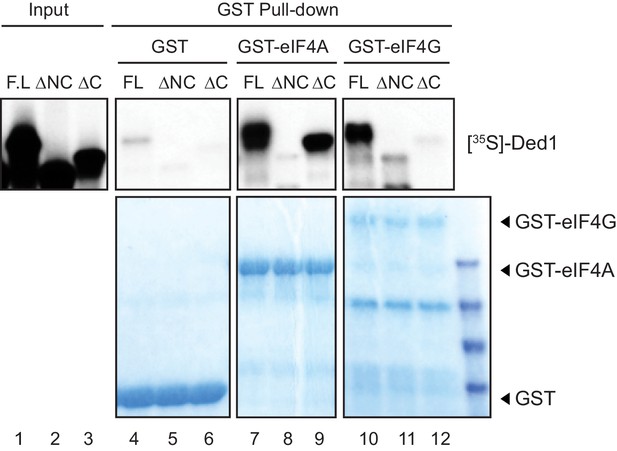
Distinct interactions of eIF4A and eIF4E with RNA helicase Ded1 stimulate translation in vivo
Figures
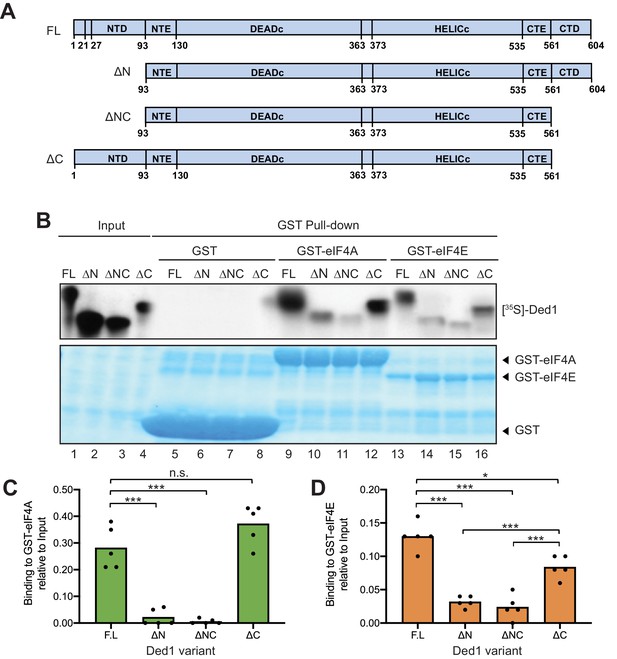
eIF4A and eIF4E interact primarily with the Ded1 N-terminus in vitro.
(A) Schema of in vitro synthesized Ded1 variants, either full length (FL), lacking either the N-terminal domain (NTD) residues 2–92 (ΔN), C-terminal domain (CTD) residues 562–604 (ΔC), or both (ΔNΔC), used in GST pull-down assays. All derivatives contain the entire N- (NTE) and C-terminal (CTE) extensions and the two RecA domains (DEADC and HELICC) responsible for RNA helicase activity. (B) The Ded1 NTD is required for strong binding to GST-tagged eIF4A and eIF4E. The amounts of [35S]-labeled FL or truncated Ded1 proteins, visualized by fluorography (upper panel), present in the reactions (Input) or pulled down by GST, GST-eIF4A, or GST-eIF4A (visualized by Coomassie Blue staining, lower panel). (C–D) Quantification of the binding reactions for GST-eIF4A (C) or GST-eIF4E (D) by ImageJ analysis of fluorograms as in (B) from five replicate pull-down assays, expressing the amounts detected in the pull-downs as the percentages of input amounts. Individual dots show results of the replicates and bar heights give the mean values. n.s: not significant; *: p<0.05; **: p<0.01; ***: p<0.001.
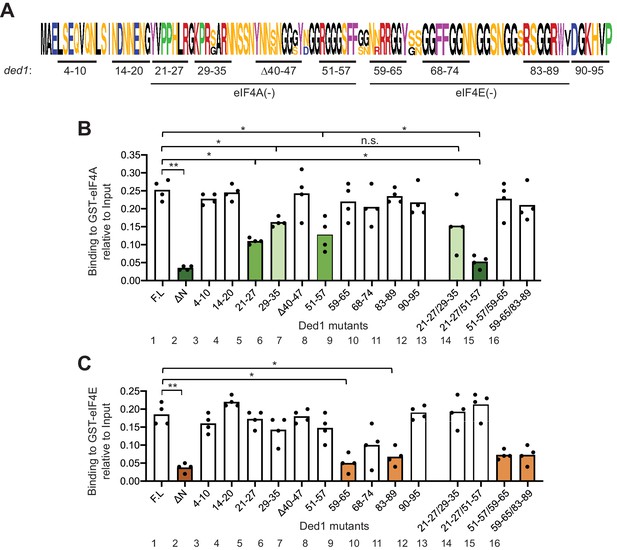
GST-eIF4A and GST-eIF4E bind to distinct non-overlapping segments of the Ded1 NTD.
(A) WebLogo of amino acid sequence conservation in the Ded1 NTD among Saccharomyces species S. cerevisiae, S. arboricola, S. kudriavzevii, S. bayanus, S. boulardii, S. mikatae, S. paradoxus, and S. pastorianus. The blocks of residues chosen for clustered alanine substitutions, or in one case deletion (Δ40), of every residue in the block are underlined and labeled by the residue positions. The locations of segments implicated in binding to eIF4A or eIF4E by results in (B–C) are indicated. (B) Multiple segments in the N-terminal portion of the Ded1-NTD promote binding to GST-eIF4A. Results of pull-down assays using GST-eIF4A and the indicated FL or mutant Ded1 proteins, determined as in Figure 1B–C except using four replicates. Differences between mean values were analyzed with an unpaired students’s t-test. n.s.: not significant, *: p<0.05, **: p<0.01. Shades of green indicate statistically significant decreases in mean values versus the FL construct, with darker shades indicating greater defects. (C) Multiple segments in the C-terminal portion of the Ded1-NTD promote binding to GST-eIF4E. Results of pull-down assays using GST-eIF4E and the indicated FL or mutant Ded1 proteins, determined as in Figure 1B–C except using four replicates. Shades of orange indicate statistically significant decreases in mean values versus the FL construct.
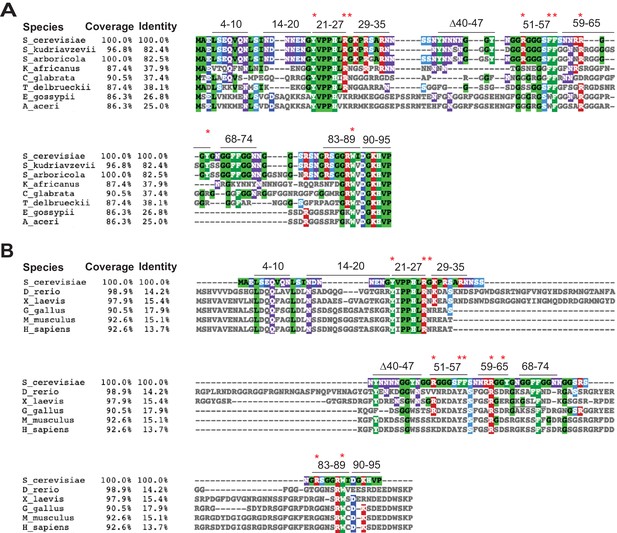
Multiple sequence alignment of the NTDs of Ded1/Ddx3 homologs and residues substituted here in the NTD of S. cerevisiae Ded1.
(A–B) Multiple sequence alignment of Ded1/Ddx3 homologs from the indicated 8 Saccharomycetaceae species (A), and 5 Animalia species and S. cerevisiae Ded1 (B), listed in column 1 (Species), showing only the N-terminal 90 residues of S. cerevisiae Ded1. Residues exhibiting conservation are colored according to the nature of the side chain: green for hydrophobic, red for basic and cyan for acidic. The percentages of residues that can be aligned with, and are identical to, the S. cerevisiae Ded1 sequence are indicated in columns 2–3, respectively. The blocks of residues subjected to clustered alanine (Ala) mutagenesis or deletion are indicated above the S. cerevisiae Ded1 sequences,; and the positions of single-residue Ala substitutions also made subsequently are shown as red asterisks. The alignments were generated by T-Coffee and visualized by MView.
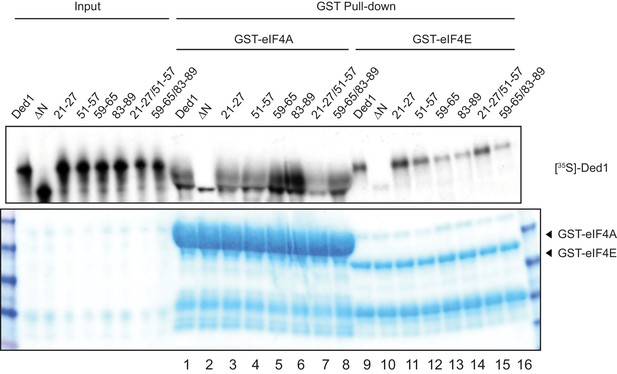
Representative GST pull-down assays of key Ded1 NTD substitution mutants.
Pull-down assays using GST-eIF4A (rows 1–8) or GST-eIF4E (rows 9–16) were conducted in parallel on a subset of Ded1 substitutions that impaired its interactions with eIF4A or eIF4E, carried out as described in Figure 1A.
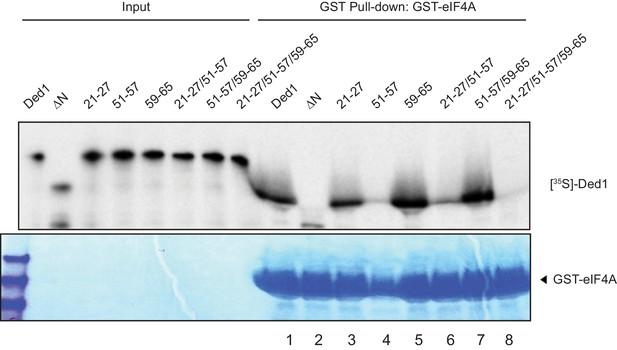
Evidence that substitution 59–65 rescues binding of the Ded1 51–57 variant, but not that of the 21-27/51-57 double mutant, to eIF4A.
GST-eIF4A pull-down assays on the indicated Ded1 variants conducted as in Figure 1B–C.
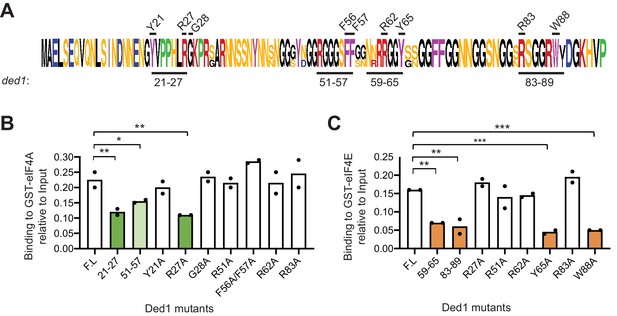
Highly conserved Ded1 residues Arg-27 and Trp-88 and moderately conserved Tyr-65 are all critical for Ded1 binding in vitro to GST-eIF4A or GST-eIF4E, respectively.
WebLogo of amino acid sequence conservation in the Ded1 NTD among the Saccharomyces species, as in Figure 2A, indicating the residues targeted for single Ala substitutions within the indicated blocks of residues subjected to clustered Ala mutagenesis in Figure 2. (B–C) Results of pull-down assays using GST-eIF4A (B) or GST-eIF4E (C) and the indicated WT (FL) or mutant Ded1 proteins, determined as in Figure 1B–C, except using two replicates.
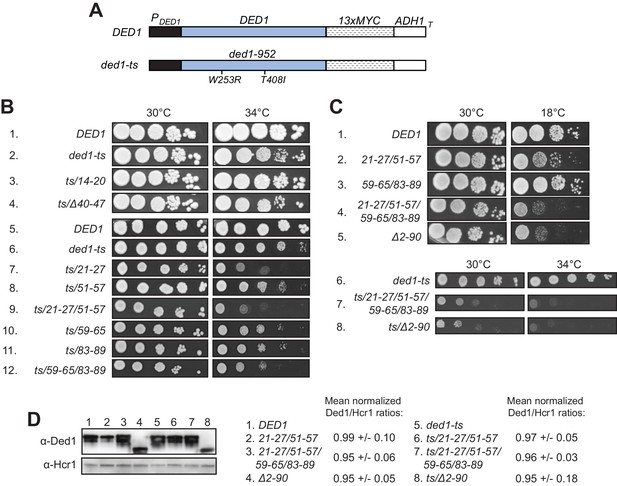
Clustered substitutions of Ded1 NTD residues that impair interaction with eIF4A or eIF4E in vitro confer growth defects in yeast.
(A) Schematics of the myc13-tagged parental DED1 and ded1-ts alleles, expressed from the native DED1 promoter (PDED1) on a single copy (sc) plasmid, used to introduce NTD mutations described in Figure 2A. (B) Mutations substituting single or double binding determinants for eIF4A (21-27; 51-57; 21-27/51-57) or eIF4E (59-65; 83-89; 59-65/83-89) display synthetic temperature sensitivities with the ded1-ts allele of differing severity. Serial dilutions of yeast strains derived from yRP2799 by plasmid-shuffling containing the indicated derivatives of the ded1-ts (rows 2–12), or WT DED1 allele (row 1), on sc LEU2 plasmids (listed in Table 3) were spotted on synthetic complete medium lacking Leu (SC-Leu) and incubated at the indicated temperatures for 2-4d. (C) Disruption of eIF4A and eIF4E binding sites concurrently confers a severe growth defect comparable to that of NTD deletion Δ2–90. Yeast strains harboring the indicated derivatives of DED1 (rows 1–5) or ded1-ts (rows 6–8) were analyzed as in (B). (D) Expression levels of the indicated mutants from (B) or (C) were assessed by Western analysis of WCEs extracted under denaturing conditions with TCA, using the indicated antibodies, following growth in SC-Leu at 18°C for DED1 derivatives and 34°C for ded1-ts derivatives. Ded1/Hcr1 ratios of the indicated derivatives of the WT DED1 or ded1-ts allele were obtained by ImageJ analysis of 3 independent experiments and are normalized to the corresponding parental allele’s Ded1/Hcr1 ratios. This particular blot was atypical in suggesting a reduced level of the ded1-ts-Δ2–90 product.
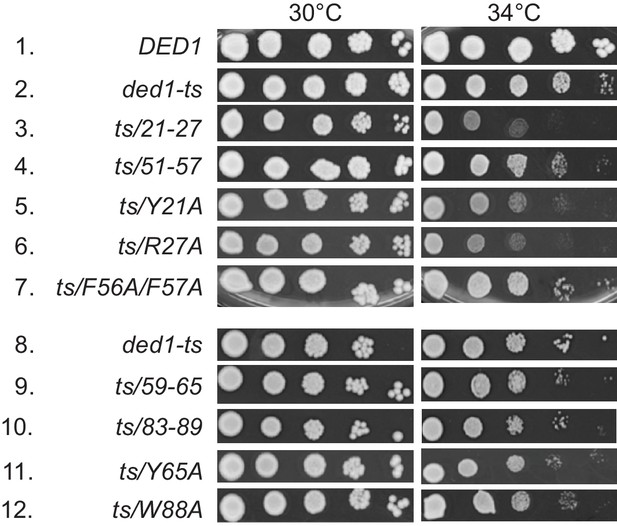
Substitutions of single Ded1 NTD residues that impair interaction with eIF4A or eIF4E in vitro confer growth defects in yeast.
Rates of colony formation of strains derived from yRP2799 containing the WT DED1 allele (row 1) or the indicated derivatives of ded1-ts (rows 2–12), analyzed as in Figure 3B.
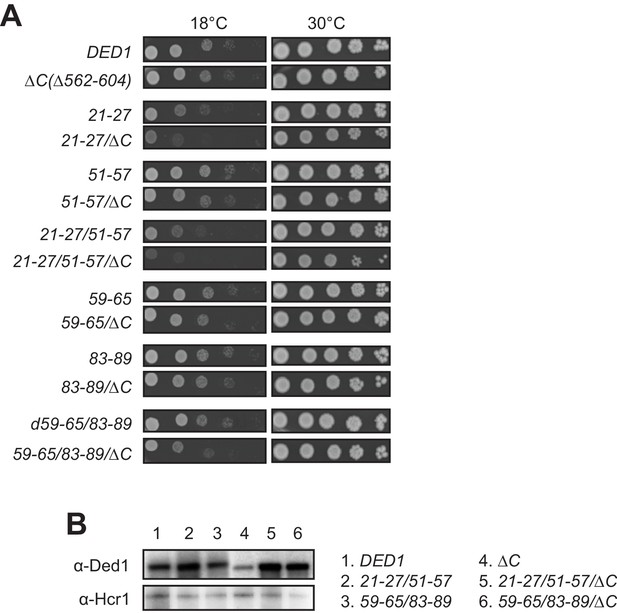
The Ded1 CTD is important for robust cell growth when NTD interactions with eIF4A or eIF4E are compromised.
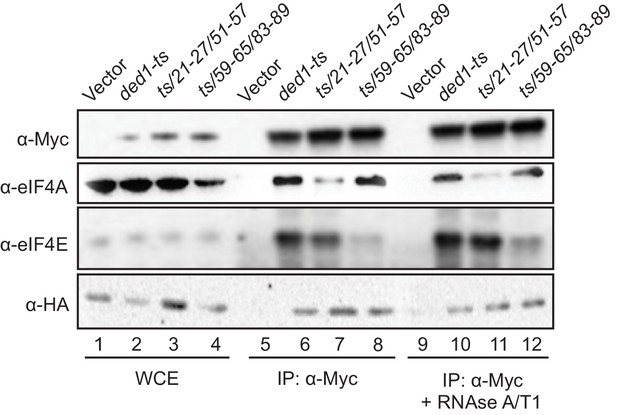
Disrupting Ded1 NTD binding determinants for eIF4A or eIF4E selectively impair association of the ded1-ts product with native eIF4A or eIF4E in yeast WCEs.
Transformants of strain H4436 (expressing HA-tagged eIF4G1 and lacking eIF4G2) harboring the indicated derivatives of (myc13-tagged) ded1-ts were cultured in SC-Leu-Trp medium and WCEs prepared under non-denaturing conditions (lanes 1–4) were immunoprecipitated with anti-myc antibodies (lanes 5–12). Aliquots corresponding to 5% of the WCEs and 50% of the resulting immune complexes, either without treatment (lanes 5–8) or following 30 min treatment with RNAse A/T1 at room temperature (lanes 9–12), were subjected to Western analysis with the indicated antibodies.
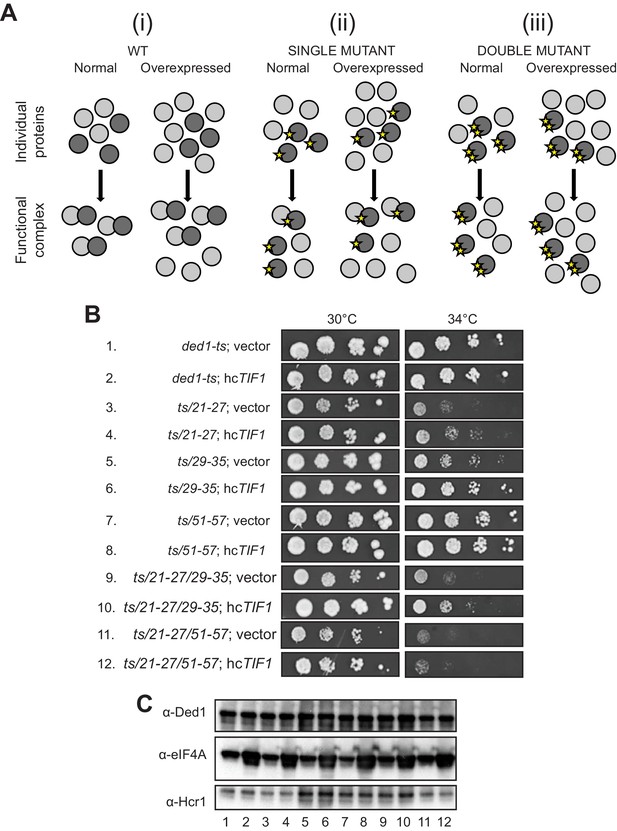
Evidence that Ded1-NTD binding determinants of eIF4A promote cell growth by enhancing eIF4A association.
(A) Schema summarizing expected outcomes for Ded1-eIF4A association based on mass action on overexpressing eIF4A (grey circles) in cells containing different ded1-ts proteins (dark grey circles), as follows: (i) otherwise WT; (ii) a ded1-ts derivative lacking a single binding determinant (single star) that only reduces binding to eIF4A, or (iii) a ded1-ts derivative lacking two binding determinants (double star) that essentially abolishes eIF4A binding. Ded1-eIF4A association depicted by overlapping the circles. (B) Derivatives of strain yRP2799 containing the indicated ded1-ts alleles harboring hcTIF1 plasmid pBAS3432 or empty vector were examined for rates of colony formation at the indicated temperatures as in Figure 3B. (C) Western blot analysis of the strains in (B) conducted as in Figure 3D using the indicated antibodies.
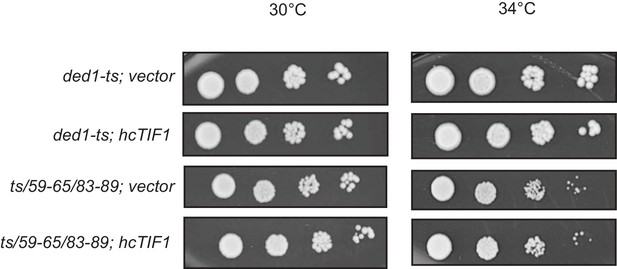
Derivatives of strain yRP2799 containing the indicated ded1-ts alleles harboring hcTIF1 plasmid pBAS3432 or empty vector were examined for rates of colony formation at the indicated temperatures as in Figure 6.
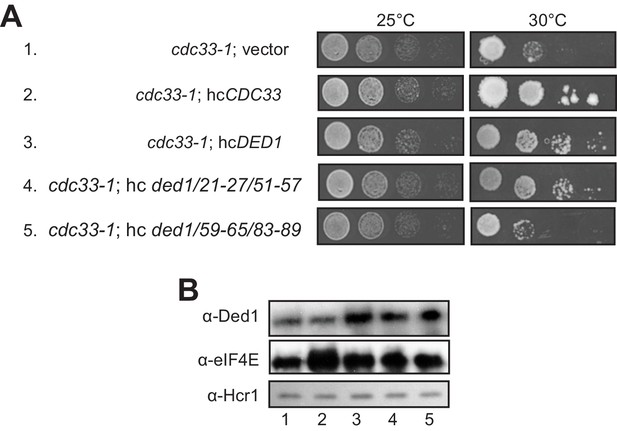
Evidence that Ded1-NTD binding determinants of eIF4E promote cell growth by enhancing eIF4E association.
(A) Transformants of cdc33-1 mutant F696 harboring empty vector YEplac195, hcCDC33 plasmid (p3351), or hc plasmids with the indicated WT (p4504) or mutant (pSG48 or pSG49) DED1 alleles were examined for rates of colony formation at the indicated temperatures as in Figure 3B. (B) Western blot analysis of the strains in (A) conducted as in Figure 3D using the indicated antibodies.
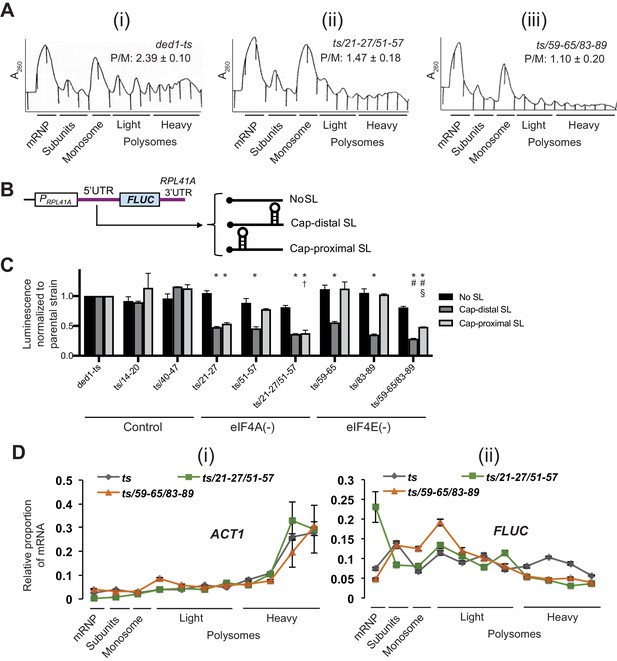
Disruption of Ded1 NTD interactions with eIF4A or eIF4E confer bulk and mRNA-specific translation defects in vivo.
(A) Polysome profiling of derivatives of strain yRP2799 containing the indicated ded1-ts alleles lacking binding determinants for eIF4A (ii) or eIF4E (iii) exhibit a decrease in bulk polysome assembly compared to the parental ded1-ts strain (i). Strains were cultured in SC-Leu medium after shifting from 30°C to 36°C for 3 hr, and WCE extracts were resolved by velocity sedimentation and scanned at 260 nm. The mean ratios of polysomes to monosomes (P/M) determined from three replicate WCEs are indicated. The mean P/M ratios for each mutant differ significantly from the WT mean ratio (both p-values<0.002), but not from each other (p=0.08). Parallel analysis of isogenic DED1+ cells revealed, as expected, a significantly greater P/M ratio (3.90 + / - 0.11) compared to that shown in the figure for ded1-ts (2.39 + / - 0.10). (B) Schema of the reporter constructs employed to interrogate Ded1-dependent translation in vivo, containing the RPL41A promoter and 3’UTR containing derivatives of the RPL41A 5’UTR harboring 23 tandem repeats of CAA nucleotides (nt) and designed to be largely unstructured (No SL, pFJZ342), or additionally containing a stem-loop insertion (of predicted ΔG of −3.7kcal/mol) located 55 nt from the 5’end (Cap-distal SL, pFJZ623), or a SL (of predicted ΔG of −8.1kcal/mol) at seven nt from the 5’end (Cap-proximal SL, pFJZ669). (C) Transformants of strains derived from yRP2799 with the indicated ded1-ts alleles and reporter constructs from (B) were cultured in SC-Leu at 34°C for two doublings and mean specific luciferase activities were determined from three independent transformants and normalized to that obtained for the reporters in the ded1-ts parental strain, which were set to unity.*: Significant differences in mean values compared to ded1-ts; †: compared to ded1-ts/51–57; #: compared to ded1-ts/59–65; §: compared to ded1-ts/83–89, as indicated by a p-value of <0.05 in an unpaired student’s t-test. The effects of substitutions in impairing binding to eIF4A, eIF4E, or neither protein (Control), are indicated at the bottom. (D) Distributions of ACT1 mRNA (i), or Cap-proximal SL reporter mRNA (ii), across sucrose gradients following velocity sedimentation of WCEs of strains from (C) containing the indicated ded1 alleles and the Cap-proximal SL reporter. qRT-PCR was conducted on total RNA purified from each fraction using primers specific for ACT1 or FLUC mRNA, and the abundance of each transcript was normalized to that of a set of ‘spike-in’ controls and plotted as a fraction of the total normalized abundance in the entire gradient. The mean values and S.E.M.s determined from three replicate gradients are plotted.
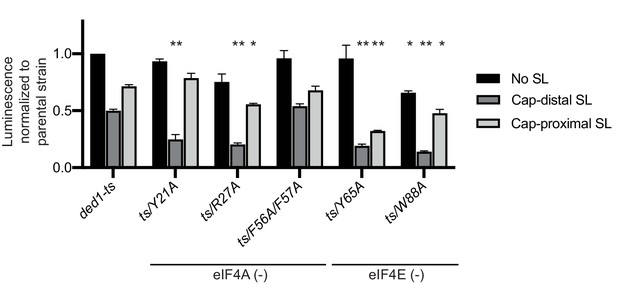
Substitutions of single Ded1 NTD residues that impair interaction with eIF4A or eIF4E in vitro confer translation initiation defects in yeast.
Analysis of FLUC reporter expression for selected ded1-ts derivatives from Figure 3—figure supplement 1 conducted as in Figure 8C, except that expression of the two SL reporters in the parental ded1-ts strain was not normalized to 1.0. Mean specific luciferase activities were determined from three independent transformants. *: p<0.05; **p<0.01. Locations of substitutions within the regions implicated in binding to eIF4A or eIF4E are indicated at the bottom.
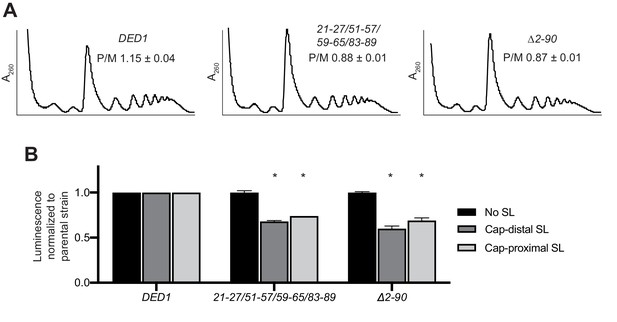
Combining four Ded1-NTD clustered substitutions that individually impair interaction with eIF4A or eIF4E in vitro is comparable to deletion of the Ded1 NTD in reducing translation initiation vivo.
(A) Analysis of bulk polysome assembly in strains derived from yRP2799 containing WT DED1 or the indicated mutant derivatives of DED1, analyzed as in Figure 8A except that cells were shifted from 30°C to 15°C for 1 hr. The mean ratios of polysomes to monosomes (P/M) determined from three replicate WCEs are indicated. (B) Analysis of FLUC reporter expression for the mutants in (A) conducted as in Figure 8C except that cells were cultured at 18°C for two doubling times. Mean specific luciferase activities were determined from three independent transformants. *: p<0.001.
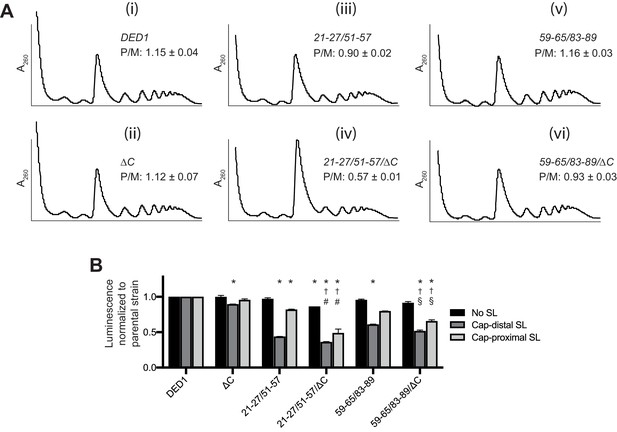
Removal of the Ded1 CTD impairs translation initiation in vivo only when NTD interactions with either eIF4A or eIF4E are compromised.
(A) Analysis of bulk polysome assembly in selected strains from Figure 4 containing WT DED1 or the indicated mutant derivatives of DED1, analyzed as in Figure 8A except that cells were shifted from 30°C to 15°C for 1 hr. The mean ratios of polysomes to monosomes (P/M) determined from three replicate WCEs are indicated. (B) Analysis of FLUC reporter expression for the mutants in (A) conducted as in Figure 8C except that cells were cultured at 18°C for two doubling times. Mean specific luciferase activities were determined from three independent transformants. *: Significant difference compared to DED1; †: compared to ∆C; #: compared to 21-27/51-57; §: compared to 59-65/83-89.
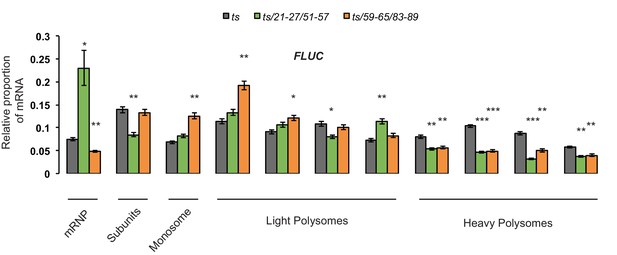
Data from Figure 8D (ii) for the cap-proximal SL FLUC mRNA was analyzed using a Student’s unpaired t-test to compare the mean FLUC mRNA proportions calculated from three replicates for each polysome fraction between the ts/21-27/51-57 and ts/59-65/83-89 mutant strain and the parental strain (ts).
The most significant changes (decreases) in FLUC mRNA distribution occur in heavy polysome fractions in both ts/21-27/51-57 and ts/59-65/83-89 mutants. FLUC mRNA accumulates significantly in the mRNP fraction and the last of the light polysome fractions in the ts/21-27/51-57 mutant; and in the monosome fraction and first two light polysome fractions in the ts/59-65/83-89 mutant. *p<0.05, **p<0.01, ***p<0.005.
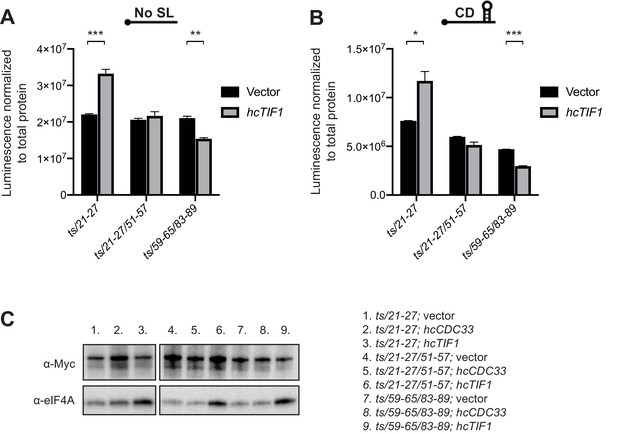
hcTIF1 mitigates the reduction in LUC reporter expression conferred by the ts/21–27 mutation but not by the ts/21-27/51-57 or ts/59-65/83-89 mutations.
(A–B) TIF1 inserted in hc plasmid pRS423 (hcTIF1) or empty vector was transformed into strains derived from yRP2799 with the indicated ded1-ts alleles and (A) No SL or (B) Cap-distal SL reporter constructs from Figure 8B. Luciferase assays were performed as in Figure 8C. Mean luminescence values were determined from three independent transformants and normalized to total protein concentration. *: p<0.05; **: p<0.01; ***: p<0.001. (C) Western blots showing eIF4A overexpression in yRP2799 derivatives expressing the indicated ded1-ts alleles and hcTIF1, compared to the same strains transformed with empty vector pRS423 or hcCDC33 examined as a negative control. Lanes 1, 3, 4, 6, 7 and 9 are representative of the Ded1 and eIF4A amounts in strains used in A and B.
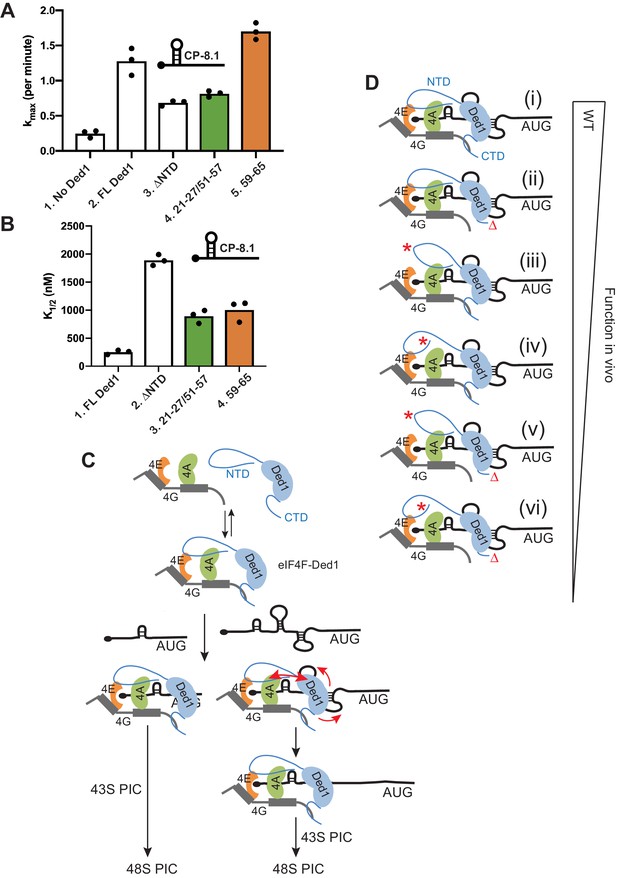
Disruption of Ded1 NTD interactions with eIF4A or eIF4E alter the kinetics of 48S assembly in the reconstituted system for a synthetic mRNA with Cap-proximal SL.
(A–B) Kinetics of 48S PIC assembly was analyzed in reactions containing reconstituted 43S PICs, a radiolabeled capped reporter mRNA containing a cap-proximal SL of predicted ΔG of −8.1kcal/mol located five nt from the 5’end (depicted schematically), and different concentrations of mutant or WT Ded1 protein. Formation of 48S PICs was detected using a native gel mobility shift assay and measured as a function of time, allowing determination of observed rates at each Ded1 concentration. The maximum rates (kmax) (A) and Ded1 concentration at the half-maximal rate (K1/2) (B) were determined from three replicate sets of assays and the individual values (black points) and mean values (bar heights) are plotted for experiments containing no Ded1, WT full-length Ded1 (FL Ded1), Ded1 lacking N-terminal residues 2–116 (ded1ΔNTD), or FL Ded1 harboring the indicated NTD substitutions that impair binding to eIF4A (green bars) or eIF4E (orange bars). (C) Model to account for the greater requirements for the Ded1-NTD interactions with eIF4A and eIF4E in stimulating translation of mRNAs with strong secondary structures versus relatively unstructured 5’UTRs. Ded1 interactions with each of the subunits of the eIF4F complex lowers the concentration of Ded1 required for its recruitment to the capped 5’ ends of all mRNAs, where it can unwind structures that impede PIC attachment or scanning to the start codon. Structured mRNAs (right) require a more stable association between Ded1 and eIF4F for maximum Ded1 recruitment and, hence, are relatively more dependent on having both eIF4A and eIF4E contacts with the Ded1 NTD intact. Unwinding stable SL structures for the latter additionally requires the enhancement of Ded1 unwinding activity conferred by its interaction with eIF4A (red arrows on the right) to achieve maximum acceleration of PIC attachment or scanning. (D) Schematic summary of the relative importance of interactions of the Ded1 NTD with eIF4A and eIF4E and the Ded1 CTD with eIF4G, in stimulating Ded1 recruitment to mRNA and its RNA helicase activity. (i) In WT cells, Ded1’s multiple interactions with the subunits of eIF4F enhance recruitment of Ded1 in complex with eIF4F to the m7G cap to form a stable activated mRNP, which can subsequently recruit the 43S PIC and efficiently scan the 5’UTR to locate the AUG codon (not depicted). Eliminating the Ded1 CTD and its direct contact to eIF4G (ii, red Δ), confers a modest reduction in Ded1 function, whereas greater reductions are conferred by substitution mutations in the Ded1 NTD (red asterisks) that impair Ded1 interaction with eIF4E (iii) or eIF4A (iv). Even greater decreases in Ded1 function are seen on combining each of the Ded1 NTD substitutions with deletion of the Ded1 CTD (v–vi).
-
Figure 9—source data 1
CP-8.1 mRNA recruitment source data.
- https://cdn.elifesciences.org/articles/58243/elife-58243-fig9-data1-v2.xlsx
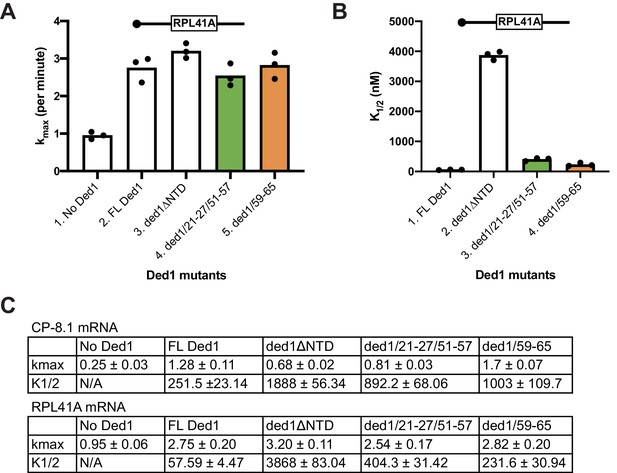
Effects of Ded1 NTD substitutions that impair eIF4A or eIF4E binding on 48S PIC assembly on RPL41A mRNA in the yeast reconstituted system.
(A–B) The analysis was conducted exactly as described in Figure 9A–B for the same panel of Ded1 proteins, except using RPL41A mRNA. (C) Summary of effects of Ded1 NTD substitutions on kmax (in units of min−1) and K1/2 (in nM) for CP-8.1 mRNA determined from the data in Figure 9A–B, and for RPL41A mRNA determined from the data in panels A-B above, as shown for three replicate sets of assays.
-
Figure 9—figure supplement 1—source data 1
RPL41A mRNA recruitment source data.
- https://cdn.elifesciences.org/articles/58243/elife-58243-fig9-figsupp1-data1-v2.xlsx
Tables
Plasmids and yeast alleles used in this study.
| Plasmid | Description | Source |
|---|---|---|
| YCplac111 | empty vector | Gietz and Sugino, 1988 |
| pSG1 | DED1-myc in YCplac111 | This study |
| pSG2 | ded1-ts-myc in YCplac111 | This study |
| pSG3 | ded1-ts/4–10-myc in YCplac111 | This study |
| pSG4 | ded1-ts/14–20-myc in YCplac111 | This study |
| pSG5 | ded1-ts/21–27-myc in YCplac111 | This study |
| pSG6 | ded1-ts/29–35-myc in YCplac111 | This study |
| pSG7 | ded1-ts/Δ40–47-myc in YCplac111 | This study |
| pSG8 | ded1-ts/51–57-myc in YCplac111 | This study |
| pSG9 | ded1-ts/59–65-myc in YCplac111 | This study |
| pSG10 | ded1-ts/68–74-myc in YCplac111 | This study |
| pSG11 | ded1-ts/83–89-myc in YCplac111 | This study |
| pSG12 | ded1-ts/90–95-myc in YCplac111 | This study |
| pSG13 | ded1-ts/21-27/29-35-myc in YCplac111 | This study |
| pSG14 | ded1-ts/21-27/51-57-myc in YCplac111 | This study |
| pSG15 | ded1-ts/59-65/83-89-myc in YCplac111 | This study |
| pSG16 | ded1-ts/21-27/51-57/59-65/83-89-myc in YCplac111 | This study |
| pSG17 | ded1-ts/Δ2–90-myc in YCplac111 | This study |
| pSG18 | ded1-ts,Δ562–604-myc in YCplac111 | This study |
| pSG19 | ded1-ts/21-27/51-57/59-65/83-89-/Δ562–604-myc in YCplac111 | This study |
| pSG20 | ded1-ts,Δ2–90,Δ562–604-myc in YCplac111 | This study |
| pSG21 | ded1-ts/Y21A-myc in YCplac111 | This study |
| pSG22 | ded1-ts/R27A-myc in YCplac111 | This study |
| pSG23 | ded1-ts/G28A-myc in YCplac111 | This study |
| pSG24 | ded1-ts/F56A/F57A-myc in YCplac111 | This study |
| pSG25 | ded1-ts/Y65A-myc in YCplac111 | This study |
| pSG26 | ded1-ts/W88A-myc in YCplac111 | This study |
| pSG27 | ded1-21-27-myc in YCplac111 | This study |
| pSG28 | ded1-51-57-myc in YCplac111 | This study |
| pSG29 | ded1-59-65-myc in YCplac111 | This study |
| pSG30 | ded1-83-89-myc in YCplac111 | This study |
| pSG31 | ded1-21-27/51-57-myc in YCplac111 | This study |
| pSG32 | ded1-59-65/83-89-myc in YCplac111 | This study |
| pSG33 | ded1-21-27/51-57/59-65/83-89-myc in YCplac111 | This study |
| pSG34 | ded1Δ2–90-myc in YCplac111 | This study |
| pSG35 | ded1Δ562–604-myc in YCplac111 | This study |
| pSG36 | ded1-21-27/Δ562–604-myc in YCplac111 | This study |
| pSG37 | ded1-51-57/Δ562–604-myc in YCplac111 | This study |
| pSG38 | ded1-59-65/Δ562–604-myc in YCplac111 | This study |
| pSG39 | ded1-83-89/Δ562–604-myc in YCplac111 | This study |
| pSG40 | ded1-21-27/51-57/Δ562–604-myc in YCplac111 | This study |
| pSG41 | ded1-59-65/83-89/Δ562–604-myc in YCplac111 | This study |
| pSG42 | ded1-21-27/51-57/59-65/83-89/Δ562–604-myc in YCplac111 | This study |
| pSG43 | ded1Δ2–90,Δ562–604-myc in YCplac111 | This study |
| p1992/YEplac195 | empty vector | Gietz and Sugino, 1988 |
| p3333/pBAS3432 | TIF1 in YEplac195 | Neff and Sachs, 1999 |
| p3351 | CDC33 in YEplac195 | de la Cruz et al., 1997 |
| p4504/YEplac195-DED1 | DED1 in YEplac195 | de la Cruz et al., 1997 |
| pSG44 | ded1-21-27/51-57 in YEplac195 | This study |
| pSG45 | ded1-59-65/83-89 in YEplac195 | This study |
| pSG46 | ded1Δ2–90 in YEplac195 | This study |
| p6053/pFJZ342 | RPL41A 5'UTR with 23 CAA repeats inserted (91nt long) in YCplac33 | Sen et al., 2015 |
| p6058/pFJZ669 | RPL41A 5'UTR with cap-proximal SL with ∆G of −8.1 kcal/mol in YCplac33 | Sen et al., 2015 |
| p6062/pFJZ623 | RPL41A 5'UTR with cap-distal SL with ∆G of −3.7 kcal/mol in YCplac33 | Sen et al., 2015 |
| p2917/pGEX-4T1 | empty vector | GE Healthcare 28954549 |
| pGEX-4G1 | TIF4631 in pGEX-4T1 | Mitchell et al., 2010 |
| pSG47 | TIF1 in pGEX-4T1 | This study |
| pSG48 | CDC33 in pGEX-4T1 | This study |
| pET22b | empty vector | EMD Millipore 69744–3 |
| p5946/pET-C-Ded1 | DED1 in pET22b | Hilliker et al., 2011 |
| pSG49 | ded1(93-604)-His in pET22b | This study |
| pSG50 | ded1(1-561)-His in pET22b | This study |
| pSG51 | ded1(93-561)-His in pET22b | This study |
| pSG52 | ded1-4-10-His in pET22b | This study |
| pSG53 | ded1-14-20-His in pET22b | This study |
| pSG54 | ded1-21-27-His in pET22b | This study |
| pSG55 | ded1-29-35-His in pET22b | This study |
| pSG56 | ded1-Δ40–47-His in pET22b | This study |
| pSG57 | ded1-51-57-His in pET22b | This study |
| pSG58 | ded1-59-65-His in pET22b | This study |
| pSG59 | ded1-68-74-His in pET22b | This study |
| pSG60 | ded1-83-89-His in pET22b | This study |
| pSG61 | ded1-90-95-His in pET22b | This study |
| pSG62 | ded1-21-27/29-35-His in pET22b | This study |
| pSG63 | ded1-21-27/51-57-His in pET22b | This study |
| pSG64 | ded1-21-27/68-74-His in pET22b | This study |
| pSG65 | ded1-51-57/59-65-His in pET22b | This study |
| pSG66 | ded1-59-65/83-89-His in pET22b | This study |
| pSG67 | ded1-Y21A-His in pET22b | This study |
| pSG68 | ded1-R27A-His in pET22b | This study |
| pSG69 | ded1-G28A-His in pET22b | This study |
| pSG70 | ded1-R51A-His in pET22b | This study |
| pSG71 | ded1-F56A/F57A-His in pET22b | This study |
| pSG72 | ded1-R62A-His in pET22b | This study |
| pSG73 | ded1-Y65A-His in pET22b | This study |
| pSG74 | ded1-R83A-His in pET22b | This study |
| pSG75 | ded1-W88A-His in pET22b | This study |
| pET-N-Ded1 | His-DED1 in pET15b | Gupta et al., 2018 |
| pET-SUMO-ded1ΔN | His-ded1Δ2–117 in pET-SUMO | Gupta et al., 2018 |
| pSG76 | His-ded1-21 in pET15b | This study |
| pSG77 | His-ded1-51 in pET15b | This study |
| pSG78 | His-ded1-59 in pET15b | This study |
| pSG79 | His-ded1-83 in pET15b | This study |
| pSG80 | His-ded1-21,51 in pET15b | This study |
| pSG81 | His-ded1-59,83 in pET15b | This study |
| pGIBS-LYS | lysA | ATCC #87482 |
| pGIBS-TRP | trpCDEF | ATCC #87485 |
| pGIBS-PHE | pheB | ATCC #87483 |
Primer sequences used in this study.
| Primer | 5’-Sequence-3’ | Source |
|---|---|---|
| SG1/Ded1-13xmyc-F | TTTTTTGCATGCCCAAAGGTGTTCTTATGTAGTGACACCGAT | This study |
| SG2/Ded1-13xmyc-R | TTTTTTGAGCTCTTACCCTGTTATCCCTAGCGGATCTGCCGG | This study |
| SG134/W253R-F | AATTTACTTATAGATCCAGGGTCAAGGCCTGCGTC | This study |
| SG135/W253R-R | GACGCAGGCCTTGACCCTGGATCTATAAGTAAATT | This study |
| SG136/T408I-F | CAATTGATCTGCCATTCTCTTAATTTCGACAAAGATCAAAGTCAAA | This study |
| SG137/T408I-R | TTTGACTTTGATCTTTGTCGAAATTAAGAGAATGGCAGATCAATTG | This study |
| SG3/Cdc33-F | TTTTTTGGATCCTCCGTTGAAGAAGTTAGCAAG | This study |
| SG4/Cdc33-R | TTTTTTCTCGAGTTACAAGGTGATTGATGGTTG | This study |
| SG132/Tif1-F | TTTTTTGGATCCATGTCTGAAGGTATTACTGA | This study |
| SG133/Tif1-R | TTTTTTCTCGAGTTAGTTCAACAAAGTAGCGA | This study |
| SG5/pETded1(93-561)-F | GGGGGTCATATGCATGTCCCAGCTCCAAGAAA | This study |
| SG6/pETded1(93-561)-R | TTTTTTCTCGAGGCCTCCGGCCTTACGGTAAT | This study |
| SG7/pETded1(93-604)-F | TTTTTTCATATGCATGTCCCAGCTCCAAGAAAC | This study |
| SG8/pETded1(93-604)-R | TTTTTTCTCGAGCCACCAAGAAGAGTTGTTTGA | This study |
| SG9/pETded1(1-561)-F | TGTGTGCATATGGCTGAACTGAGCGAACAAGTG | This study |
| SG10/pETded1(1-561)-R | TTTTTTCTCGAGGCCTCCGGCCTTACGGTAAT | This study |
| SG41/pET4-s | GTGAGGAGGAACATAACCATTCTCGTTGTTGTCGTTGATGCTTAAAGCTGCCGCTGCTGCGGCCGCTTCAGCCATATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTT | This study |
| SG42/pET4-as | AAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGCTGAAGCGGCCGCAGCAGCGGCAGCTTTAAGCATCAACGACAACAACGAGAATGGTTATGTTCCTCCTCAC | This study |
| SG43/y4-s | GGAGGAACATAACCATTCTCGTTGTTGTCGTTGATGCTTAAAGCTGCCGCTGCTGCGGCCGCTTCAGCCATAATATGAAATGCTTTTCTTGTTGTTCTTACGGA | This study |
| SG44/y4-as | TCCGTAAGAACAACAAGAAAAGCATTTCATATTATGGCTGAAGCGGCCGCAGCAGCGGCAGCTTTAAGCATCAACGACAACAACGAGAATGGTTATGTTCCTCC | This study |
| SG45/14 s | CTTGGTTTTCCTCTTAAGTGAGGAGGAACATAAGCAGCCGCGGCGGCGGCGGCGATGCTTAAATTTTGCACTTGTTCGCTCAGTTCAGCCA | This study |
| SG46/14-as | TGGCTGAACTGAGCGAACAAGTGCAAAATTTAAGCATCGCCGCCGCCGCCGCGGCTGCTTATGTTCCTCCTCACTTAAGAGGAAAACCAAG | This study |
| SG47/21 s | TTGCTACTGTTATTTCTGGCACTTCTTGGTTTTCCTGCTGCGGCAGCAGCAGCAGCACCATTCTCGTTGTTGTCGTTGATGCTTAAATTTTGCACTTG | This study |
| SG48/21-as | CAAGTGCAAAATTTAAGCATCAACGACAACAACGAGAATGGTGCTGCTGCTGCTGCCGCAGCAGGAAAACCAAGAAGTGCCAGAAATAACAGTAGCAA | This study |
| SG49/29 s | GTTGTAGCCGCCGTTGTTGTTATTGTAGTTGCTACTGTTAGCTGCTGCAGCTGCTGCTGCTCCTCTTAAGTGAGGAGGAACATAACCATTCTCGTTGTTG | This study |
| SG50/29-as | CAACAACGAGAATGGTTATGTTCCTCCTCACTTAAGAGGAGCAGCAGCAGCTGCAGCAGCTAACAGTAGCAACTACAATAACAACAACGGCGGCTACAAC | This study |
| SG51/51 s | ACCACCACGACGGTTGTTGCTAGCGGCGGCGGCAGCGGCAGCGCCACCGTTGTAGCCGCCGTTG | This study |
| SG52/51-as | CAACGGCGGCTACAACGGTGGCGCTGCCGCTGCCGCCGCCGCTAGCAACAACCGTCGTGGTGGT | This study |
| SG53/68 s | CGTTAGATCTGCTGCCACCGTTGGCTGCAGCGGCGGCAGCAGCGTTGCCGTAACCACCACGACGG | This study |
| SG54/68-as | CCGTCGTGGTGGTTACGGCAACGCTGCTGCCGCCGCTGCAGCCAACGGTGGCAGCAGATCTAACG | This study |
| SG55/83 s | TTGGAGCTGGGACATGTTTGCCATCGGCCGCTGCAGCAGCAGCAGCGCCGTTAGATCTGCTGCCACCGTTGT | This study |
| SG56/83-as | ACAACGGTGGCAGCAGATCTAACGGCGCTGCTGCTGCTGCAGCGGCCGATGGCAAACATGTCCCAGCTCCAA | This study |
| SG57/90 s | GATCTCGGCCTTTTCGTTTCTTGCAGCTGCGGCAGCTGCGGCAGCGATCCATCTACCACCAGAACGG | This study |
| SG58/90-as | CCGTTCTGGTGGTAGATGGATCGCTGCCGCAGCTGCCGCAGCTGCAAGAAACGAAAAGGCCGAGATC | This study |
| SG105/40del-s | AAATAACAGTAGCAACAACGGTGGCCGTGGCG | This study |
| SG106/40del-as | CGCCACGGCCACCGTTGTTGCTACTGTTATTT | This study |
| SG303/59 s | TTCCACCGAAGAAACCACCGTTGCCGGCAGCAGCAGCAGCGGCGGCGCTAAAGAAGCTGCCACCGCCACGGC | This study |
| SG304/59-as | GCCGTGGCGGTGGCAGCTTCTTTAGCGCCGCCGCTGCTGCTGCTGCCGGCAACGGTGGTTTCTTCGGTGGAA | This study |
| SG305/59on51-s | CACCGAAGAAACCACCGTTGCCGGCAGCAGCAGCAGCGGCGGCGCTAGCGGCGGCGGCAGCGGCAG | This study |
| SG306/59on51-as | CTGCCGCTGCCGCCGCCGCTAGCGCCGCCGCTGCTGCTGCTGCCGGCAACGGTGGTTTCTTCGGTG | This study |
| SG195/R27A-s | CTTCTTGGTTTTCCTGCTAAGTGAGGAGGAACATAACCATTCTCG | This study |
| SG196/R27A-as | CGAGAATGGTTATGTTCCTCCTCACTTAGCAGGAAAACCAAGAAG | This study |
| SG197/R51A-s | CTAAAGAAGCTGCCAGCGGCAGCGCCACCGTTGTAGCC | This study |
| SG198/R51A-as | GGCTACAACGGTGGCGCTGCCGCTGGCAGCTTCTTTAG | This study |
| SG199/R62A-s | GAAACCACCGTTGCCGTAAGCAGCAGCACGGTTGTTGCTAAAGAAG | This study |
| SG200/R62A-as | CTTCTTTAGCAACAACCGTGCTGCTGCTTACGGCAACGGTGGTTTC | This study |
| SG201/R83A-s | CCATCGATCCATCTACCAGCAGCAGCGCCGTTAGATCTGCTGCC | This study |
| SG202/R83A-as | GGCAGCAGATCTAACGGCGCTGCTGCTGGTAGATGGATCGATGG | This study |
| SG161/ydelC-s | CCGTAAGGCCGGAGGCGGTGAACAAAAGCTAAT | This study |
| SG162/ydelC-as | ATTAGCTTTTGTTCACCGCCTCCGGCCTTACGG | This study |
| SG265/y2-90del-s | CTGGGACATGTTTGCCATCCATAATATGAAATGCTTTTCTTGTTGTTC | This study |
| SG266/y2-90del-as | GAACAACAAGAAAAGCATTTCATATTATGGATGGCAAACATGTCCCAG | This study |
| SG233/Y21A-s | AAGTGAGGAGGAACAGCACCATTCTCGTTGTTGTCGTTGATGC | This study |
| SG234/Y21A-as | GCATCAACGACAACAACGAGAATGGTGCTGTTCCTCCTCACTT | This study |
| SG235/F56F57-s | CACGACGGTTGTTGCTAGCGGCGCTGCCACCGCCACGGC | This study |
| SG236/F56F57-as | GCCGTGGCGGTGGCAGCGCCGCTAGCAACAACCGTCGTG | This study |
| SG241/Y65A-s | CCACCGTTGCCGGCACCACCACGACGGTTGT | This study |
| SG242/Y65A-as | ACAACCGTCGTGGTGGTGCCGGCAACGGTGG | This study |
| SG243/W88A-s | CATGTTTGCCATCGATCGCTCTACCACCAGAACGGC | This study |
| SG244/W88A-as | GCCGTTCTGGTGGTAGAGCGATCGATGGCAAACATG | This study |
| SG249/G28A-s | TGGCACTTCTTGGTTTTGCTCTTAAGTGAGGAGGA | This study |
| SG250/G28A-as | TCCTCCTCACTTAAGAGCAAAACCAAGAAGTGCCA | This study |
| FZ158/Fluc-F | ATG GAA GAC GCC AAA AAC ATA AAG | Sen et al., 2015 |
| FZ159/Fluc-R | TTA CAA TTT GGA CTT TCC GCC CTT | Sen et al., 2015 |
| act1-F | TGTGTAAAGCCGGTTTTGCC | Zeidan et al., 2018 |
| act1-R | GATACCTCTCTTGGATTGAGCTTC | Zeidan et al., 2018 |
Yeast strains used in this study.
| Strain | Description | Source |
|---|---|---|
| F2041/yRP2799 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pRP1560 (DED1 URA3) | Hilliker et al., 2011 |
| H3666 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 DED1-myc13 HIS3 | Dong et al., 2005 |
| F729/BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| H4436* | MATa ade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::leu2hisG tif4632::ura3 pEP88 (TIF4631-HA-Bam TRP1 CEN4) | Park et al., 2011 |
| F694 | MATa cdc33::LEU2 ade2 his3 leu2 trp1 ura3 p(CDC33, TRP1, ARS/CEN4) | Altmann and Trachsel, 1989 |
| F695 | MATa cdc33::LEU2 ade2 his3 leu2 trp1 ura3 p(cdc33-4-2, TRP1, ARS/CEN4) | Altmann and Trachsel, 1989 |
| F696 | MATa cdc33::LEU2 ade2 his3 leu2 trp1 ura3 p(cdc33-1, TRP1, ARS/CEN4) | Altmann and Trachsel, 1989 |
| SGY1 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG1 (DED1-myc LEU2) | This study |
| SGY2 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG2 (ded1-ts-myc LEU2) | This study |
| SGY3 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG3 (ded1-ts/4–10-myc LEU2) | This study |
| SGY4 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG4 (ded1-ts/14–20-myc LEU2) | This study |
| SGY5 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG5 (ded1-ts/21–27-myc LEU2) | This study |
| SGY6 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG6 (ded1-ts/29–35-myc LEU2) | This study |
| SGY7 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG7 (ded1-ts/Δ40–47-myc LEU2) | This study |
| SGY8 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG8 (ded1-ts/51–57-myc LEU2) | This study |
| SGY9 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG9 (ded1-ts/59–65-myc LEU2) | This study |
| SGY10 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG10 (ded1-ts/68–74-myc LEU2) | This study |
| SGY11 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG11 (ded1-ts/83–89-myc LEU2) | This study |
| SGY12 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG12 (ded1-ts/90–95-myc LEU2) | This study |
| SGY13 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG13 (ded1-ts/21-27/29-35-myc LEU2) | This study |
| SGY14 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG14 (ded1-ts/21-27/51-57-myc LEU2) | This study |
| SGY15 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG15 (ded1-ts/59-65/83-89-myc LEU2) | This study |
| SGY16 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG16 (ded1-ts/21-27/51-57/59-65/83-89-myc LEU2) | This study |
| SGY17 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG17 (ded1-ts,Δ2–90-myc LEU2) | This study |
| SGY18 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG18 (ded1-ts,Δ562–604-myc LEU2) | This study |
| SGY19 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG19 (ded1-ts/21-27/51-57/59-65/83-89-myc/Δ562–604-myc LEU2) | This study |
| SGY20 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG20 (ded1-ts,Δ2–90,Δ562–604-myc LEU2) | This study |
| SGY21 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG21 (ded1-ts/Y21A-myc LEU2) | This study |
| SGY22 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG22 (ded1-ts/R27A-myc LEU2) | This study |
| SGY23 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG23 (ded1-ts/G28A-myc LEU2 | This study |
| SGY24 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG24 (ded1-ts/F56A/F57A-myc LEU2) | This study |
| SGY25 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG25 (ded1-ts/Y65A-myc LEU2) | This study |
| SGY26 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG26 (ded1-ts/W88A-myc LEU2) | This study |
| SGY27 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG27 (ded1-21-27-myc LEU2) | This study |
| SGY28 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG28 (ded1-51-57-myc LEU2) | This study |
| SGY29 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG29 (ded1-59-65-myc LEU2) | This study |
| SGY30 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG30 (ded1-83-89-myc LEU2) | This study |
| SGY31 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG31 (ded1-21-27,51–57-myc LEU2) | This study |
| SGY32 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG32 (ded1-59-65,83–89-myc LEU2) | This study |
| SGY33 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG33 (ded1-21-27/51-57/59-65/83-89-myc-myc LEU2) | This study |
| SGY34 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG34 (ded1Δ2–90-myc LEU2) | This study |
| SGY35 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG35 (ded1Δ562–604-myc LEU2) | This study |
| SGY36 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG36 (ded1-21-27/Δ562–604-myc LEU2) | This study |
| SGY37 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG37 (ded1-51-57/Δ562–604-myc LEU2) | This study |
| SGY38 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG38 (ded1-59-65/Δ562–604-myc LEU2) | This study |
| SGY39 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG39 (ded1-83-89/Δ562–604-myc LEU2) | This study |
| SGY40 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG40 (ded1-21-27/51-57/Δ562–604-myc LEU2) | This study |
| SGY41 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG41 (ded1-59-65/83-89/Δ562–604-myc LEU2) | This study |
| SGY42 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG42 (ded1-21-27/51-57/59-65/83-89-myc/Δ562–604-myc LEU2) | This study |
| SGY43 | MATa his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 ded1Δ::kanMX4 pSG43 (ded1-Δ2–90,Δ562–604-myc LEU2) | This study |
-
*The plasmid in this strain contains TIF4631-HA under the control of the TIF4632 promoter and transcription terminator, as described previously (Tarun and Sachs, 1996) modified to insert a BamHI site at the start codon (Park et al., 2011).