Interplay between bacterial deubiquitinase and ubiquitin E3 ligase regulates ubiquitin dynamics on Legionella phagosomes
Figures
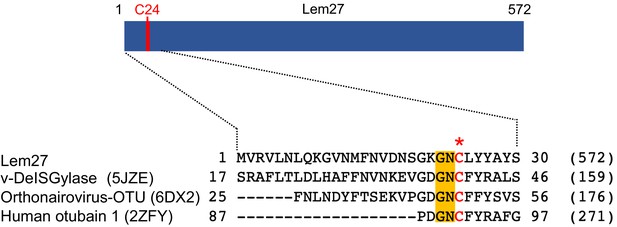
Sequence alignment of Lem27 with proteases involved in removing ubiquitin or ISG15 from modified proteins.
The protein sequence of Lem27 was used as a search query for HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred) analysis. Proteins retrieved by the search known to have protease activity relevant to ubiquitin were aligned manually. The PDB codes for these proteins are in the parentheses. The cysteine residue critical for catalysis in red letter was indicated by an asterisk. Conserved residues adjacent to the catalytic cysteine were highlighted by an orange background. The numbers at the ends of the sequences indicate the positions of the residues in the proteins and the numbers in the parentheses are the lengths of the proteins.
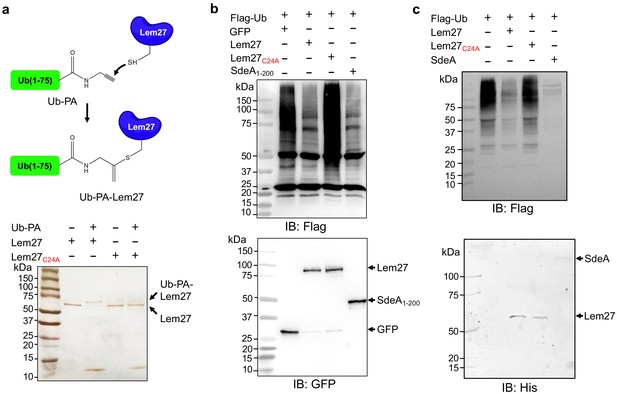
Lem27 is a deubiquitinase whose activity requires Cys24.
(a) The formation of a covalent conjugate between Lem27 and the DUB inhibitor Ub-PA. A diagram showing the chemical reaction between the reactive end of Ub-PA and the side chain of Cys24 from Lem27 (upper panel). Lem27 or Lem27C24A was incubated with Ub-PA and the products resolved by SDS-PAGE were detected by silver staining (lower panel). Note the molecular weight shift of Lem27 after reacting with Ub-PA and the inability of Lem27C24A to cause such shift. (b) Lem27 interferes with protein ubiquitination in cells. HEK293T cells were transfected to coexpress Flag-Ub and GFP-Lem27, GFP-Lem27C24A , or GFP-SdeA1-200. Proteins modified by Flag-Ub were detected by immunoblotting with a Flag-specific antibody (upper panel). The expression of the DUBs and their mutants were detected with GFP antibodies by immunoblotting. (c) Recombinant Lem27 removes ubiquitin from modified proteins. Ubiquitinated proteins isolated by immunoprecipitation from cells transfected to express Flag-Ub were incubated with His6-Lem27, His6-Lem27C24A, or His6-SdeA. Ubiquitination signals were detected by immunoblotting with a Flag-specific antibody (upper panel); recombinant proteins used in the reactions were detected with a His6-specific antibody (lower panel).
Lem27 impacts the association of ubiquitinated proteins on the LCV.
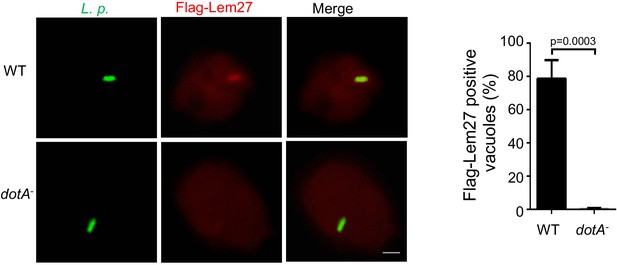
(a) Lem27 is injected into host cells during L. pneumophila infection. U937 cells infected with bacterial strains expressing 4xFlag-Lem27 were lysed with saponin and detected for the proteins of interest in soluble and insoluble fractions, respectively. Note that the protein was expressed comparably in these two strains but it was detected only in cells infected with the strain harboring a functional Dot/Icm system. (b–c) Quantitation of the association of 4xFlag-Lem27 with the LCV and representative images of ubiquitin decorated phagosomes. U937 macrophages infected with the indicated bacterial strains for 2 hr were subjected to immunostaining and the number of vacuoles stained positive by the Flag antibody was determined (b). At least 150 vacuoles were scored for each sample and similar results were obtained in three independent experiments. Data shown were mean ±s.e. Images (c) were acquired with an Olympus IX-83 fluorescence microscope. Bar: 2 μm. (d–f) Lem27 regulates the association of ubiquitinated proteins on the LCV. Macrophages infected with the indicated L. pneumophila strains were immunostained to identify the bacterial vacuoles followed by staining with the FK1 ubiquitin antibody. The percentage of ubiquitin positive vacuoles was scored by counting at least 150 intracellular bacterial (d) and the intensity of the ubiquitin staining signal of the scored vacuoles was measured (e). Results shown were mean ± s.e. from three independent experiments. Representative images of the association of ubiquitin with LCVs containing relevant bacterial strains (f). Images were acquired with an Olympus IX-83 fluorescence microscope. Bar: 2 μm.
The association of Lem27 with L. pneumophila phagosomes in D. discoideum D. discoideum cells seeded on coverslips were infected with wild-type L. pneumophila or the dotA mutant expressing.
Flag-Lem27 for 2 hr. Samples were fixed and subjected to immunostaining to identify the LCVs and Flag-Lem27. Samples were analyzed by visual inspection under a fluorescence microscope to acquire representative images of the LCV positive or negative for Flag-Lem27 (left panel) and to determine the rates of vacuoles stained positive for the protein (right panel) by counting at least 150 phagosomes each sample. Data shown were from three independent experiments. Bar: 2 μm.
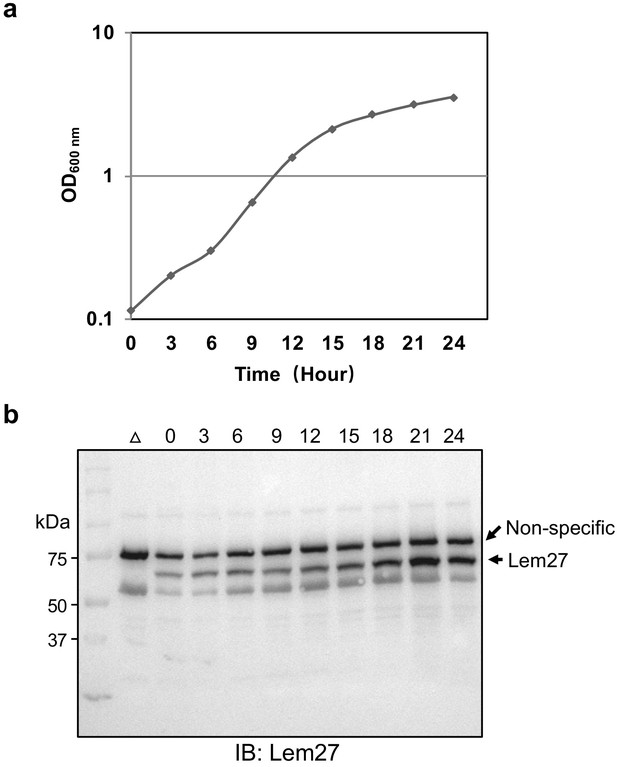
Expression of lem27 in L. pneumophila grown at different phases.
The L. pneumophila strain Lp02 was grown at 37°C in a shaker, cell growth was monitored by measuring the OD600 and continued for 24 hr. The expression level of lem27 was monitored throughout the culture by withdrawing equal amounts of cells at the indicated times. Total proteins resolved by SDS-PAGE were detected by immunoblotting with Lem27-specific antibodies. The ∆lem27 mutant grown at the post-exponential phase was included to identify the protein band representing Lem27. The band above the target protein recognized nonspecifically by the Lem27 antibodies was used as a loading control.
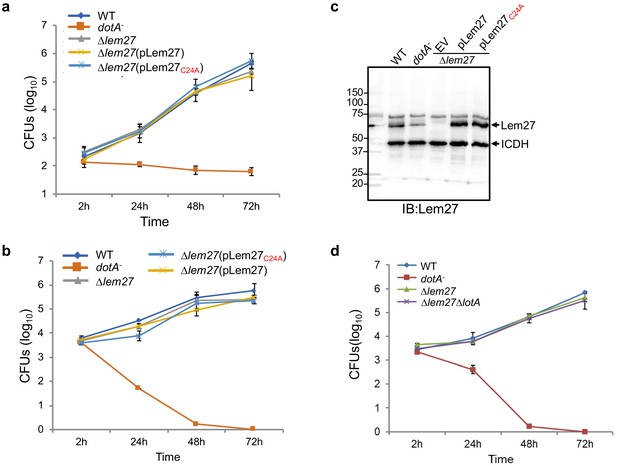
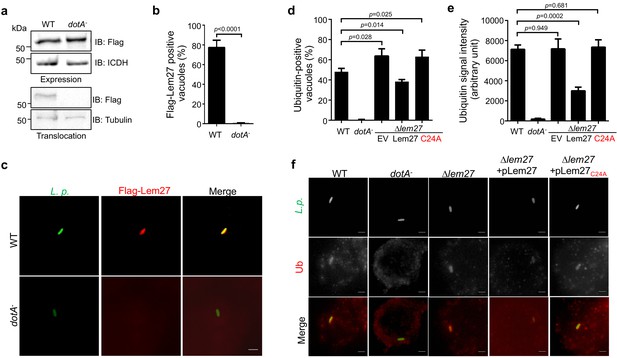
Intracellular growth of L. pneumophila mutants lacking one or more bacterial deubiquitinase genes in host cells.
(a–c) RAW264.7 (a) or D. discoideum (b) cells seeded in 24-well plates were infected with the indicated bacterial strains at an MOI of 0.05, extracellular bacteria were removed by washing 2 hr after uptake. At the indicated time points, samples were lysed with 0.2% saponin and the lysates were plated on CYE plates after proper dilutions. Bacterial colonies were counted to calculate the total CFUs in each sample. Data shown are mean ± s.e. from three samples each strain. Similar results were obtained in three independent experiments. The expression of lem27 in L. pneumophila strains used for intracellular growth experiments. Equal amounts of bacterial cells from strains used for infection in a panel were lysed and proteins resolved by SDS-PAGE was detected by immunoblotting with antibodies specific for Lem27. The metabolic enzyme isocitrate dehydrogenase (ICDH) was probed as a loading control (c). (d) A mutant lacking both lem27 and lotA is not defective in intracellular replication in D. discoideum. The experiments were performed as described in panels a and c with the indicated L. pneumophila strains and the total CFUs in each sample at the indicated time were determined. Data shown are mean ± s.e. from three samples each strain and similar results were obtained in three independent experiments.
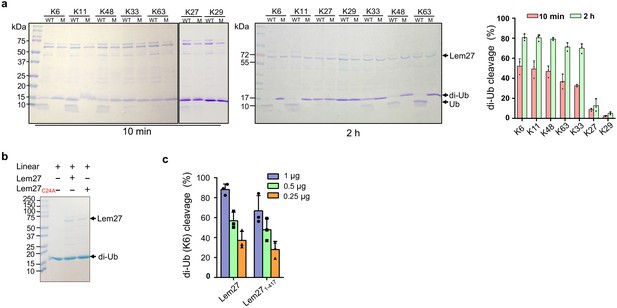
Substrate preference of Lem27.
(a) Diubiquitin linked by K6, K11 or K48 was the preferred substrates for Lem27. A representative image of diubiquitin digestion by Lem27 in which recombinant Lem27 or Lem27C24A was incubated with the indicated diubiquitin for 10 min (left panel) or 2 hr (middle panel). The cleavage of the substrates was detected by Coomassie brilliant blue (CBB) staining after SDS-PAGE. The percentage of cleavage for each tested diubiquitin was calculated from three independent experiments (right panel). (b) Lem27 cannot cleave linear diubiquitin. Recombinant Lem27 or Lem27C24A was incubated with linear diubiquitin for 2 hr and proteins in the reactions were separated by SDS-PAGE and detected by CBB staining. (c) A Lem27 deletion mutant lacking 155 residues from its carboxyl end is active. Reactions containing K6-linked diubiquitin and the indicated amounts of Lem27 or Lem271-417 was allowed to proceed for 10 min prior to SDS-PAGE and CBB staining. The percentage of cleavage shown were from three independent experiments. Data shown were mean ±s.e.
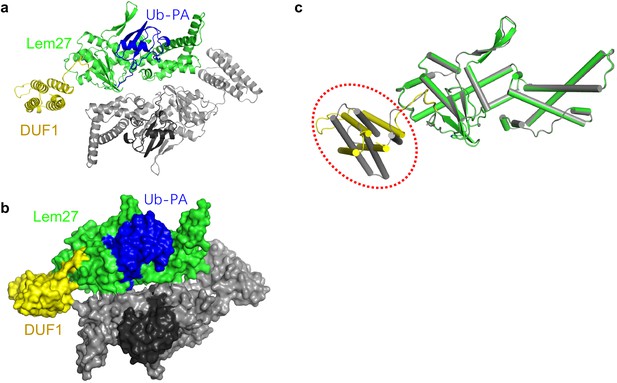
The overall structure of the Lem271-417 -ubiquitin complex in one asymmetric unit.
(a–b) The ribbon mode (a) and surface mode (b) of the structure in one asymmetric unit. (c) Superimposition of the two Lem271-417 in one asymmetric cell, an approximate 35o rotation of the DUF1 domain is circled.
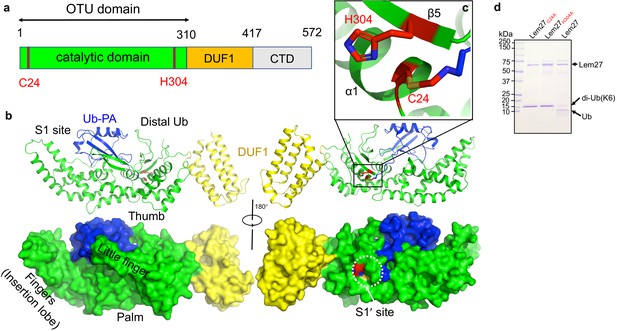
The structure of the covalent complex formed by Lem271-417 and Ub-PA.
(a) Domain organization of full-length Lem27, the DUB module (residues 1–310), the DUF1 (residues 315–417) and the carboxyl terminus domain (CTD, not available in the structure) are shown. (b) Two different views of the overall structure of Lem271-417-Ub-PA. The ribbon mode of Lem271-417-Ub-PA binary complex (upper panel) and surface model (lower panel). Distinct portions of the hand-shaped structure and the S1 and S1' sites were indicated. (c) The configuration of the catalytic center formed by Cys24 and His304 of Lem27. The side chain of Cys24 has formed a chemical bond with Ub-PA. (d) Residues Cys24 and His304 are essential for the enzymatic activity of Lem27. His6-Lem27, His6-Lem27C24A, or His6-Lem27H304A was incubated with K6-type diubiquitin and the protein products were detected by CBB staining after SDS-PAGE. Similar results were obtained in at least three independent experiments.
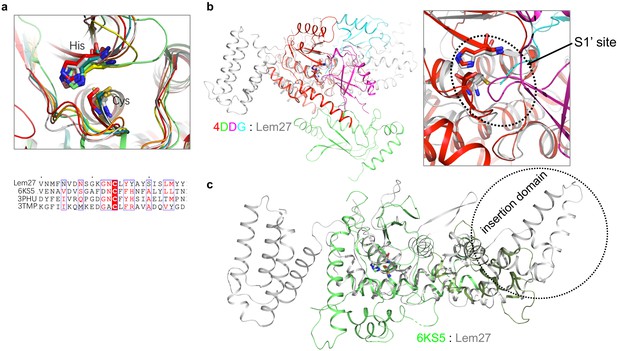
Structural comparison between Lem27 and several DUBs.
(a) Superimposition of the catalytic residues (Cys24 and His304) of Lem27 in grey with the OTU domain-containing proteins listed in Table S1 (upper panel) and the alignment of the indicated sequences around the catalytic Cys residues (lower panel). (b) Superimposition of Lem27 in grey and OTUB1/UbcH5b ~ Ub/Ub (PDB ID: 4DDG) (left panel). Zooming in configuration of the catalytic residues of Lem27 and human OTUB1 in the S1 site (right panel). (c) Superimposition of Lem27 in grey and our previously solved structure of Ceg23 (PDB ID: 6KS5) in green. Note that the insertion domain associated Ceg23 is considerably shorter than that of Lem27.
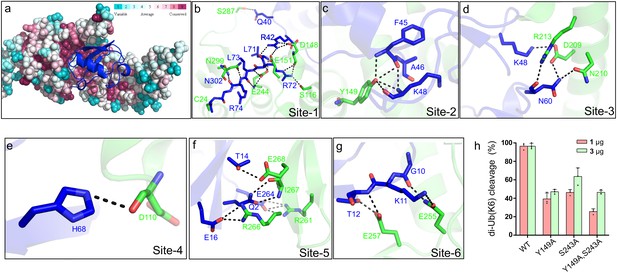
Molecular recognition of ubiquitin by the DUB module of Lem27.
(a) Ubiquitin shown in ribbon mode binds to Lem27 DUB (shown in surface mode) via the vinylthioether bond formed by Cys24 and Ub-PA. Analysis by the ConSurf Server (http://consurftest.tau.ac.il/) shows that the ubiquitin binding sites on the palm of hand-shaped Lem27 are highly conserved. (b) The carboxyl terminus of ubiquitin inserts into the catalytic cleft of Lem27 DUB, which is mainly stabilized by backbone interactions, Lem27 residues involved in interaction with ubiquitin are shown as sticks. (c) Tyr149 of Lem27 specifically binds Phe45 and the Lys48 backbone of ubiquitin. (d) A hydrogen bond net is formed by Lys148, Asn60 of ubiquitin and Arg213, Asp209 and Asn210 of Lem27. (e) The hydrogen formed by His68 of ubiquitin and Asp110 of ubiquitin. (f) The interaction between the β-hairpin of Lem27 and ubiquitin, Arg266, Ile267, Glu268 of β1 of Lem27 and Arg261 of β2 are shown as sticks, the residues in ubiquitin are also shown as sticks. (g) The interaction between the β core of ubiquitin and Lem27. (h) Several residues involved in binding ubiquitin are important for substrate cleavage by Lem27. Data shown were mean ± s.e. as the percentage of cleavage of K6 diubiquitin by Lem271-147 from three independent experiments.
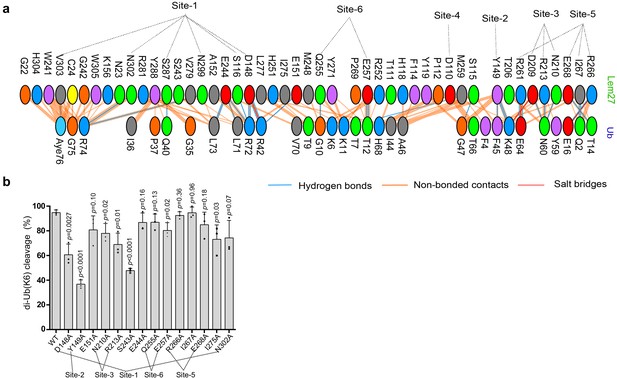
Lem27 makes extensive contacts with ubiquitin by numerous amino acids.
(a) Each oval represents one residues in Lem27 (top) and ubiquitin (bottom), the contact site described in Figure 6 are also indicated. Color codes: positively charged residues (H, His; K, Lys; R, Arg) are colored in blue; negatively charged residues (D, Asp; E, Glu) are colored in red; the neutral residues (S, Ser; T, Thr; N, Asn; Q, Gln) are colored in green; aliphatic residues (A, Ala; V, Val; L, Leu; I, Ile; M, Met) are colored in grey; aromatic residues(F, Phe; Y, Tyr; W, Trp) are colored in magenta, Pro and Gly are colored in orange; cysteine is colored in yellow. (b) Mutagenesis analysis of Lem27 residues involved in interacting with ubiquitin. A series of 14 mutants in the indicated residues were constructed in Lem271-417. Each protein was purified and assayed for the ability to cleave K6-linked diubiquitin. Results shown were the percentage of the activity wild-type protein. Data shown are mean ±s.e. from three independent experiments.
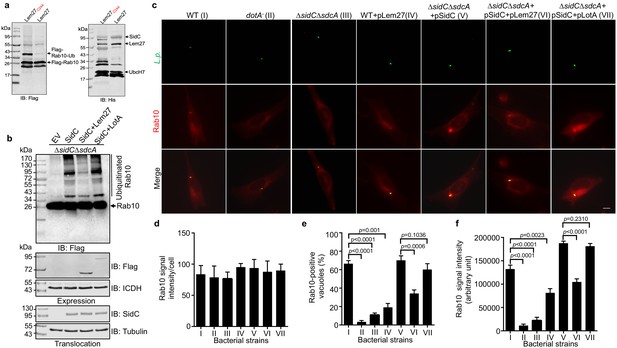
Lem27 regulates Rab10 ubiquitination and its recruitment to the LCV induced by the SidC family of E3 ubiquitin ligases.
(a) SidC-induced Flag-Rab10 ubiquitination was reversed by Lem27. Recombinant Lem27 or its catalytically inactive mutant Lem27C24A was added to completed SidC-catalyzed ubiquitination reactions. Proteins resolved by SDS-PAGE were detected by immunoblotting with the Flag-specific antibody (left panel) or with His6-specific antibody (right panel). Note that Lem27 removes ubiquitin from modified Rab10 and self-modified SidC (His6-SidC was used for the ubiquitination reaction). (b) Lem27 but not LotA reduces Rab10 ubiquitination in infected cells. HEK293T cells transfected to express 4xFlag-Rab10 and the FcγII receptor for 24 hr were infected with opsonized bacteria of the indicated strains for 2 hr prior to detection by immunoblotting with Flag-specific antibody. Note that infection by the strain coexpressing Lem27 led to reduction in ubiquitinate Rab10 (3rd sample). The expression of Flag-Lem27 and Flag-LotA was examined in bacterial cells used for infection with the metabolic enzyme isocitrate dehydrogenase as loading controls (middle two panels) and the translocation of SidC into infected cells by each of the testing strains were detected in saponin-solubilized infected cells with tubulin as loading controls (lower two panels). (c-f) Lem27 but not LotA interferes with Rab10 recruitment to the LCV. A macrophage cell line stably expressing mCherry-Rab10 was infected with the indicated L. pneumophila strains (strain I to VII) for 2 hr and the cells were fixed for immunostaining to identify the bacterial vacuoles. Images were acquired using an Olympus IX-83 fluorescence microscope (c). Bar: 5 μm. The average intensity of mCherry-Rab10 signals per cell in each sample (d), the rates of Rab10-positive LCVs (e) and the intensity of Rab10 signals (f) on the LCVs formed by each bacterial strain were determined. For each sample at least 150 vacuoles were measured. Data shown were mean ±s.e. from three independent experiments.
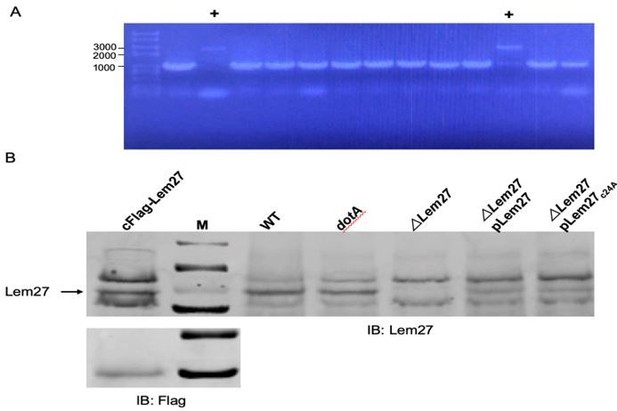
Flag-tagged Lem27 expressed from a knockin L. pneumophila strain can be detected by Lem27 antibodies but not by Flag antibody.
A construct harboring Flag-lem27 on a suicide plasmid was used to introduce tagged lem27 into the ∆lem27 strain make the knockin strain in which Flag-Lem27 will be expressed from its cognate promoter. A. Bacteria harboring Flag-lem27 were identified by colony PCR. The two strains harboring knocked in gene were marked by “+”. B. Expression of Lem27 or Flag-Lem27 in relevant L. pneumophila strains. Total protein lysates of the indicated strains resolved by SDS-PAGE were detected by our Lem27-specific antibodies (top panel). Note the presence of a band in all strains expressing Lem27, including strain cFlag-Lem27, with the exception of strain ∆lem27 (5th lane). Flag-Lem27 was not detected by the Flag antibody (lower panel).
Tables
Data collection and refinement statistics.
| Data collection | SeMet Lem27(1-417)-Ub-PA |
|---|---|
| Wavelength (Å) | 0.9792 |
| Space group | P 1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 66.75,118.95, 84.28 |
| α, β, γ (°) | 90.00, 98.28, 90.00 |
| Resolution (Å) | 31.95–2.43 (2.52–2.43) |
| No. of reflections | 49000 (4827) |
| Rmerge | 0.116 (0.661) |
| Rpim | 0.049 (0.324) |
| I/σI | 9.20 (2.10) |
| Completeness (%) | 99.83 (99.42) |
| Redundancy | 6.6 (5.10) |
| Refinement | |
| Resolution (Å) | 31.950–2.43 (2.52–2.43) |
| No. reflections | 48979 (4824) |
| Rwork/Rfree (%) | 22.27 (33.48)/22.69 (33.46) |
| Total no. of atoms | 7947 |
| Ramachandran plot | |
| Wilson B-factor (Å) | 49.28 |
| Favoured (%) | 96.03 |
| Allowed (%) | 3.77 |
| Outliers (%) | 0.21 |
-
Values in parentheses are for the highest resolution shell.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (L. pneumophila Lp02) | Lp02∆lem27 | This paper | N/A | A lem27 deletion mutant of strain Lp02 |
| Bacterial strain, strain background (L. pneumophila Lp02) | Lp02∆lem27∆lotA | This paper | N/A | A lem27 and lotA deletion mutant of strain Lp02 |
| Bacterial strain, strain background (L. pneumophila Lp02) | Lp02∆sidC∆sdcA | Hsu et al., 2014 | N/A | A sidC and sdcA deletion mutant of strain Lp02 |
| Strain, strain background (L. pneumophila) | Lp02∆sidC∆sdcA(pZL199) | This paper | N/A | Strain Lp02∆sidC∆sdcA expressing SidC |
| Strain, strain background (L. pneumophila) | Lp02∆sidC∆sdcA(pZL199 and pLem27) | This paper | N/A | Strain Lp02∆sidC∆sdcA expressing SidC and Lem27 |
| Strain, strain background (L. pneumophila) | Lp02∆sidC∆sdcA(pZL199 and pLotA) | This paper | N/A | Strain Lp02∆sidC∆sdcA expressing SidC and LotA |
| Cell line (Human) | HEK293T | ATCC | CRL-1573 | |
| Cell line (Human) | Hela | ATCC | CCL-2 | |
| Cell line (Mouse) | RAW264.7 | ATCC | TIB:71 | |
| Cell line (Human) | U937 | ATCC | CRL-1593.2 | |
| Cell line (Mouse) | MLE | ATCC | CRL-2110 | |
| Antibody | Mouse monoclonal ANTI-FLAG antibody M2 | Sigma | Cat. #: F1804 | WB (1: 3000) IF (1: 200) |
| Antibody | Mouse monoclonal ANTI-GFP antibody | Sigma | Cat. #: SAB5300167 | WB (1:5000) |
| Antibody | Mouse monoclonal ANTI-His antibody | Sigma | Cat. #: H1029 | WB (1: 10,000) |
| Antibody | Mouse monoclonal Anti-HA antibody | Santa Cruz | Cat. #: sc-7392 | WB (1: 1000) |
| Antibody | Rabbit polyclonal Anti-ICDH antibody | Xu et al., 2010 | N/A | WB (1: 20,000) |
| Antibody | Mouse monoclonal Anti-tubulin antibody | DSHB | E7 | WB (1: 10,000) |
| Antibody | Rabbit polyclonal Anti-Lem27 antibodies | This paper | N/A | WB (1:500) |
| Antibody | Rabbit polyclonal Anti-SidC antibodies | Luo and Isberg, 2004 | N/A | WB (1:10000) |
| Antibody | Rabbit polyclonal Anti- L. pneumophila antibodies | Xu et al., 2010 | N/A | IF (1:10,000) |
| Antibody | Mouse monoclonal Anti-FKI antibody | Enzo Life Science | Prod. No. BML-PW8805 | IF (1:1,000) |
| Peptide, recombinant protein | 3XFlag Peptide | Sigma-Aldrich | Cat. #: F4799 | |
| Commercial assay or kit | Quikchange kit | Agilent | Cat. #: 600670 | |
| Commercial assay or kit | TransStart Fast Pfu DNA Polymerase | TransGen, Beijing, China | Cat. #: AP221-03 | |
| Chemical compound, drug | Ubiquitin-Propargylamine(Ub-PA) | Boston Biochem | Cat. #: U-214 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K6-linked) Protein, CF | Boston Biochem | Cat. #: UC-11B-025 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K11-linked) Protein, CF | Boston Biochem | Cat. #: UC-40B-025 | |
| Chemical compound, drug | Recombinant Human Di-Ub/Ub2 WT Chains (K27-linked), CF | Boston Biochem | Cat. #: UC-61B-025 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K29-linked) Protein, CF | Boston Biochem | Cat. #: UC-81B-025 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K33-linked) Protein, CF | Boston Biochem | Cat. #: UC-101B-025 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K48-linked) Protein, CF | Boston Biochem | Cat. #: UC-200B-025 | |
| Chemical compound, drug | AQUApure Di-Ub Chains (K63-linked) Protein, CF | Boston Biochem | Cat. #: UC-300B-025 | |
| Chemical compound, drug | Antibiotic G418 | Sigma-Aldrich | Cat. #: G8168 | |
| Software, algorithm | HHpred | Soding et al., 2005 | ||
| Software, algorithm | Adobe Photoshop CS3 Extended | Adobe | ||
| Software, algorithm | Graphpad | graphpad.com | ||
| Other | Anti-HA Magnetic Beads | MedChemExpress (MCE) | Cat. #: HY-K0201 | |
| Other | Anti-flag M2 affinity gel | Sigma | Cat. #: A2220 |
Additional files
-
Supplementary file 1
DALI search results against the PDB using the Lem271-417 structure.
- https://cdn.elifesciences.org/articles/58114/elife-58114-supp1-v3.docx
-
Supplementary file 2
Bacterial strains, plasmids and primers used in this study.
- https://cdn.elifesciences.org/articles/58114/elife-58114-supp2-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58114/elife-58114-transrepform-v3.docx





