This paper | GSE149922 | |
| Cell line (human) | 293T AAVS1 tetbxb1 clone 4 | PMID:28335006 | ||
| Cell line (human) | HEK293 VKOR activity reporter | PMID:24297869 | ||
| Recombinant DNA reagent | PX458 | Addgene | Cat#48138; RRID:Addgene_48138 |
Additional files
-
Supplementary file 1
The seven replicates of VAMP-seq performed with cells recombined and sorted for each.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp1-v1.csv
-
Supplementary file 2
The six replicates of the activity assay performed with cells recombined and sorted for each.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp2-v1.csv
-
Supplementary file 3
Evolutionary couplings VKOR model.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp3-v1.docx
-
Supplementary file 4
ITASSER homology VKOR model.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp4-v1.zip
-
Supplementary file 5
Variants found in humans that cause warfarin sensitivity or resistance, and references in which they were first reported.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp5-v1.csv
-
Supplementary file 6
Human variants abundance and activity scores.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp6-v1.csv
-
Supplementary file 7
Names and sequences for oligos used in this paper.
- https://cdn.elifesciences.org/articles/58026/elife-58026-supp7-v1.csv
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58026/elife-58026-transrepform-v1.docx