Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria
Figures
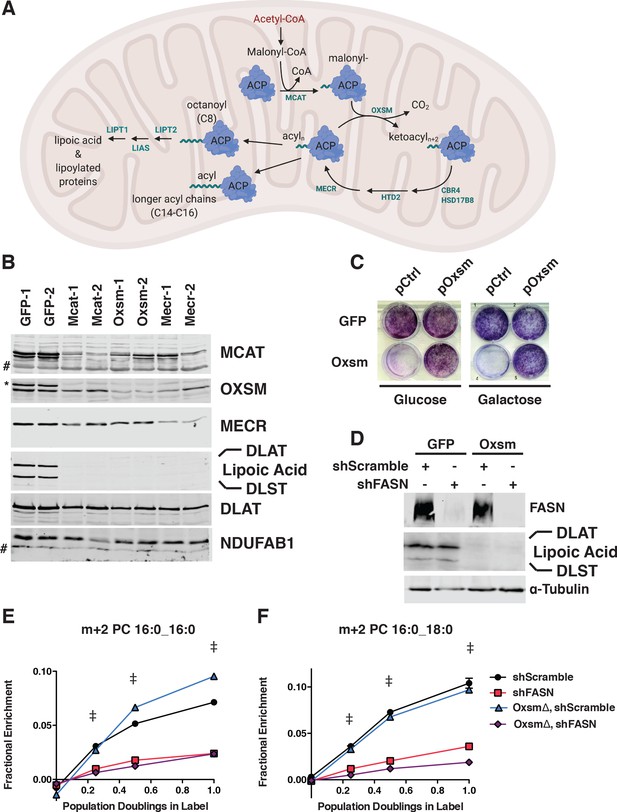
MtFAS is an essential pathway in mammalian skeletal myoblasts but does not contribute to synthesis of major cellular lipids.
(A) Schematic of the mitochondrial fatty acid synthesis pathway and downstream lipoic acid synthesis. (B) Crude isolated mitochondrial fractions from duplicate single cell clones of Mcat, Oxsm, and Mecr mutants, compared with GFP control clonal cell lines, were separated via SDS-PAGE and immunoblotted for the indicated targets. *=Lipoic acid band (reprobe of earlier blot). #=non specific bands C. GFP control and Oxsm mutant cells were infected with retroviral control plasmid (pCtrl) or a plasmid expressing Oxsm off the CMV promoter (pOxsm), plated at equal densities in normal growth medium with either 4.5 g/L glucose or 10 mM galactose and grown for 3 or 4 days, respectively, then stained with crystal violet. (D) Whole cell lysates from stable cell lines generated by infecting Oxsm mutant cells (OxsmΔ) or GFP controls with shRNA constructs targeting FASN (shFASN) or scramble control (shScramble) were separated by SDS-PAGE and immunoblotted for FASN, lipoylated proteins, or tubulin. (E-F) Stable cell lines created in (D) were incubated with U13C-glucose for the indicated number of doublings, harvested, lipids extracted, and analyzed via LC-MS. Shown are quantitation of m+2 isotopologues for two representative phospholipid species, PC 16:0_16:0 and PC 16:0_18:0. †=p < 0.001, ‡=p < 0.0001, error bars are SEM.
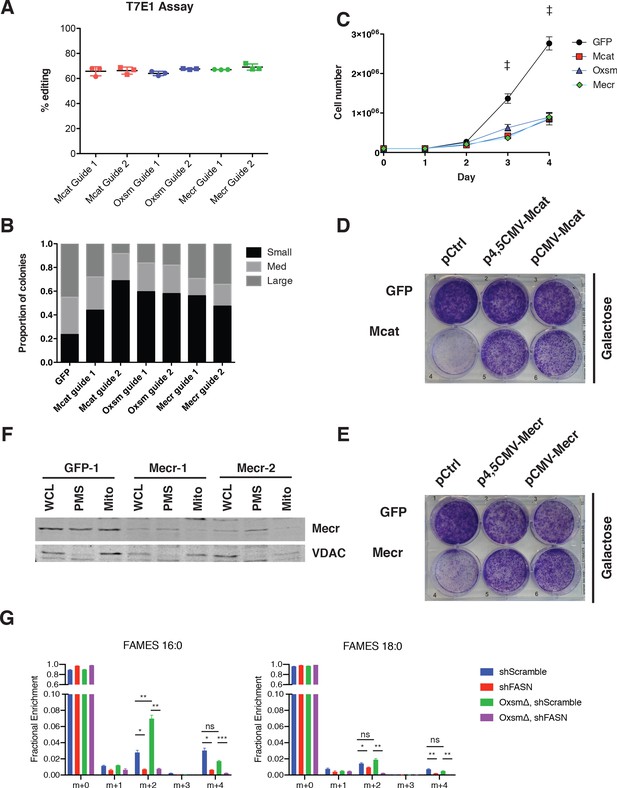
mtFAS is required for growth of cultured skeletal myoblasts but does not significantly contribute to cellular fatty acids.
(A) Quantification of T7E1 assay for editing efficiency in bulk transfected C2C12 cells with sgRNAs targeting the indicated mtFAS genes. (B) Quantification of single cell clone colony size 7 days after single cell sort. Small clones = <150 cells, Med = 150–500 cells, Large = >500 cells. (C) Cells were plated at 100,000 cells/plate in 10 cm dishes and counted every 24 hr. #=p < 0.01, all groups compared with control, †=p < 0.001, all groups compared with control, error bars SEM. (D-E) GFP control, Mcat, and Mecr mutant cells were infected with retroviral control plasmid (pCtrl) or plasmids expressing Mcat (D) or Mecr (E) off the truncated (Δ4,5CMV) or full CMV promoters as indicated. Cells were plated at equal densities in normal growth medium without glucose and with 10 mM galactose and grown for 4 days, then stained with crystal violet. (F) Sub-cellular fractionation of Mecr mutant cell lines and control. Whole cell lysate (WCL), post mitochondrial supernatant (PMS), and mitochondrial lysate (Mito) was isolated from each cell line indicated, normalized for total protein via BCA assay, and immunoblotted for Mecr or VDAC as a mitochondrial marker control. (G) Control (GFP) or Oxsm mutant (OxsmΔ) cells were stably infected with shRNA targeting FASN (shFASN) or control (shScramble) then incubated with U13C-glucose for one doubling were harvested, lipids extracted and saponified, and analyzed via FAMES analysis. Shown are quantitation of c16:0 and c18:0 fatty acids. ns = non significant, *=p < 0.05, #=p < 0.01, †=p < 0.001, error bars are SEM.
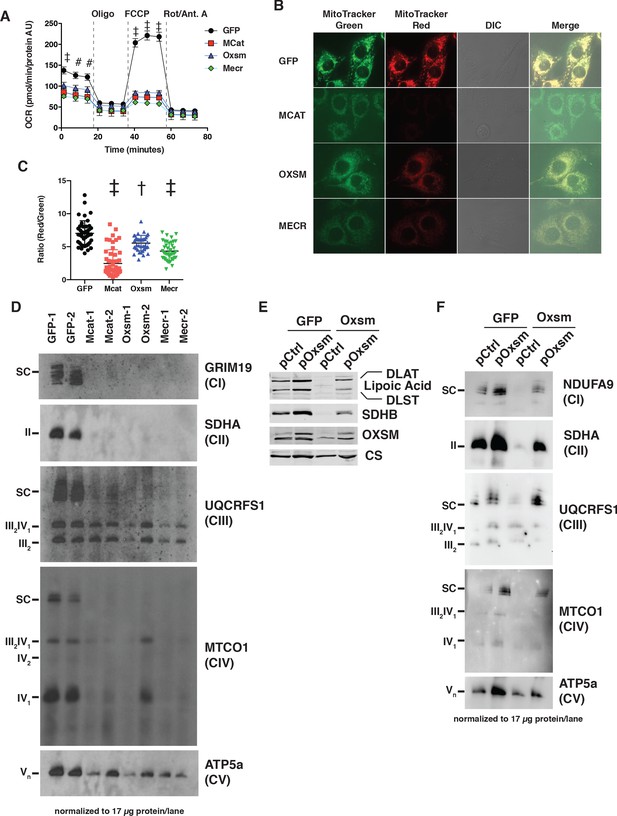
MtFAS mutants display profound loss of mitochondrial respiration and ETC complexes.
(A) Cells from three clones of each of the indicated genotypes were seeded in eight wells of a 96-well seahorse plate and allowed to adhere overnight, then equilibrated and treated with the indicated drugs following standard mitochondrial stress test protocols from the manufacturer to determine Oxygen Consumption Rate (OCR). #=p < 0.01, †=p < 0.001, ‡=p < 0.0001 all comparisons are to GFP control, error bars are SEM. (B) Cells of the indicated genotype were seeded in chambered coverglass slides, stained with Mitotracker Red and Mitotracker Green, and imaged. (C) Ratio of MitoTracker Red to MitoTracker Green fluorescence of cells from 30 fields of view quantified using Fiji ImageJ. *=p < 0.05, †=p < 0.001, ‡=p < 0.0001 all comparisons are to GFP control, error bars are SD. (D) Mitochondrial lysates generated from the indicated cell lines were normalized for total protein by BCA assay, incubated with 1% digitonin, then separated by blue-native PAGE and immunoblotted with the indicated antibodies.
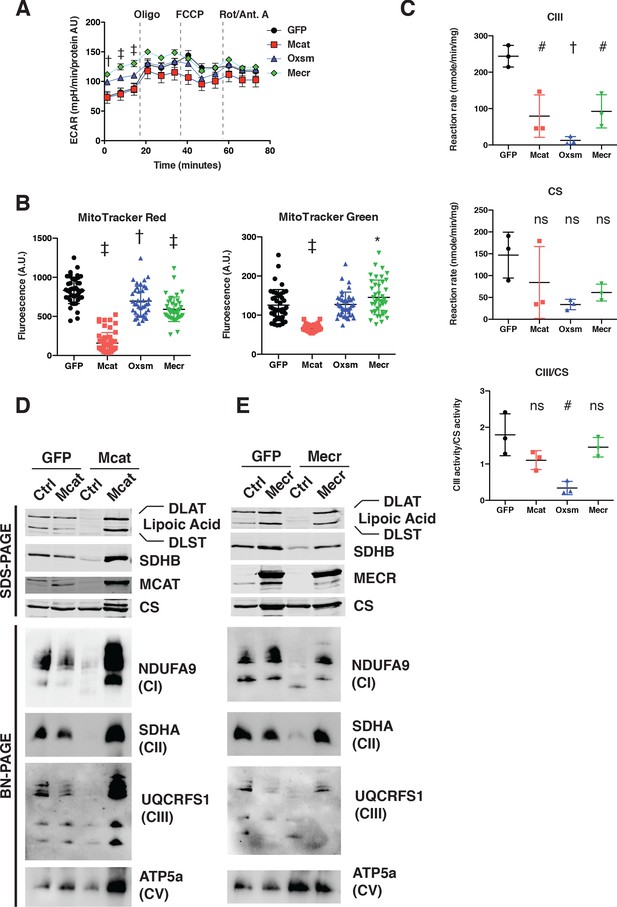
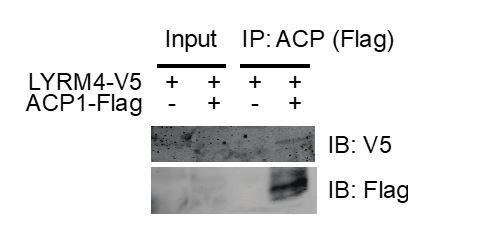
ETC complex assembly and activity is regulated by interaction of LYRM proteins with ACP.
(A) Cells from three clones of each of the indicated genotypes were seeded in eight wells of a 96-well seahorse plate and allowed to adhere overnight, then equilibrated and treated with the indicated drugs following standard mitochondrial stress test protocols from the manufacturer to determine ExtraCellular Acidification Rate (ECAR). (B) Cells of the indicated genotype were seeded in chambered coverglass slides, stained with Mitotracker Red and Mitotracker Green, and imaged. Fluorescence of cells from 30 fields of view was quantified using Fiji ImageJ. *=p < 0.05, †=p < 0.001, ‡=p < 0.0001 all comparisons are to GFP control, error bars are SD. (C) Crude mitochondrial fractions were isolated from triplicate biological samples of mtFAS mutant and control cells using differential centrifugation. Isolated mitochondria were normalized for protein content via BCA assay and analyzed for Bc1-complex activity and citrate synthase activity (CS). ns = non significant, #=p < 0.01, †=p < 0.001, error bars are SD.
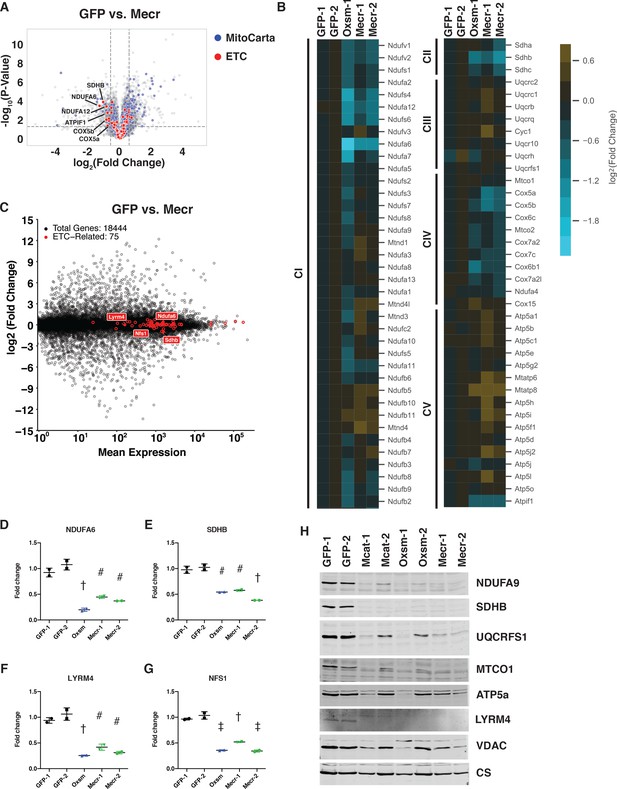
Posttranslational loss of ETC components in mtFAS mutants is specific to LYR proteins and their targets.
(A-B) Duplicate samples from the indicated cell lines were grown under proliferative conditions and subjected to TMT labeling and quantitative proteomics analysis. (A) Volcano plot of compiled Mecr clones vs. GFP controls showing all proteins (gray), mitochondrial proteins (blue), and electron transport chain subunits (ETC, red). Dashed gray lines indicate cutoffs for significance at -log10(p-value) = 1.3 and log2(Fold Change) = +/- 0.59. (B) Heatmap depicting log2(Fold Change) of OXPHOS subunits in the indicated cell lines. (C) Quadruplicate samples from mtFAS mutant cells and controls were grown under proliferative conditions. Total RNA was isolated, used as input for mRNA library prep, and sequenced. Resulting data were aligned to the mouse genome and analyzed for differential expression. ETC subunit-encoding transcripts are shown in red vs. all other transcripts (gray). (D-G) Relative abundance of the indicated LYR proteins or their targets in the indicated cell lines from the quantitative proteomics experiment described in (A-B) #=p < 0.01, †=p < 0.001, ‡=p < 0.0001, error bars are SD. All statistical comparisons shown are between mtFAS mutants and GFP-1 clone; p-values when compared with GFP-2 clone were similar or smaller than when compared with GFP-1 clone. (H) Crude mitochondrial lysates generated from the indicated cell lines by differential centrifugation were normalized for total protein by BCA assay, separated by SDS-PAGE, and immunoblotted with the indicated antibodies.
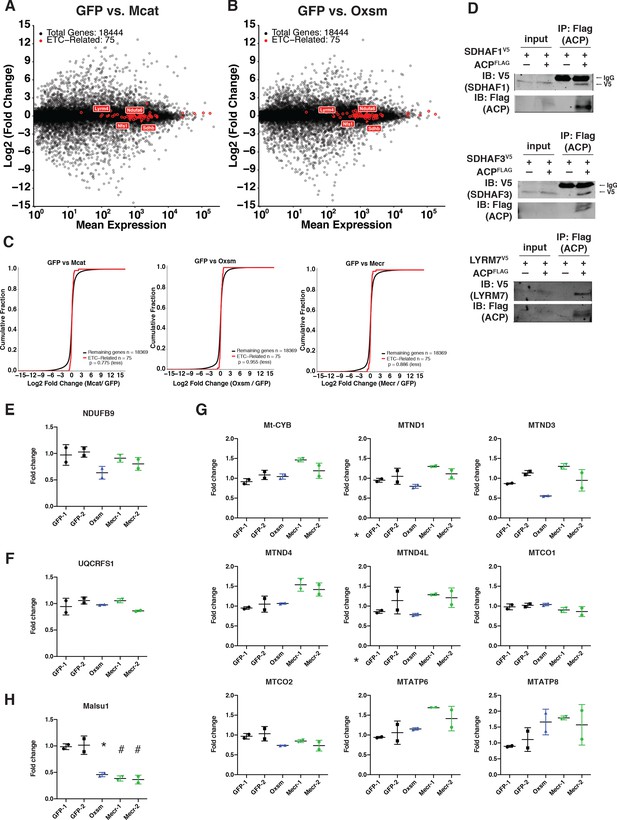
Transcription of ETC subunit encoding genes and translation of mitochondrially encoded proteins are unaffected in mtFAS mutants.
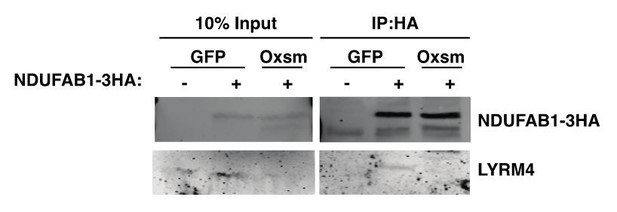
(A-C) Quadruplicate samples from mtFAS mutant cells and controls were grown under proliferative conditions. Total RNA was isolated, used as input for mRNA library prep, and sequenced. Resulting data were aligned to the mouse genome and analyzed for differential expression. ETC subunit encoding transcripts are shown in red vs. all other transcripts (black). (C) Cumulative distribution plots showing no difference in abundance between ETC encoding transcripts vs. all other genes for the indicated mtFAS mutant vs. GFP control, p>0.05, Wilcoxon rank-sum test. (D) HEK293T cells were transiently transfected with human NDUFAB1-Flag or empty vector control along with the indicated LYRM-V5 constructs, grown for 48 hr after transfection, harvested, and crude mitochondrial lysates were prepared by differential centrifugation. Flag immunoprecipitation was performed and eluates were blotted with the indicated antibodies alongside 10% of input. (E-H) Duplicate samples from the indicated cell lines were grown under proliferative conditions and subjected to TMT labeling and quantitative proteomics analysis. Relative abundance of the indicated LYR protein (E), LYR target (F), mitochondrially encoded proteins (G), and MALSU1 (H). *=p < 0.05, #=p < 0.01, error bars are SD. All statistical comparisons shown are between mtFAS mutants and GFP-1 clone.
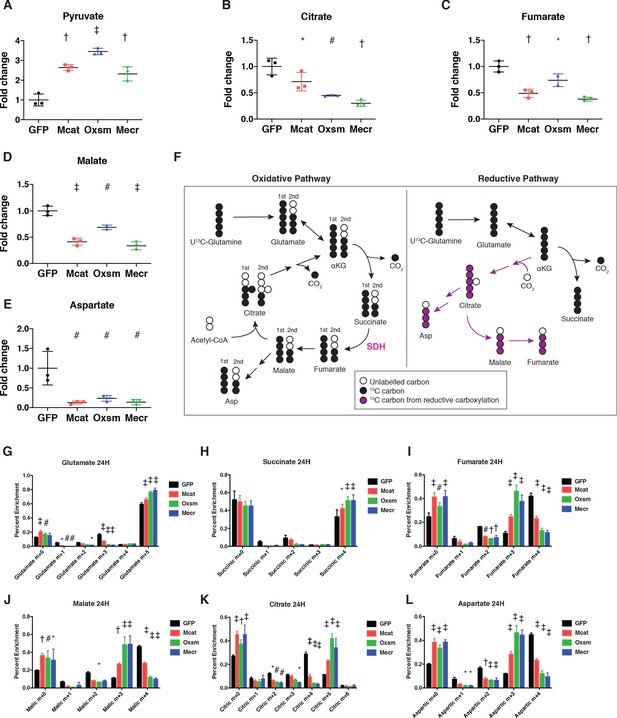
Impairment of mtFAS promotes switch from oxidative to reductive mitochondrial metabolism.
(A–E) Triplicate biological samples from mtFAS mutant cell lines or GFP control were grown under standard proliferative conditions in high glucose medium (25 mM) and harvested for steady-state metabolomics analysis by LC-MS. Shown are relative pool sizes for the indicated metabolites. *=p < 0.05, #=p < 0.01, †=p < 0.001, ‡=p < 0.0001, error bars are SD. (F) Schematic of isotopomeric labeling of TCA cycle intermediates upon feeding with U13C-glutamine. Black circles indicate 13C carbons derived from labeled glutamine via oxidative TCA cycle flux (left). Purple circles indicate 13C carbons derived from labeled glutamine via reductive carboxylation (right). (G-L) Triplicate biological samples of the indicated genotype were labeled for 24 hr with U13C-glutamine, harvested, and analyzed via GC-MS for the indicated metabolites and their isotopologues. *=p < 0.05, #=p < 0.01, †=p < 0.001, ‡=p < 0.0001, error bars are SD.
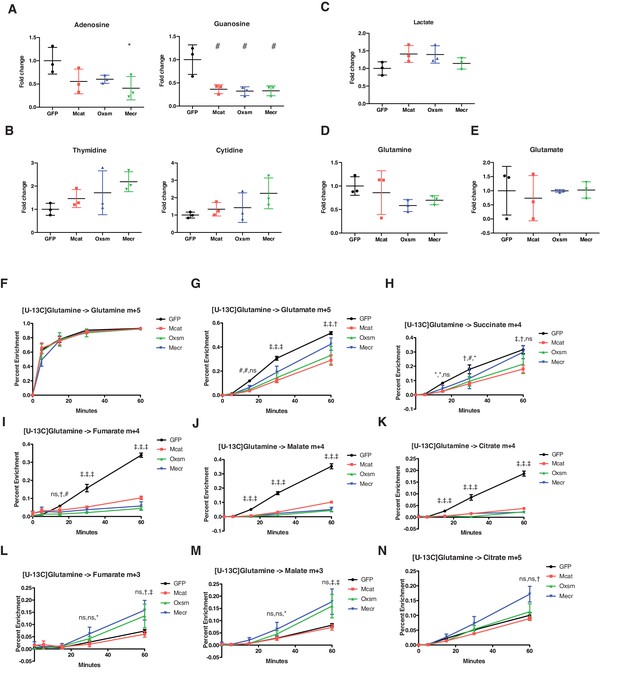
Impairment of mtFAS causes changes in purine abundance and TCA cycle flux.
(A–E) Triplicate biological samples from mtFAS mutant cell lines or GFP control were grown under standard proliferative conditions in high glucose media (25 mM) and harvested for steady-state metabolomics analysis by LC-MS. Shown are relative pool sizes for the indicated metabolites. *=p < 0.05, #=p < 0.01, error bars are SD. (F-N) Triplicate biological samples of the indicated genotype were labeled with U13Cglutamine for 5 min, 15 min, 30 min, or 1 hr, harvested, and analyzed via GC-MS for the indicated metabolite isotopologues. *=p < 0.05, #=p < 0.01, †=p < 0.001, ‡=p < 0.0001, error bars are SD. Symbols indicate statistical comparison to GFP control in the order Mcat, Oxsm, Mecr.
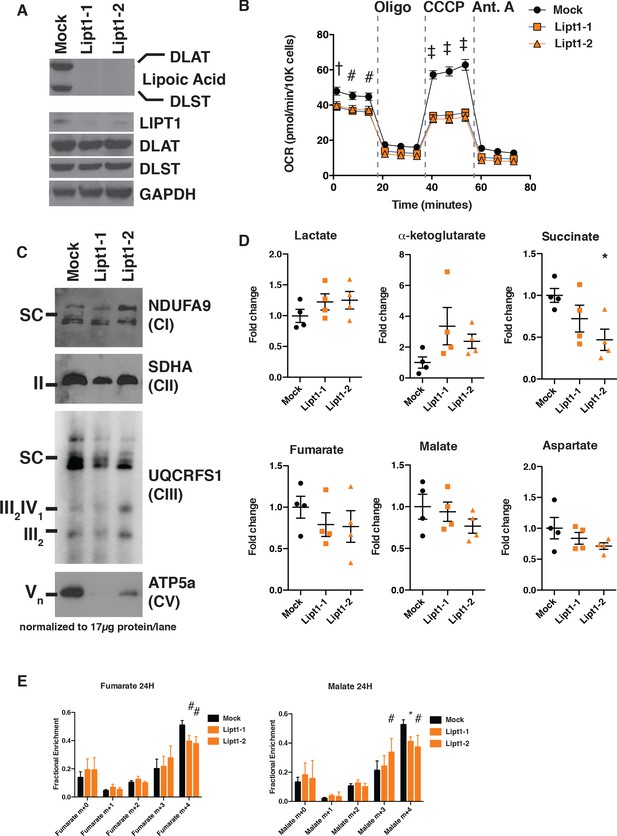
MtFAS mutant phenotypes are not recapitulated by loss of lipoic acid alone.
(A) Immunoblot for the indicated proteins in whole cell lysates from Lipt1 mutant and control cell lines. (B) In triplicate experiments, cells of each of the indicated clones were seeded in eight wells of a 96-well seahorse plate and allowed to adhere overnight, then equilibrated and treated with the indicated drugs following standard mitochondrial stress test protocols from the manufacturer to determine Oxygen Consumption Rate (OCR). #=p < 0.01, †=p < 0.001, ‡=p < 0.0001 all comparisons are to GFP control, error bars are SEM. (C) Mitochondrial lysates generated from the indicated cell lines by differential centrifugation were normalized for total protein by BCA assay, incubated with 1% digitonin, then separated by BN-PAGE and immunoblotted with the indicated antibodies. (D) Four biological replicates from mtFAS mutant cell lines or GFP control were grown under standard proliferative conditions and harvested for steady-state metabolomics analysis by LC-MS. Shown are relative pool sizes for the indicated metabolites. *=p < 0.05, error bars are SD. (E) Four biological samples of the indicated genotype were labeled for 24 hr with U13Cglutamine, harvested, and analyzed via GC-MS for the indicated metabolites and their isotopologues. *=p < 0.05, #=p < 0.01, †=p < 0.001, error bars are SD.
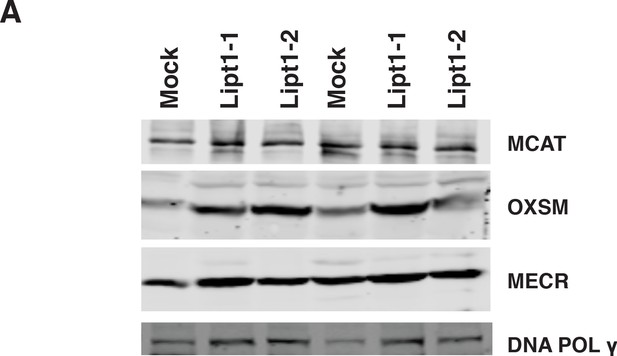
Expression of mtFAS proteins is unchanged in Lipt1 mutant cell lines.
(A) Crude isolated mitochondrial proteins were isolated by SDS-PAGE and immunoblotted for MCAT, OXSM, and MECR, along with DNA polymerase gamma.
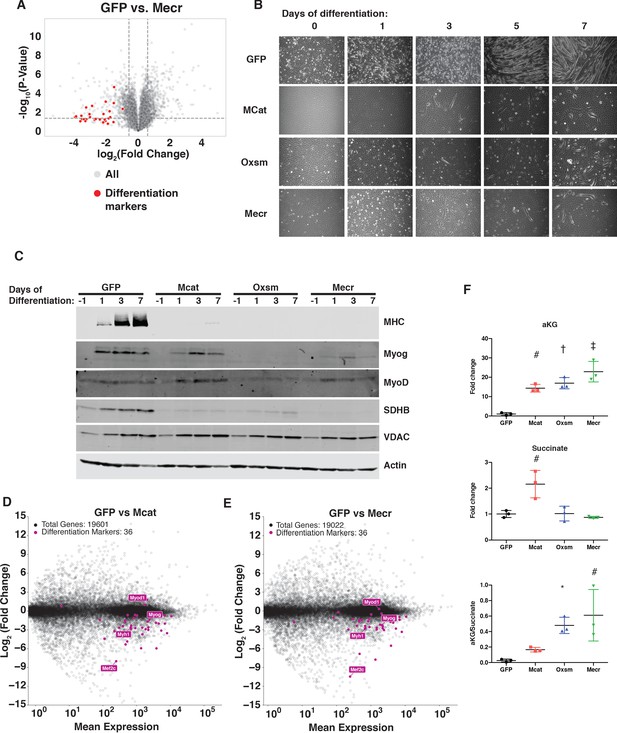
MtFAS is required for skeletal myoblast differentiation.
(A) Volcano plot of quantitative proteomics experiment showing Mecr mutant samples (n = 4) vs. GFP (n = 4) controls; all proteins (gray), and proteins associated with the differentiated skeletal muscle lineage (red). Dashed lines indicate cutoffs for significance at -log10(p-value) = 1.3 and log2(Fold Change) = +/- 0.59. (B-C) Cells of the indicated genotypes were plated and grown for 24 hr under proliferative conditions (day −1), then switched to 2% horse serum differentiation medium at 95% confluency (day 0). Medium was replaced daily for the indicated number of days. Plates were imaged (B) or whole cell lysates were collected at the indicated time points, separated by SDS-PAGE, and immunoblotted for the indicated proteins (C). (D-E) Quadruplicate samples from mtFAS mutant cells and controls were grown under proliferative conditions to 95% confluency then switched to 2% horse serum differentiation medium for 24 hr. Total RNA was isolated, used as input for mRNA library prep, and sequenced. Resulting data were aligned to the mouse genome and analyzed for differential expression. Transcripts for differentiation related proteins from panel (A) are shown in purple vs. all other transcripts (gray). (F) Relative steady state pool sizes of the indicated metabolites in mtFAS mutant cell lines vs. GFP controls grown in proliferative conditions from steady-state LC-MS metabolomics analysis. *=p < 0.05, #=p < 0.01, †=p < 0.001, ‡=p < 0.0001, error bars are SD.
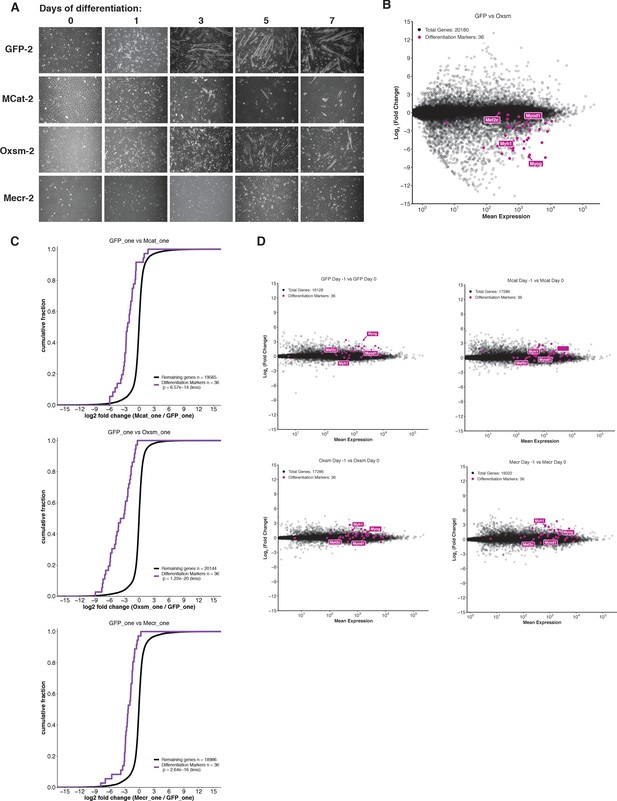
Skeletal myoblast differentiation is delayed in mtFAS mutant cell lines.
(A) Cells of the indicated genotypes were plated and grown for 24 hr under proliferative conditions (day −1), then switched to 2% horse serum differentiation medium at 95% confluency (day 0). Medium was replaced daily for the indicated number of days. Plates were imaged at the indicated time points. (B-C) Quadruplicate samples from mtFAS mutant cells and controls were plated and allowed to adhere overnight, then harvested at Day −1 (proliferative,~70% confluency), Day 0 (~95% confluent), or Day one after switching to 2% horse serum differentiation medium for 24 hr. Total RNA was isolated, used as input for mRNA library prep, and sequenced. Resulting data were aligned to the mouse genome and analyzed for differential expression. Transcripts for differentiation related proteins from panel (A) are shown in purple vs. all other transcripts (gray). (B) Oxsm mutant vs. GFP controls after 24 hr in differentiating media. (C) Cumulative distribution plots of differentiation related transcripts vs. all other genes for the indicated genotypes, p>0.05, Wilcoxon rank-sum test. (D) Change in expression of transcripts from Day −1 to Day 0 for mtFAS mutant cell lines and GFP controls.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| cell line (M. musculus) | C2C12 | ATCC | #CRL-1772, RRID:CVCL_0188 | |
| cell line (H. sapiens) | HEK293T | ATCC | #CRL-11268, RRID:CVCL_1926 | |
| antibody | Anti-MCAT (mouse monoclonal) | Santa Cruz | #sc-390858, RRID:AB_2827536 | WB: (1:100) |
| Antibody | Anti-OXSM (rabbit polyclonal) | Thermo Fisher Scientific | #PA5-32132, RRID:AB_2549605 | WB: (1:1000) |
| Antibody | Anti-MECR (rabbit polyclonal) | Proteintech | #51027–2-AP, RRID:AB_615013 | WB: (1:1000) |
| Antibody | Anti-Lipoic Acid (rabbit polyclonal) | Abcam | #ab58724, RRID:AB_880635 | WB: (1:1000) |
| Antibody | Anti-DLAT (rabbit monoclonal) | Abcam | #ab172617, RRID:AB_2827534 | WB: (1:1000) |
| antibody | Anti-NDUFAB1 (rabbit polyclonal) | Abcam | #ab96230, RRID:AB_10686984 | WB: (1:1000) |
| Antibody | Anti-Flag (rabbit polyclonal) | Sigma-Aldrich | #F7425, RRID:AB_439687 | WB: (1:1000) |
| Antibody | Anti-V5 (rabbit polyclonal) | Abcam | #ab9116, RRID:AB_307024 | WB: (1:2,000) |
| Antibody | Anti-GRIM19 (mouse monoclonal) | Abcam | #ab110240, RRID:AB_10863178 | WB: (1:1,000) |
| Antibody | Anti-SDHA (mouse monoclonal) | Abcam | #ab14715, RRID:AB_301433 | WB: (1:10,000) |
| Antibody | Anti-UQCRFS1 (mouse monoclonal) | Abcam | #ab14746, RRID:AB_301445 | WB: (1:1000) |
| Antibody | Anti-MTCO1 (mouse monoclonal) | Abcam | #ab14705, RRID:AB_2084810 | WB: (1:1000) |
| Antibody | Anti-ATP5a (mouse monoclonal) | Abcam | #ab14748, RRID:AB_301447 | WB: (1:1000) |
| Antibody | Anti-NDUFA9 (mouse monoclonal) | Abcam | #ab14713, RRID:AB_301431 | WB: (1:1000) |
| Antibody | Anti-SDHB (mouse monoclonal) | Abcam | #ab14714, RRID:AB_301432 | WB:(1:2,000) |
| Antibody | Anti-LYRM4 (rabbit polyclonal) | Thermo-Fisher | #PA5-56448 RRID:AB_2643635 | WB:(0.4 µg/mL) |
| Antibody | Anti-VDAC (rabbit polyclonal) | Cell Signaling | #4866, RRID:AB_2272627 | WB: (1:1000) |
| Antibody | Anti-CS (rabbit polyclonal) | Abcam | #ab96600, RRID:AB_10678258 | WB: (1:1000) |
| Antibody | Anti-Lipoic Acid (rabbit polyclonal) | Millipore | #437695, RRID:AB_212120 | WB: (1:1000) |
| Antibody | Anti-LIPT1 (rabbit polyclonal) | Sigma-Aldrich | #AV48784, RRID:AB_185290 | WB: (1:1000) |
| Antibody | Anti-DLAT (mouse monoclonal) | Cell Signaling | #12362, RRID:AB_279789 | WB: (1:1000) |
| Antibody | Anti-DLST (rabbit polyclonal) | Cell Signaling | #5556, RRID:AB_106951 | WB: (1:1000) |
| Antibody | Anti-GAPDH (rabbit monoclonal) | Cell Signaling | #8884 RRID:AB_11129865 | WB: (1:2000) |
| Antibody | Anti-MHC (mouse monoclonal) | DSHB | #SC-71, RRID:AB_2147165 | WB: (0.2 µg/ml) |
| Antibody | Anti-Myogenin (mouse monoclonal) | DSHB | #F5D, RRID:AB_2146602 | WB: (0.5 µg/ml) |
| Antibody | Anti-MyoD (mouse monoclonal) | DSHB | #D7F2, RRID:AB_1157912 | WB: (0.5 µg/ml) |
| Antibody | Goat Anti-Mouse IgG (H and L) Antibody Dylight 800 Conjugated | Rockland | #610-145-002-0.5, RRID:AB_10703265 | WB:(1:10,000) |
| Antibody | Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 680 | Invitrogen | #A10043, RRID:AB_2534018 | WB:(1:10,000) |
| Antibody | Goat Anti-Mouse IgG (H+L) Antibody, Alexa Fluor 680 Conjugated | Invitrogen | #A21057, RRID:AB_141436 | WB:(1:10,000) |
| Antibody | Donkey Anti-Rabbit IgG (H and L) Antibody Dylight 800 Conjugated | Rockland | #611-145-002-0.5, AB_11183542 | WB:(1:10,000) |
| Antibody | Goat anti-Mouse IgG (H+L), Superclonal Recombinant Secondary Antibody, HRP | Thermo Fisher Scientific | #A28177, RRID:AB_2536163 | WB:(1:10,000) |
| recombinant DNA reagent | pMcat CRISPR sgRNA guide 1 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | GCGCGTCGCAATGAGCGCTC |
| recombinant DNA reagent | pMcat CRISPR sgRNA guide 2 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CGAGGCCGCGCACCGGGTAC |
| recombinant DNA reagent | pOxsm CRISPR sgRNA guide 1 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CTAGTGACACCACTTGGCGT |
| recombinant DNA reagent | pOxsm CRISPR sgRNA guide 2 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CGTTGGGACTCAACTAGTTT |
| recombinant DNA reagent | pMecr CRISPR sgRNA guide 1 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | AGGCTTGGTACCGCCACGGC |
| recombinant DNA reagent | pMecr CRISPR sgRNA guide 2 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CGTGGCGGTACCAAGCCTCG |
| recombinant DNA reagent | pLipt1 CRISPR sgRNA guide 1 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CACCGCTTCTAGATGTATGTGGTCG |
| recombinant DNA reagent | Lipt1 CRISPR sgRNA guide 2 (plasmid) | This paper | sgRNA cloned in pLentiCRISPRv2 | CACCGCTCCTTCTGTCGTCATCGGC |
| recombinant DNA reagent | pLentiCRISPRv2 (plasmid) | Addgene | #52961 RRID:Addgene_52961 | |
| recombinant DNA reagent | pLKO.1 shFASN (plasmid) | The Broad Institute | #shRNA TRCN0000075703 | |
| recombinant DNA reagent | pshScramble (plasmid) | Addgene | #1864, RRID:Addgene_1864 | |
| recombinant DNA reagent | psPAX2 (plasmid) | Addgene | #12259, RRID:Addgene_12259 | |
| recombinant DNA reagent | pDM2.G (plasmid) | Addgene | #12260, RRID:Addgene_12260 | |
| recombinant DNA reagent | pQXCIP mtDSRed (plasmid) | This paper | mtDSRed in pQXCIP backbone | |
| recombinant DNA reagent | pQXCIP-Δ4,5CMV-mMcat (plasmid) | This paper | Mouse Mcat with truncated CMV promoter in pQXCIP backbone | |
| recombinant DNA reagent | pQXCIP-CMV-mMcat (plasmid) | This paper | Mouse Mcat with full CMV promoter in pQXCIP backbone | |
| recombinant DNA reagent | pQXCIP-CMV-mOxsm (plasmid) | This paper | Mouse Oxsm with full CMV promoter in pQXCIP backbone | |
| recombinant DNA reagent | pQXCIP-Δ4,5CMV-mMecr (plasmid) | This paper | Mouse Mecr with truncated CMV promoter in pQXCIP backbone | |
| recombinant DNA reagent | pQXCIP-CMV-mMecr (plasmid) | This paper | Mouse Mecr with full CMV promoter in pQXCIP backbone | |
| sequence-based reagent | Mcat CRISPR Ver Fwd | This paper | PCR primers | GACCGACATGCAACTGCAAATAG |
| sequence-based reagent | Mcat CRISPR Ver Rev | This paper | PCR primers | GGCCAGTGAAGCCACAAAGA |
| sequence-based reagent | Oxsm CRISPR Ver Fwd | This paper | PCR primers | CAACCATGTTGTCAAAATGCTTG |
| sequence-based reagent | Oxsm CRISPR Ver Rev | This paper | PCR primers | GGTCTGAAACAGCAAAGCAGTTTC |
| sequence-based reagent | Mecr CRISPR Ver Fwd | This paper | PCR primers | GCTGTCGCGGACGAATG |
| sequence-based reagent | Mecr CRISPR Ver Rev | This paper | PCR primers | GTCGGAAGCATCCACTGAGAC |
| commercial assay or kit | TruSeq Stranded mRNA Library Prep kit | Illumina | #20020595 | |
| commercial assay or kit | TruSeq RNA UD Indexes | Illumina | #20022371 | |
| commercial assay or kit | NovaSeq S1 reagent Kit | Illumina | #20027465 | |
| commercial assay or kit | Kapa Library Quant Kit | Kapa Biosystems | #KK4824 | |
| commercial assay or kit | Pierce BCA Assay | Thermo | #23225 | |
| commercial assay or kit | D1000 ScreenTape assay | Agilent Technologies | 5067–5583 | |
| chemical compound, drug | [U-13C]glutamine | Cambridge Isotopes | #CLM-1822 | |
| chemical compound, drug | [U-13C]glucose | Cambridge Isotopes | #CLM-1396 | |
| chemical compound, drug | Digitonin | GoldBio | #D-180–2.5 | |
| chemical compound, drug | SPLASH Lipidomix | Avanti Polar Lipids | #330707 | |
| software, algorithm | Agilent Mass Hunter Qual B.07.00 | Agilent | https://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter/masshunter-software | |
| software, algorithm | Agilent Mass Hunter Quant B.07.00 | Agilent | https://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter/masshunter-software | |
| software, algorithm | Lipid Annotator | Agilent | https://www.agilent.com/en/products/software-informatics/mass-spectrometry-software/data-analysis/mass-profiler-professional-software | |
| software, algorithm | MetaboAnalyst 4.0 | Xia and Wishart, 2016 | http://www.metaboanalyst.ca | |
| Other | Lipofectamine 2000 transfection reagent | Invitrogen | 11668019 |
Additional files
-
Supplementary file 1
Table of all steady-state metabolites measured in mtFAS mutants and controls.
All steady-state metabolites measured via LCMS. Values shown are average fold change from mean GFP abundance.
- https://cdn.elifesciences.org/articles/58041/elife-58041-supp1-v2.docx
-
Supplementary file 2
Proteins associated with skeletal muscle differentiation are decreased in abudance in mtFAS mutants.
Relative expression of proteins associated with skeletal muscle differentiation from SL-TMT experiment. Values are arbitrary units.
- https://cdn.elifesciences.org/articles/58041/elife-58041-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58041/elife-58041-transrepform-v2.docx