Modular output circuits of the fastigial nucleus for diverse motor and nonmotor functions of the cerebellar vermis
Figures
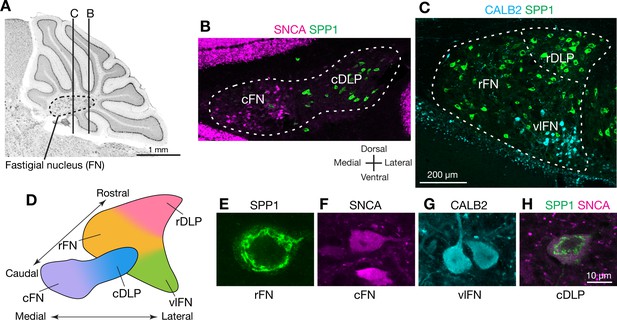
Anatomically distinct cell types in the fastigial nucleus.
(A) The fastigial nucleus (FN) in a sagittal section of the mouse cerebellar vermis identified with Nissl staining. Vertical lines indicate the location of sections in panels B and C. (B) Double immunostaining for alpha synuclein (SNCA) and osteopontin (SPP1) in a coronal section of the FN (n = 3 males). SNCA immunopositive neurons (green) are prominent in the caudal FN (cFN). SPP1 immunopositive neurons (magenta) are distributed throughout the caudal portion of the dorsolateral protuberance (cDLP). (C) Double immunostaining for calretinin (CALB2) and SPP1 in a more rostral section of the FN (n = 3 males). Intensely CALB2+ neurons (cyan) are densely clustered in the ventrolateral FN (vlFN). SPP1+ neurons (red) are distributed throughout the rostral FN (rFN) and the rostral portion of the DLP (rDLP). (D) Five subregions of the FN can be delineated by the distribution of neurons expressing SNCA, CALB2, and/or SPP1: (1) the cFN (purple) comprises SNCA+ neurons; (2) the cDLP (blue) comprises neurons which co-express SNCA and SPP1; (3) the vlFN (green) comprises neurons that express CALB2 and SNCA; (4, 5) the rDLP (pink) and rFN (yellow) each comprise exclusively SPP1+ neurons. (E–H) High magnification images of immunohistochemically revealed neurons located in the rFN (E, SPP1), cFN (F, SNCA), vlFN (G, CALB2) and cDLP (H, SPP1 and SNCA). Note considerable difference in the sizes of these neurons. Scale bar in C applies to B and C. Scale bar in H applies to E-H.
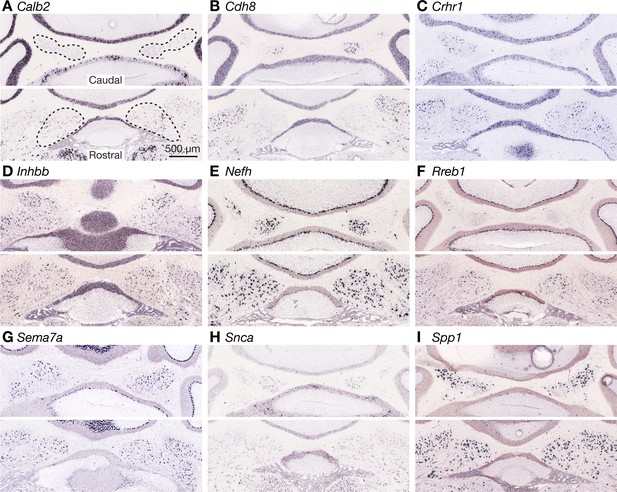
Expression of marker candidate genes for fastigial cell types.
(A–I) Expression of marker candidate genes for molecularly distinct fastigial neurons. The images are from the in situ hybridization database available at Allen Brain Atlas (Lein et al., 2007). The experiment IDs are, 79556662 for Calb2 (A), 79568026 for Cdh8 (B), 297 for Crhr1 (C), 73636087 for Inhbb (D), 74512048 for Nefh (E), 72340134 for Rreb1(F), 955 for Sema7a (G), 79908848 for Snca (H), and 70436740 for Spp1 (I). Each panel consists of two representative coronal sections showing caudal (top) and rostral (bottom) areas of the FN. Contour of the FN are drawn with dotted line in (A). Scale bar in A applies to A-I.
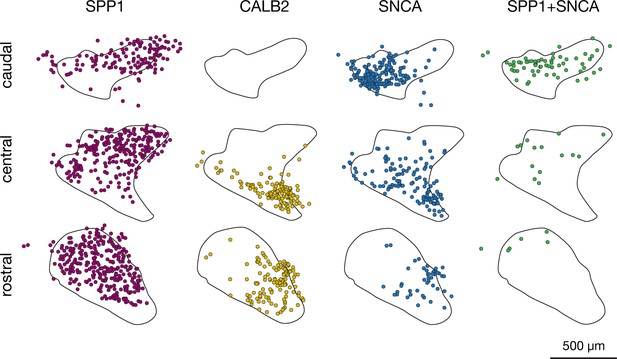
Distribution of molecularly distinct fastigial neurons that were identified with immunostaining for SPP1, CALB, and SNCA.
Individual neurons that were immunostained for 1) SPP1, 2) CALB2, 3) SNCA, or doubly immunostained for 4) SPP1 and SNCA, were mapped from serial sections (interval 120 µm). Mapping results from n = 3 male mice for each set were grouped into caudal, central, and rostral levels and superimposed onto drawings of the fastigial nucleus. Total numbers of neurons identified and mapped from three mice each are 922, 242, 334, and 88 for SPP1, CALB2, SNCA, and SPP1+SNCA, respectively.
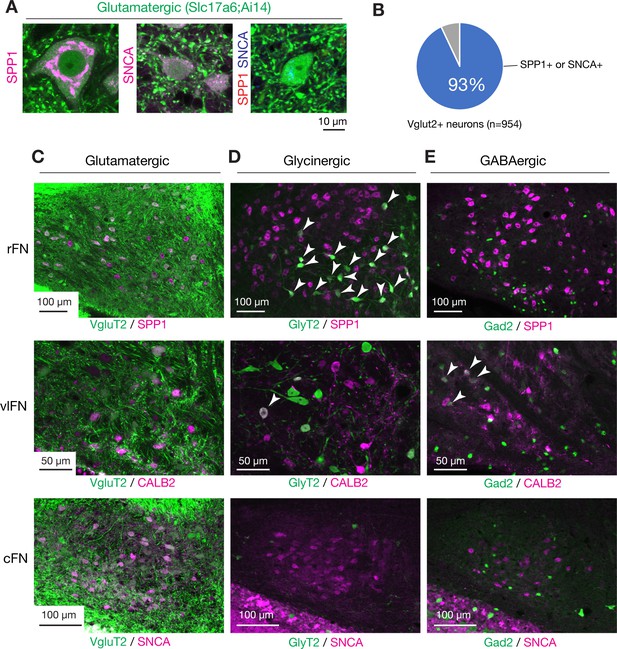
Overlap of fastigial cell type markers with glutamatergic, glycinergic, and GABAergic neurons.
(A) Fastigial marker expressions in glutamatergic neurons. Panels show high magnification images of glutamatergic neurons (green, visualized in VgluT2-Cre;Ai14 mouse) with immunostaining for SPP1 and/or SNCA. Examples show subtypes of glutamatergic neurons, which are VgluT2+/SPP1+ (left), VgluT2+/SNCA+ (center), and VgluT2+/SPP1-/SNCA- (right). Immunostaining signals are shown in magenta in the left and center panels, and double immunostaining signals in the right panel are shown in red (SPP1) and blue (SNCA). (B) Chart to show coexpression of glutamatergic marker and fastigial cell-type markers. The majority of VgluT2+ neurons examined (93%, 887/954, three mice) were colabeled with either SPP1 or SNCA. (C–E) Marker expressions in glutamatergic, glycinergic, or GABAergic fastigial neurons. Panels show immunostaining at the levels of rFN (top), vlFN (center), and cFN (bottom) in reporter mouse lines for transmitter type identification. (C) Shows examples for glutamatergic neurons, in which SPP1, CALB2, and SNCA are colabeled with 92% (43/47), 95% (19/20), and 90% (37/41) of fluorescently labeled neurons in VgluT2-Cre;Ai14 line, at rFN, vlFN, and cFN, respectively. (D) Shows examples for glycinergic neurons, in which SPP1, CALB2, and SNCA are colabeled with 100% (16/16; arrowheads), 9% (1/11; arrowhead), and 0% (0/3) of fluorescently labeled neurons in GlyT2-EGFP line, at rFN, vlFN, and cFN, respectively. (E) Shows examples for GABAergic neurons, in which SPP1, CALB2, and SNCA are colabeled with 0% (0/37), 11% (4/38; arrowheads), and 0% (0/12) of fluorescently labeled neurons in Gad2-nls-mCherry line, at rFN, vlFN, and cFN, respectively. Scale bar in A applies to all panels in A.
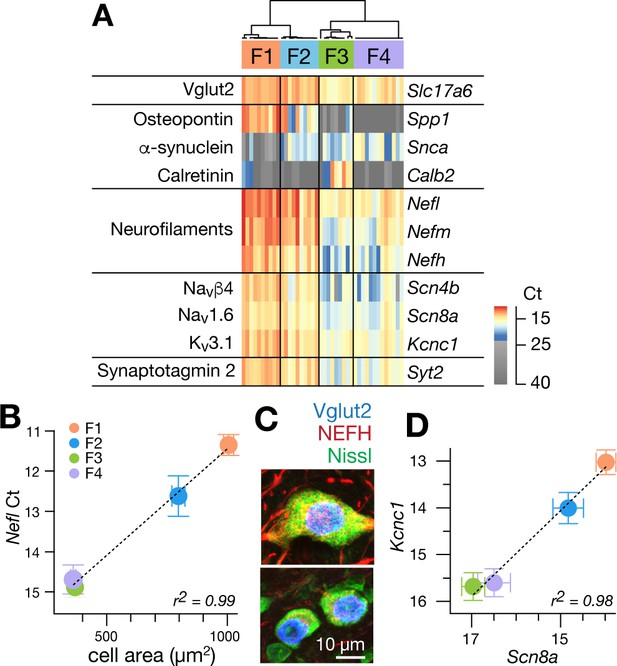
Single cell gene expression analyses confirm molecularly distinct cell types.
(A) Heatmap representation of quantitative gene expression profiles obtained via single-cell qPCR for individual excitatory fastigial neurons that express Slc17a6 (=VgluT2). Expression levels in Ct (cycle threshold in qPCR) are color coded, where insignificant expression (<5 copies of transcript, corresponding to Ct of 23.45) is shown in grey scale. Columns and rows correspond to individual neurons (n = 42) and genes examined, respectively. Clustering analysis for expression of Spp1, Snca, and Calb2 confirms four major types of fastigial neurons immunohistochemically identified, which are termed F1-F4 as shown in the dendrogram. Neurons included are from 7 (6 wildtype and 1 YFP-16) male mice. (B) Positive correlation of Nefl expression in the molecularly defined cell types with cell body area measured from the corresponding neurons immunohistochemically identified (n = 210). Plots are color-coded for the cell-types as indicated in A (F1, orange; F2, blue; F3, green; F4, purple). Note that smaller Ct values indicate greater expression levels. Population averaged data for each cell type are plotted. Error bars represent SEM. (C) NEFH immunostaining of glutamatergic fastigial neurons identified by nuclear-localized GFP (blue) in VgluT2-Cre;SUN1-sfGFP line. Size of the somata is identified with Nissl staining (green). Immunoreactivity for NEFH (red) is higher in the large glutamatergic neurons (top) than in the small glutamatergic neurons (bottom). (D) Linear correlation between expression levels of Scn8a vs Kcnc1. Color-code of the cell-type is the same as B. Plotted are population averaged data for each cell type. Error bars represent SEM.
-
Figure 2—source data 1
Raw data of single-cell qPCR with excitatory fastigial neurons on selected genes Gene expression levels (in qPCR Ct) in the individual neurons are organized in columns.
Ct values were determined with a common threshold for all the qPCR reactions (see Methods). Left columns of the table show the genes tested and primer/probe sequences used for the qPCR reaction. The qPCR probes which include minor groove binders (MGB, Applied Biosystems) are also indicated. Note that Lys, Trp, Phe, and Thr are spike-in control RNA (1000, 100, 20, and five copies, respectively).
- https://cdn.elifesciences.org/articles/58613/elife-58613-fig2-data1-v2.xlsx
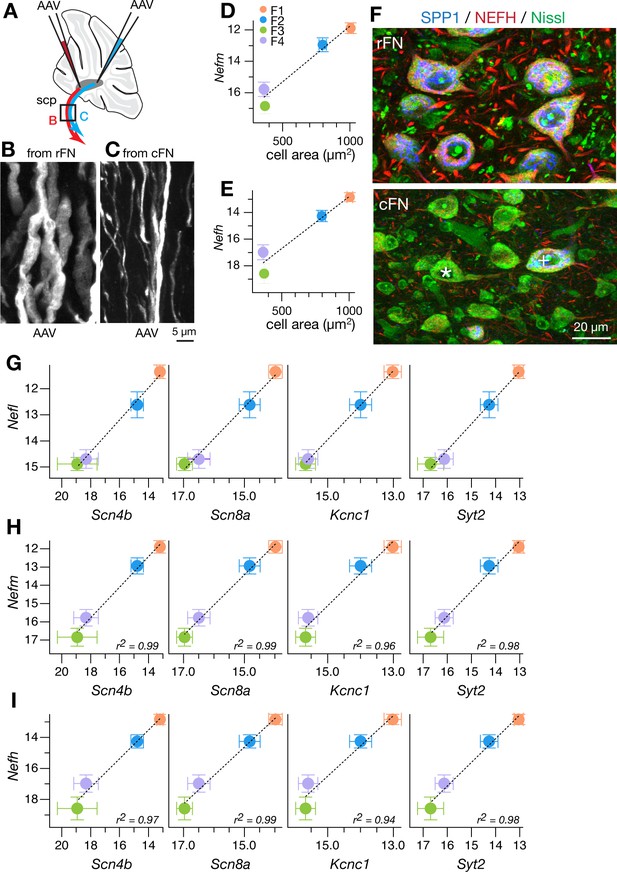
Correlation of neurofilament gene expression with axonal caliber and the expression of ion channels and synaptic molecules.
(A–C) High magnification analysis of fastigial axons that originate from rostral or caudal parts of the FN. As illustrated in (A), small, localized injections of AAV9.RFP to rostral and caudal parts of the FN anterogradely labeled subsets of fastigial axons, which were then imaged at the level of the superior cerebellar peduncle (scp). (B) Shows a case of rostral FN injection (n = 2 males), where majority of labeled axons had large axonal calibers. In contrast, (C) shows a case of caudal FN injection (n = 2 males), where the calibers of the labeled axons were significantly smaller than those in (B). Because the rostral and caudal parts of the FN are dominated by NEFH-rich and -poor neurons, respectively, these results are in accordance with a positive correlation between neurofilament expression level and axonal diameter previously reported (Hoffman et al., 1987). (D and E) Expression levels (in qPCR Ct) of Nefm (D) or Nefh (E) in the molecularly defined cell types are plotted against cell body areas of corresponding cell types measured from Nissl images in combination with marker immunostaining (n = 210). Plots are color-coded for the cell-type as indicated in inset (F1, orange; F2, blue; F3, green; F4, purple; this color-code also applies to E and G-I of this Figure). Population averaged data for each cell type are plotted. Error bars represent SEM. (F) Double immunostaining for NEFH and SPP1 of the fastigial nucleus with Nissl counterstaining. Immunoreactivity against NEFH was greater in the large SPP1+ neurons in rFN (top) than in the small SPP1- neurons in cFN (e.g. asterisk) (bottom). The medium to large SPP1+ neurons (indicated with '+') that were scattered in cFN showed greater NEFH immunoreactivity than neighboring small neurons (bottom). (G–I) Linear positive correlation between expression levels (in qPCR Ct) of Nefl (F), Nefm (G) or Nefh (H) vs Scn4b, Scn8a, Kcnc1, and Syt2. Plotted are population averaged data for each cell type. Error bars represent SEM. Scale bar in C applies to B and C. Scale bar in F applies to both images in F.
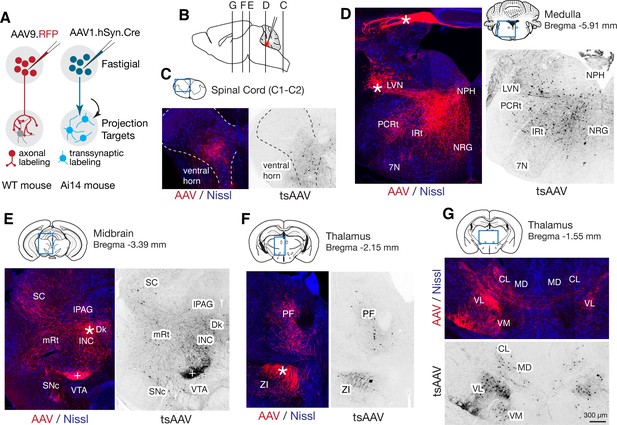
Pan-fastigial AAV injections reveal widespread fastigial output.
(A) Schematics of AAV injection experiments to label fastigial axons and terminals (left) and to transsynaptically label postsynaptic neuronal somata (right). (B) Schematic of a sagittal view of the mouse brain to show approximate levels of the coronal sections in C-G. Tracer injection to the FN is illustrated in red. (C–G) Representative low magnification confocal images obtained from pan-fastigial injections of AAVs (AAV, AAV9.RFP; tsAAV, AAV1.hSyn.Cre). Contralateral side to the injection site is shown. Blue rectangles in the drawings of the sections indicate imaged areas. In these paired images, the left image shows labeled fastigial axons and terminals in red and counterstained with Nissl in blue; the right image shows transsynaptically labeled neurons in black. Asterisks indicate axonal bundles. Results from these axonal labeling and transsynaptic tracing experiments were consistent across mice (n = 6 males for axonal labeling; n = 4 male and n = 3 female for transsynaptic labeling. (C) Shows fastigial projections to the cervical spinal cord, dense at the lamina VII-VIII, at the level of C1-C2. Dotted lines circumscribe the gray matter. (D) Shows fastigial projections to the medulla. Labeled fastigial axons crossed the midline within the cerebellum, exit via the cerebellar peduncle and provide dense projections to the vestibular and reticular nuclei, including the lateral vestibular nucleus (LVN), nucleus reticularis gigantocellularis (NRG), intermediate reticular nucleus (IRt), and parvocellular reticular nucleus (PCRt), with additional collateralization to the nucleus prepositus hypoglossi (NPH) and facial nucleus (7N). (E) Shows fastigial projections to the midbrain. Labeled fastigial axons innervate perioculomotor nuclei including the interstitial nucleus of Cajal (INC), lateral periaqueductal gray (lPAG), and nucleus of Darkschewitsch (Dk), and more laterally located nuclei including the superior colliculus (SC), mesencephalic reticular nucleus (mRt), and substantia nigra pars compacta (SNc). Innervation of the ventral tegmental area (VTA) is very sparse. Note that the dense labeling in the red nucleus (+) derived from axons from the anterior interpositus that were labeled by AAV that leaked from the injection center in the FN (Figure 3—figure supplement 1B, bottom). Injections specifically localized to FN subregions did not significantly label the red nucleus (Figure 5—figure supplement 1). (F and G) Fastigial projections to the thalamus. Labeled fastigial terminals and transsynaptically labeled somata are distributed at the PF, CL, MD, VM, and VL thalamic nuclei and the zona incerta (ZI). Fastigial axons projecting to the contralateral thalamus traverse the midline at the level of G, and innervate the ipsilateral thalamus. Scale bar in G applies to all confocal images in C-G. Abbreviations, 7N, facial nucleus; CL, centrolateral thalamic nucleus; Dk, nucleus of Darkschewitsch; INC, interstitial nucleus of Cajal; IRt, intermediate reticular nucleus; lPAG, lateral periaqueductal gray; LVN, lateral vestibular nucleus; MD, mediodorsal thalamic nucleus; mRt, mesencephalic reticular nucleus; NRG, nucleus reticularis gigantocellularis; NPH, nucleus prepositus hypoglossi; PCRt, parvocellular reticular nucleus; PF, parafascicular thalamic nucleus; SC, superior colliculus; SNc, substantia nigra pars compacta; VL, ventrolateral thalamic nucleus; VM, ventromedial thalamic nucleus; VTA, ventral tegmental area; ZI, zona incerta.
-
Figure 3—source data 1
Fastigial projection targets identified by localized anterograde tracer injections.
Rows list the brain regions and specific nuclei that were anterogradely labeled in pan-fastigial tracer injections. Color-coded columns show distinct projections from each of the fastigial subregions identified by localized anterograde injections of AAV and/or BDA to the rFN (n = 2 males and n = 2 females), rDLP (n = 2 males and n = 1 female), cDLP (n = 2 males), vlFN (n = 3 males), and cFN (n = 3 males). Significant vs sparse terminal innervations are indicated with + vs ±. Absence of labeled terminals is indicated with –. Sparse projections are listed separately at the bottom.
- https://cdn.elifesciences.org/articles/58613/elife-58613-fig3-data1-v2.xlsx
-
Figure 3—source data 2
List of tracers, coordinates, injection volume, and mice used for tracing experiments.
Experimental parameters used in the tracing experiments, including tracers, coordinates, injection volumes, injection side (left or right), and mouse sex. These parameters were associated with 'Injection Site'. AP, ML, DV coordinates are given in mm. For the tracer injections with angled approach, AP and ML correspond with the location of craniotomy before tilting the manipulator or stage, and DV corresponds with the distance from the surface of the brain to the target in the angled setting.
- https://cdn.elifesciences.org/articles/58613/elife-58613-fig3-data2-v2.xlsx
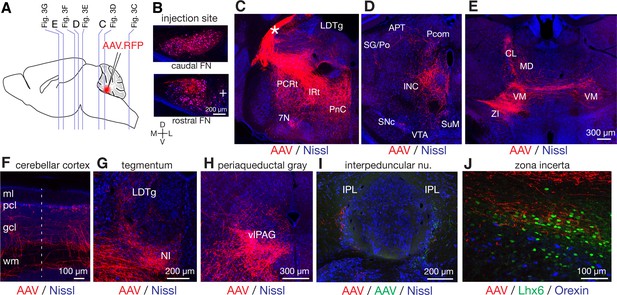
Fastigial projection targets revealed with AAV- mediated anterograde tracing.
(A) Schematic of AAV injection experiments to examine fastigial output projections. Vertical lines in a sagittal scheme indicates approximate rostrocaudal levels of images in C-E of this figure and in Figure 3. (B) Representative coronal section images of injection sites. Two images are from the same case, showing that the labeled injection site (red) covered the entire fastigial nucleus with little spread to the neighboring anterior interpositus nucleus (indicated with '+'). (C–E) Fastigial axonal innervation (red) of reticular formation, midbrain, and thalamus are shown with Nissl counterstaining (blue). (C) Shows a labeled axonal bundle exiting the cerebellum from the contralateral cerebellar peduncle (asterisk) which then innervated the pontine reticular nucleus (PnC), intermediate reticular nucleus (IRt), and parvocellular reticular nucleus (PCRt). Labeled axonal innervation was also found at laterodorsal tegmental nucleus (LDTg) and a confined part of the facial nucleus (7N). (D) Shows significant axonal innervation of perioculomotor area including the interstitial nucleus of Cajal (INC) and sparse innervation of the anterior pretectal nucleus (APT), nucleus of posterior commissure (Pcom), and suprageniculate thalamic nucleus/posterior thalamic group (SG/Po). The labeled axonal terminals also reached the supramammillary region (SuM) and a medial part of the substantia nigra pars compacta (SNc). (E) Shows fastigial projections to the thalamic nuclei including ventromedial (VM), centrolateral (CL), and mediodorsal (MD) nuclei and to the zona incerta (ZI). The panel also shows labeled fastigial axons traversing the midline to project to the thalamic nuclei ipsilateral to the injection side (right in the image). (F) Fastigial projections to the bilateral cerebellar cortex with ipsilateral predominance. The image shows that the projections terminate as mossy fibers at or beneath the Purkinje cell layer (pcl), as described previously for nucleocortical projections from other cerebellar nuclei (Houck and Person, 2015; Gao et al., 2016; Low et al., 2018). Dotted line indicates the midline. The result is in accordance with a previous study in cats and monkeys (Tolbert et al., 1978). (G) Fastigial projections to the nucleus incertus (NI) and laterodorsal tegmental nucleus (LDTg). (H) Fastigial projections to the ventrolateral periaqueductal gray (vlPAG). (I) Fastigial projections to the interpeduncular nucleus. A double injection case is shown in which red and green AAVs were injected in the right and left FN, respectively. Resultant labeling showed sparse but consistent axonal projections to lateral interpeduncular nucleus (IPL). (J) Fastigial projections to the zona incerta (ZI). Labeled fastigial axons (red) within the Lhx6+ neuronal clusters (green, visualized with Lhx6 reporter mouse; n = 2 females), which demarcates the ZI, are shown. No clear fastigial projections to the lateral hypothalamus, which was identified with clustered orexinergic neurons that are visualized with immunostaining for orexin (blue). Scale bar in E applies to C-E. Abbreviations, 7N, facial nucleus; APT, anterior pretectal nucleus; CL, centrolateral thalamic nucleus; gcl, granule cell layer; IRt, intermediate reticular nucleus; INC, interstitial nucleus of Cajal; IPL, lateral interpeduncular nucleus; LDTg, laterodorsal tegmental nucleus; MD, mediodorsal thalamic nucleus; ml, molecular layer; NI, nucleus incertus; pcl, Purkinje cell layer; Pcom, nucleus of posterior commissure; PCRt, parvocellular reticular nucleus; PF, parafascicular thalamic nucleus; PnC, caudal pontine reticular nucleus; SG/Po, suprageniculate thalamic nucleus/posterior thalamic group; SNc, substantia nigra, pars compacta; SuM, supramammillary region; VM, ventromedial thalamic nucleus; vlPAG, ventrolateral periaqueductal gray; VTA, ventral tegmental area; wm, cerebellar white matter; ZI, zona incerta.
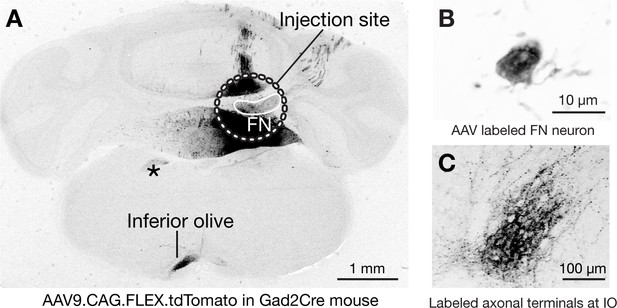
Cre-dependent anterograde tracing of GABAergic neuronal projections from the fastigial nucleus.
(A) Example low magnification image of Cre-dependent anterograde tracing experiments with Gad2Cre mouse. The image shows injection site and resultant axonal labeling in the brainstem. The injections site (circumscribed with a dotted white line) of Cre-dependent AAV, AAV9.CAG.FLEX.tdTomato, entirely included the FN (circumscribed with a white line). Significant labeling (black) in the injection site but outside of the FN reflects labeled Purkinje cells which also express Gad2 and thus labeled with this injection strategy. The labeled GABAergic output projections (black) in the extracerebellar regions are identified only in the inferior olive (IO) (n = 2 males). Asterisk indicates labeled axonal path. (B) A high magnification example of a fastigial neuron labeled by the injection. (C) Labeled GABAergic axonal terminals innervating the IO.
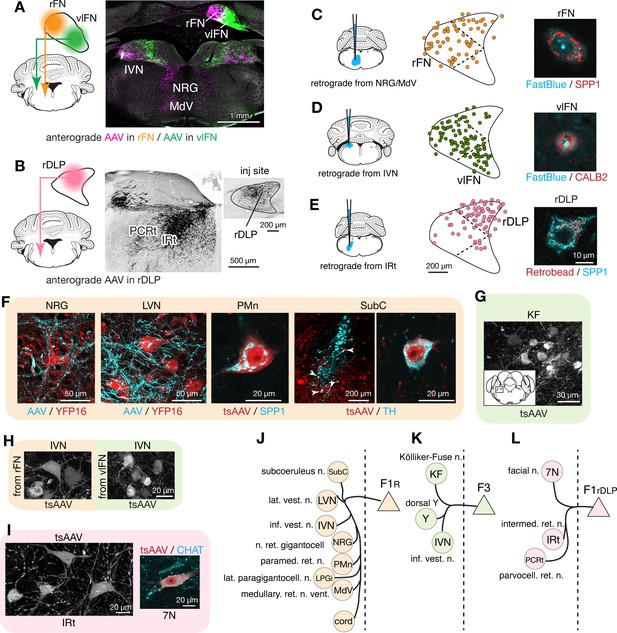
Segregated output channels from rostral parts of the FN.
(A and B) AAV-mediated anterograde tracing with injections localized to subregions of the rostral fastigial nucleus that distinguish projections from F1R, F3, and F1rDLP neurons. (A) Shows a dual AAV injection in the rFN (F1R region, AAV9.RFP, magenta in confocal image) and vlFN (F3 region, AAV9.GFP, green in confocal image) and the resultant labeling in the inferior vestibular nucleus (IVN) and medulla. Results were consistent across four mice (n = 2 male and n = 2 female) for rFN projections, and across three male mice for vlFN projections. (B) Shows an AAV9.RFP injection in the rDLP (F1rDLP region, inset) and the resultant labeling (black in confocal image) in the IRt and PCRt. Results were consistent across three mice (n = 2 male and n = 1 female). (C–E) Retrograde tracing experiments to confirm segregation among rostral fastigial output projections. Injections were made into NRG/MdV (C: n = 3 males), IVN (D: n = 3 males), and IRt (D: n = 2 males). (C) Shows retrograde tracer (FastBlue, cyan in the right image) injection to the NRG/MdV. Retrogradely labeled neurons are mapped onto the rFN (middle) and overlapped with SPP1+ (red in the right image) neurons of large cell bodies, indicating that F1R neurons project to the NRG/MdV. Similarly, (D and E) Show results of retrograde labeling from the IVN (FastBlue) and IRt (retrobead). The localization of labeled cells and immunoreactivity for CALB2 and SPP1 indicate that F3 and F1rDLP neurons project to the IVN and IRt, respectively. Dotted lines in the middle panels indicate approximate borders for the fastigial subregions. (F) Key projection targets of F1R neurons. AAV and tsAAV indicate labeling by AAV9.RFP and AAV1.hSyn.Cre, respectively. Panels show dense fastigial axonal innervation (cyan) of large neurons in the NRG and LVN, which are fluorescently labeled in YFP16 mouse line (red). Also shown are transsynaptically labeled SPP1+ PMn neuron and a TH+ SubC neuron (white arrowheads) (axonal tracing, n = 2 males and n = 2 females; transsynaptic tracing, n = 3 females). (G) F3 neurons project to the Kölliker-Fuse (KF) nucleus, as demonstrated by anterograde transsynaptic labeling from the vlFN. Inset shows location of the labeled KF neurons (axonal tracing, n = 2 males; transsynaptic tracing, n = 2 males). (H) F1R and F3 differentially innervate large and small neurons in the IVN, respectively, as demonstrated by anterograde transsynaptic labeling from the rFN and vlFN. (I) Projections of F1rDLP neurons to IRt and CHAT+ facial nucleus neurons, demonstrated by anterograde transsynaptic tracing from the rDLP (axonal tracing, n = 2 males and n = 1 female; transsynaptic tracing, n = 3 females). (J–L) Summary of the major output targets of F1R, F3, and F1rDLP, respectively. Dotted vertical line indicates midline. Targets are arranged rostro-caudally from top to bottom. Scale bars in the middle and right panels in E applies to similar panels in C and D. Scale bar in H applies to both left and right images in H. Figure 3—source data 1 contains a complete list of projection targets. Abbreviations, 7N, facial nucleus; AAV, adeno associated virus; cord, spinal cord; IRt, intermediate reticular nucleus; IVN, inferior vestibular nucleus; KF, Kölliker-Fuse nucleus; LPGi, lateral paragigantocellular nucleus; LVN, lateral vestibular nucleus; MdV, medullary reticular nucleus, ventral; NRG, nucleus reticularis gigantocellularis; PCRt, parvocellular reticular nucleus; PMn, paramedian reticular nucleus; SubC, subcoeruleus nucleus; tsAAV, anterograde transsynaptic labeling with AAV; Y, dorsal group Y.
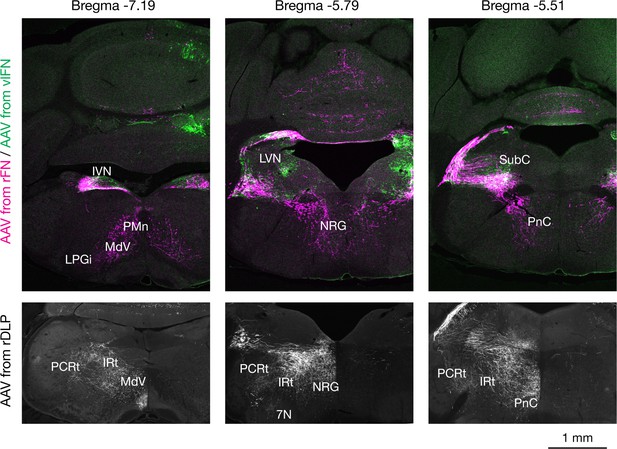
Representative results of anterograde tracing from injections localized to rFN, vlFN, or rDLP.
AAV9 injection cases that labeled axons of the right FN are shown in three rostrocaudal levels. Top row shows labeled axons from the rFN (magenta) and vlFN (green) in a double injection case. F1R projections are revealed to innervate subcoeruleus region (SubC), lateral vestibular nucleus (LVN), and ventromedial regions of reticular nuclei including the pontine reticular nucleus caudal part (PnC), nucleus reticularis gigantocellularis (NRG), medullary reticular nucleus ventral part (MdV), paramedian reticular nucleus (PMn), and lateral paragigantocellular nucleus (LPGi). F3 projections are localized to dorsal regions including inferior vestibular nucleus (IVN) and SubC. Because the glycinergic projection neurons are intermingled with F1R and F3, axons projecting to ipsilateral brainstem are also labeled. Bottom row shows labeled axons from the rDLP. F1rDLP projections are prominent to dorsolateral regions of the reticular nuclei, including PnC, NRG, intermediate reticular nucleus (IRt), and parvocellular reticular nucleus (PCRt) but not to vestibular nuclei. Scale bar applies to all panels.
Segregated output channels from caudal parts of the FN.
(A) AAV-mediated anterograde tracing with localized injections to the cFN and cDLP subregions of the FN revealed distinct projection targets of F2 and F4 neurons in the thalamus. A coronal thalamic section in the middle shows a case of cFN injection (territory of F4 neurons), with labeled neurons (black) in the MD, CL, and VM nuclei, but only sparsely in the VL nucleus. A similar thalamic section in the right shows a case of cDLP injection (territory of F2 neurons), with labeled neurons predominantly in the VL nucleus, but only sparsely to the MD, CL and VM nuclei. (B and C) Retrograde tracing experiments to confirm segregation among caudal fastigial output projections. In (B), retrogradely labeled neurons from the injections to the VM nucleus (n = 3 males) are mapped onto the cFN (middle); high magnification analysis shows colabeling of the retrograde tracer (retrobead, cyan) with SNCA immunoreactivity (magenta) in small cFN neurons (right), indicating that F4 neurons project to the VM nucleus. Similarly, (C) demonstrates that F2 neurons, which are localized in the cDLP and are SPP1+, project to the VL thalamus (n = 2 males and n = 1 female). (D) Anterograde transsynaptic tracing (by AAV1.hSyn.Cre, labeled as tsAAV) including the cDLP labeled neurons in the VL and PF thalamic nuclei, superior colliculus (SC), interstitial nucleus of Cajal (INC), and caudal pontine reticular nucleus (PnC) (n = 3 males and n = 2 females). (E) Anterograde transsynaptic tracing from the cFN labeled neurons in the CL, MD, and VM thalamic nuclei, posterior hypothalamus (PH), nucleus incertus (NI), substantia nigra pars compacta (SNc), laterodorsal tegmental nucleus (LDTg), and pedunculotegmental nucleus (PTg) (n = 3 males and n = 2 females). Fastigial axons and terminals anterogradely labeled by AAV9.RFP (labeled as AAV) contacted with TH+ SNc neuron (red) and Chat+ LDTg neurons (red), which were identified by TH immunostaining and ChatCre;Ai3 line, respectively. Anterograde transsynaptic tracing (tsAAV) confirmed the innervation of cFN neurons onto catecholaminergic neurons in the SNc (TH+, magenta) (n = 3 males and n = 1 female) and cholinergic neurons in the LDTg and PTg (CHAT+, green) (n = 3 males). (F–G) Summary of the major output targets of F2 (F) and F4 neurons (G). Dotted vertical line indicates midline. Targets are arranged rostro-caudally from top to bottom. Scale bars in the middle and right panels in C applies to similar panels in B. Figure 3—source data 1 contains a complete list of projection targets. Abbreviations, cord, spinal cord; CL, centrolateral thalamic nucleus; INC, interstitial nucleus of Cajal; LDTg, laterodorsal tegmental nucleus; MD, mediodorsal thalamic nucleus; mRt, mesencephalic reticular nucleus; NPH, nucleus prepositus hypoglossi; NI, nucleus incertus; NRG, nucleus reticularis gigantocellularis; PAG, periaqueductal gray; PF, parafascicular thalamic nucleus; PH, posterior hypothalamus; PnC, pontine reticular nucleus, caudal; PTg, pedunculotegmental nucleus; SC, superior colliculus; SNc, substantia nigra, pars compacta; VL, ventrolateral thalamic nucleus; VM, ventromedial thalamic nucleus; ZI, zona incerta.
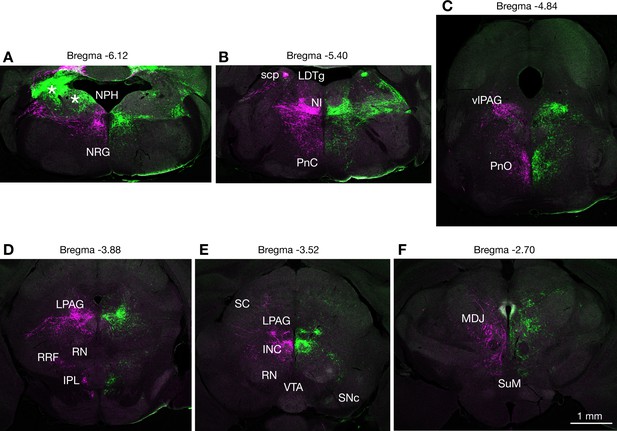
Representative results of anterograde tracing from injections localized to caudal regions of the fastigial nucleus.
An AAV9 injection case that hit cFN and cDLP (F2 and F4 regions) of both sides is shown (green from the left and magenta from the right). These two injections resulted in a highly reproducible pattern. At medullary and pontine levels (A–C), labeled axons are prominent in the dorsomedial regions of the reticular nuclei, including nucleus reticularis gigantocellularis (NRG), pontine reticular nucleus caudal part (PnC), and pontine reticular nucleus oral part (PnO). They also innervate more dorsal nuclei, including the nucleus prepositus hypoglossi (NPH), nucleus incertus (NI), laterodorsal tegmental nucleus (LDTg), and ventrolateral periaqueductal gray (vlPAG). Asterisks in (A) are Purkinje cell axons that were labeled from virus spread from the injection site. At midbrain levels (D–E), these fastigial axons strongly innervated lateral periaqueductal gray (LPAG), and perioculomotor nuclei including interstitial nucleus of Cajal (INC). Significant projections are revealed at interpeduncular nucleus lateral subnucleus (IPL), retrorubral field (RRF) and intermediate/deep layers of the superior colliculus (SC). Projection to substantia nigra pars compacta is sparse at its caudalmost level (E). No obvious projections are detectable in the red nucleus (RN; D,E) and ventral tegmental area (VTA; E). At a prethalamic level (F), the projections are revealed in the nuclei of mesodiencephalic junction (MDJ) and supramammillary region (SuM). Scale bar applies to all panels.
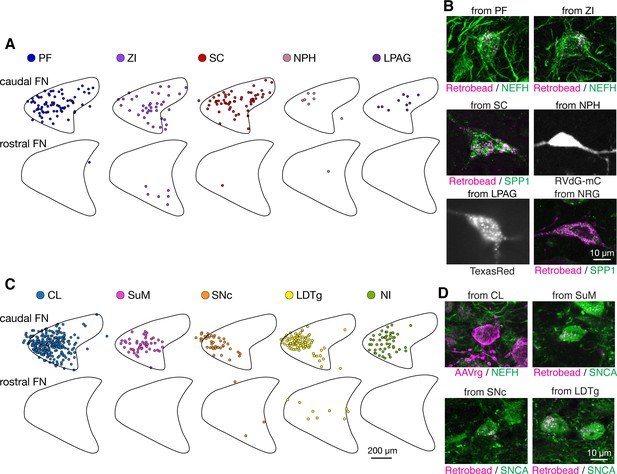
Analysis of retrograde tracing revealing discrete projections of F2 and F4.
(A) Mapping of retrograde tracing results from injections to the parafascicular (PF) thalamus (n = 5 males and n = 3 females), zona incerta (ZI) (n = 3 males), superior colliculus (SC) (n = 3 males), nucleus prepositus hypoglossi (NPH) (n = 3 females), and lateral periaqueductal gray (LPAG) (n = 2 males and n = 2 females). Each dot indicates an individual labeled neuron. Labeled fastigial neurons were roughly evenly distributed at both the cFN and cDLP. (B) Examples of retrogradely labeled fastigial neurons that are large in size and/or either SPP1+ or NEFH-rich, which thus are identified as F2 neurons. Retrograde tracing cases from PF, ZI, SC, NPH, LPAG, and nucleus reticularis gigantocellularis (NRG) are shown. (C) Mapping of retrograde tracing results from injections to centrolateral (CL) thalamus (n = 2 males and n = 2 females), supramammillary region (SuM) (n = 2 males), substantia nigra pars compacta (SNc) (n = 3 males), laterodorsal tegmental nucleus (LDTg) (n = 2 males), and nucleus incertus (NI) (n = 2 females). Distribution of these labeled fastigial neurons was significantly biased to the cFN. (D) Examples of retrogradely labeled fastigial neurons that are small in size and either SNCA+ or NEFH-poor, which thus are identified as F4 neurons. Retrograde tracing cases from CL, SuM, SNc, and LDTg are shown. Abbreviations, CL, centrolateral thalamic nucleus; LDTg, laterodorsal tegmental nucleus; lPAG, lateral periaqueductal gray; NI, nucleus incertus; NRG, nucleus reticularis gigantocellularis; NPH, nucleus prepositus hypoglossi; PF, parafascicular thalamic nucleus; SC, superior colliculus; SNc, substantia nigra, pars compacta; SuM, supramammillary region; ZI, zona incerta.
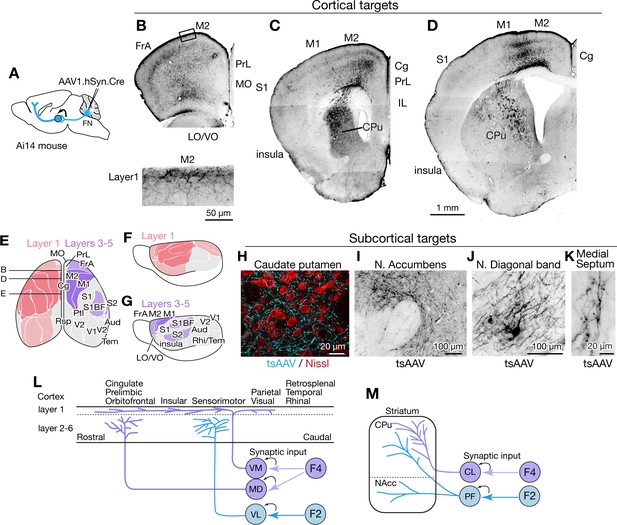
Fastigial disynaptic input to the forebrain.
(A) Schematic of anterograde transsynaptic tracing from the FN to identify fastigial disynaptic input to forebrain areas. AAV1.hSyn.Cre injected in the FN is transsynaptically transported to postsynaptic neurons in which Cre recombinase drives reporter (tdTomato) expression in Ai14 mice, thereby axonal projections of those postsynaptic neurons to the forebrain areas are robustly labeled. (B–D) Disynaptic projections from the FN to the cortical areas. Panels show example coronal sections of the forebrain in which labeled axons (black) of the fastigial postsynaptic neurons show a characteristic distribution. Fastigial disynaptic input to layer one was widely distributed over the M2 (B–D), frontal association cortex (FrA) (B), lateral/ventral orbital cortex (LO/VO) (B), prelimbic cortex (PrL) (B and C), S1 (C and D), insular cortex (C and D), cingulate cortex (Cg) (C and D), and M1 (D). Inset in (B) is high magnification of layer 1 of the M2 (region circumscribe by rectangle in upper image), showing dense terminal arborization at layer 1. Fastigial disynaptic input also terminates at deeper layers of the FrA (B), LO/VO (B), M2 (B–D), and M1 (D). Note no labeled axons in all layers of medial orbital cortex (MO) (B) and infralimbic cortex (IL) (C), and deeper layers of insular cortex (C), S1 (C and D), and Cg (D). (E–G) Color-coded illustration of the cortical areas that receive disynaptic input from the FN to layer 1 (pink) and layers 3–5 (purple). Darker colors indicate denser input. Dorsal (E) and lateral (F and G) views are shown. Fastigial disynaptic input to layer one broadly covers the cortical areas including the FrA, PrL, LO/VO, M1, M2, S1, barrel field (S1BF), S2, insular cortex, parietal cortex (Ptl), and visual cortices (V1 and V2), but excluding MO, IL, retrosplenial cortex (Rsp), auditory cortex (Aud), and rhinal/temporal cortex (Rhi/Tem) (gray). Input to layers 3–5 (purple) is restricted in the FrA, LO/VO, M2, M1, S1BF, and S2. In (E), approximate levels of the sections in (B–D) are also shown. Results were consistent across n = 4 male and n = 4 female mice. (H–K) Disynaptic fastigial projection targets in subcortical forebrain areas, revealed by axonal terminals transsynaptically labeled with AAV1.hSyn.Cre (tsAAV). (H) The axonal terminal arborization of fastigial postsynaptic neurons (cyan) at proximity of neurons of caudate putamen (red, Nissl). (I) The labeled axonal terminals at the accumbens nucleus. (J) The labeled axonal terminals and tertiary infected neuron at the nucleus of diagonal band, a basal forebrain structure. (K) Sparse but consistent labeling at the medial septum. (L) Schematics summarizing disynaptic fastigial input to the cortex from F2 and F4 neurons. These circuits are likely to be comprised of two separate pathways, as the F2 and F4 neurons target different thalamic neurons that give rise discrete thalamocortical projections. F4 neurons provide synaptic input to the ventromedial (VM) and mediodorsal (MD) thalamic neurons. Axons of postsynaptic VM neurons target layer 1 of the widespread areas of the cortex (Kyuhou and Kawaguchi, 1985; Noda and Oka, 1985; Kuramoto et al., 2015). Axons of postsynaptic MD neurons target prelimbic, cingulate, and orbitofrontal cortices (Kuramoto et al., 2017). F2 neurons provide synaptic input to the ventrolateral (VL) thalamic neurons which then project to the deeper layers of the sensorimotor cortex (Jones, 2009; Kuramoto et al., 2009). (M) Schematics summarizing disynaptic fastigial input to the striatum from F2 and F4 neurons. These circuits are also likely to be comprised of two separate pathways, as the F2 and F4 neurons target different thalamic neurons that give rise discrete thalamostriatal projections. F2 neurons provide synaptic input to the parafascicular (PF) thalamic neurons that project to both the dorsal (caudate putamen; CPu) and ventral striatum (accumbens nucleus; NAcc) (Sadikot and Rymar, 2009). F4 neurons provide synaptic input to the centrolateral (CL) thalamic neurons that project to the dorsal striatum. Scale bar in D applies to low magnification images in B-D. Abbreviations, Aud, auditory cortex; Cg, cingulate cortex; CL, centrolateral thalamic nucleus; CPu, caudate putamen; FrA, frontal association cortex; IL, infralimbic cortex; LO, lateral orbital cortex; M1 and M2, primary and secondary motor cortex; MD, mediodorsal thalamic nucleus; MO, medial orbital cortex;; PF, parafascicular thalamic nucleus; PrL, prelimbic cortex; Ptl, parietal association cortex; Rhi, ecto-/peri-/ento-entorhinal cortex; Rsp, retrosplenial cortex; S1 and S2, primary and secondary sensory cortex; S1BF, barrel field of primary sensory cortex; Tem, temporal association cortex; V1 and V2, primary and secondary visual cortex; VL, ventrolateral thalamic nucleus; VM, ventromedial thalamic nucleus; VO, ventral orbital cortex.
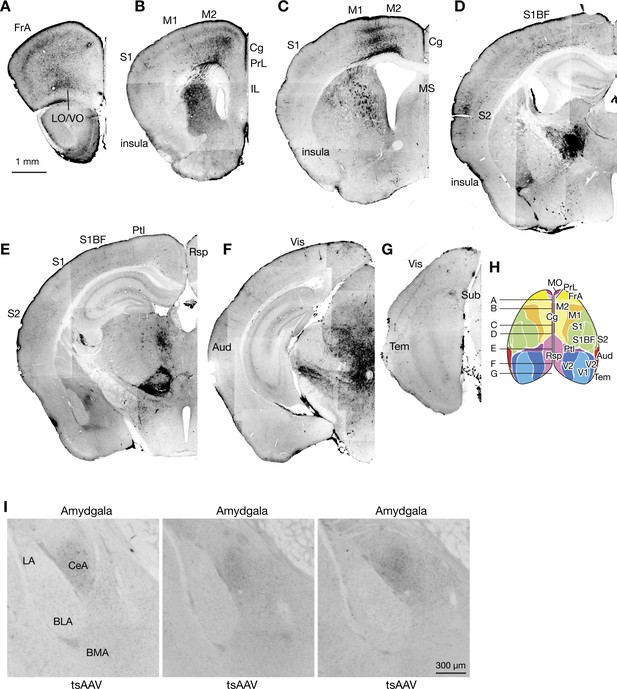
Overview of disynaptic fastigial input to the forebrain.
(A–G) Widespread cortical labeling obtained from AAV1-mediated anterograde transsynaptic tracing from the fastigial nucleus. These cortical labeling most likely reflect projections from thalamic neurons that are postsynaptic to the fastigial nucleus. Approximate rostrocaudal levels are indicated in (H). (H) Schematic of the dorsal view of the cortex that are color-coded for functional areas, which was redrawn from Watson et al., 2012. (I) No mono- or di-synaptic fastigial input to the amygdala. Photos are taken at similar rostrocaudal levels from three representative AAV1 transsynaptic tracing cases in which injections were localized in the FN. Abbreviations, Aud, auditory cortex; BLA, basolateral amygdala; BMA, basomedial amygdala; CeA, central amygdala; Cg, cingulate cortex; FrA, frontal association cortex; IL, infralimbic cortex; LA, lateral amygdala; LO, lateral orbital cortex; M1 and M2, primary and secondary motor cortex; MO, medial orbital cortex; PrL, prelimbic cortex; Ptl, parietal association cortex; Rsp, retrosplenial cortex; S1 and S2, primary and secondary sensory cortex; S1BF, barrel field of primary sensory cortex; Tem, temporal association cortex; V1 and V2, primary and secondary visual cortex; VO, ventral orbital cortex.
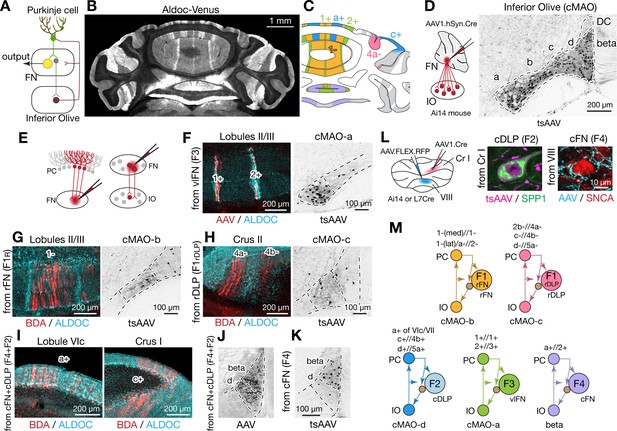
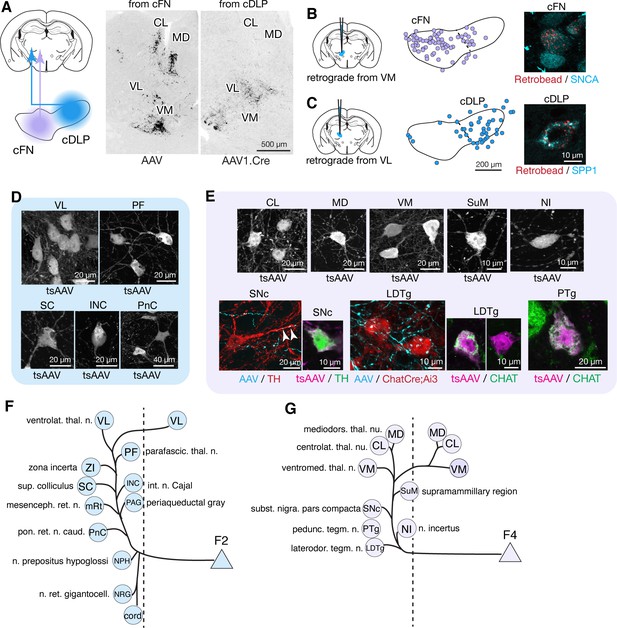
Modular connectivity of the fastigial subregions with specific PC and IO neurons.
(A) Schematic illustrating circuit connections within the canonical olivo-cerebellar loop. Neurons in a subnucleus of inferior olive (IO, garnet) innervate a specific set of Purkinje cells (PCs, green) in the cerebellar cortex, as well as their target subregions in the cerebellar nuclei. GABAergic neurons in the cerebellar nuclei (grey) in turn project back to the original IO subnucleus, closing the circuit loop between the IO and cerebellum. In the cerebellar nuclei, excitatory output neurons (yellow) and GABAergic nucleoolivary neurons which share presynaptic Purkinje cells reside in the same subregions. (B) Cerebellar coronal section from the Aldoc-venus mouse demonstrates alternating stripes of Aldoc-positive and -negative PCs. (C) PC stripes shown in (B), color-coded to indicate connections with specific fastigial cell types. Stripe terminology corresponds with Sugihara and Shinoda, 2004. (D) IO neurons that are postsynaptic to the FN. Schematic illustrates anterograde transsynaptic pan-FN injections of AAV1.hSyn.Cre (tsAAV) to label postsynaptic IO neurons. Right image shows clusters of labeled IO neurons (black) in subnuclei a-d of the caudal medial accessory olive (cMAO-a, b, c, and d) and the beta subnucleus, with no neurons labeled in the dorsal cap (DC) (n = 5 males and n = 3 females). (E) Schematics illustrating localized retrograde and anterograde tracer injections into the FN to identify presynaptic PCs and postsynaptic IO neurons, respectively. (F–H) Retrogradely labeled PCs and anterogradely and transsynaptically labeled IO neurons from localized tracer injections to vlFN, rFN, and rDLP. (F) Shows results of retrograde infection of AAV1.Cre injected into F3 region (n = 2 males), in which labeled PCs with AAV1.Cre (red) are located in the 1+ and 2+ stripes and are immunopositive for ALDOC (cyan; left panel); transsynaptically labeled IO neurons (black, right panel) from the same injections are located in cMAO-a (n = 2 males). Similarly, (G) Shows results of BDA injections into rFN (F1R region; n = 2 males and n = 1 female), in which labeled PCs with BDA (red) are located at 1- stripe and are immunonegative for ALDOC (cyan, left panel); transsynaptically labeled IO neurons from injections of AAV1.Cre into rFN are located at cMAO-b (black, right panel) (n = 3 females). (H) Shows results of BDA injections into rDLP (F1rDLP region; n = 2 males), in which labeled PCs with BDA (red) are located at 4a-/4b- stripes and are immunonegative for ALDOC (cyan) (left panel); transsynaptically labeled IO neurons from injections of AAV1.Cre into rDLP are located at cMAO-c (black, right panel) (n = 3 females). (I) Retrogradely labeled PCs from BDA injections that hit both cFN and cDLP (F4 and F2 regions). Labeled PCs (red) are located in the a+ and c+ stripes and are immunopositive for ALDOC (cyan) (n = 4 males and n = 2 females). (J) Anterogradely labeled fastigial axons innervating cMAO-d and beta from an injection that included both the cFN and cDLP (n = 6 males and n = 2 females). (K) Transsynaptically labeled beta neurons from an injection that hit cFN but not cDLP (n = 2 males). (L) Anterograde tracing experiments to identify PC projections to caudal portions of FN. Left schematic illustrates injection experiments, in which AAV1.hSyn.Cre was injected to the Crus I (magenta) of Ai14 mice to transsynaptically label FN neurons (n = 2 males), and Cre-dependent AAV, AAV.FLEX.RFP, was injected to lobule VIII (cyan) of L7Cre mice to label PC axons innervating the FN (n = 2 males). In the middle panel, transsynaptically labeled neurons (magenta) from the Crus I injections labeled neurons in cDLP confirmed as F2 by their medium size and SPP1-immunopositivity. In the right panel, PC terminals (cyan) from injections in lobule VIII contacted SNCA+ (red) small neurons in the cFN (F4 neurons). (M) Diagrams schematize five olivo-cerebellar-fastigial loops identified in this study. Arrows from brown circles indicate GABAergic nucleoolivary projections. Specific olivocerebellar projections, Purkinje cell projections, and nomenclature of the cerebellar cortical stripes are based on Sugihara and Shinoda, 2004, Sugihara and Shinoda, 2007; Voogd and Ruigrok, 2004; Sugihara et al., 2009. Abbreviations, BDA, biotinylated dextran amine; cMAO-a, b, c, and d, caudal medial accessory olive, subnucleus a, b, c, and d; Cr I, crus I; Cr II, crus II; DC, dorsal cap of Kooy; FN, fastigial nucleus; IO, inferior olive; PC, Purkinje cell.
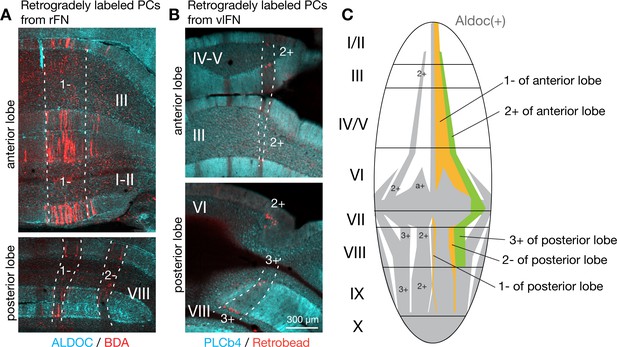
Convergent Purkinje cell projections from multiple lobules to fastigial subregions.
(A) Coronal section images from the same retrograde tracing case in which PCs projecting to the rFN are labeled. BDA was injected to the rFN as a retrograde tracer. Labeled PCs (red) were distributed across the anterior (top) and posterior lobes (bottom) but confined within the same cerebellar module (1-//1-/2-), which was identified by immunonegativity for ALDOC (cyan) and mediolateral level of those PCs. (B) Similarly, images from a retrograde tracing case with retrobead or BDA from the vlFN show that the retrogradely labeled PCs spread across the anterior and posterior lobes but were confined within the same module (2+//3+) (n = 2 males), as identified by immunonegativity for PCLb4 (cyan), which is expressed in subsets of PCs in a complemental pattern to Aldoc (Sarna et al., 2006), and the mediolateral level of those PCs. A tracing case with retrobead is shown. (C) Schematic illustration of the distribution of PCs upstream to the rFN (orange) or vlFN (green). PCs projecting to these fastigial subregions are localized in longitudinal stripes of the cerebellar cortex, which correspond to those demarcated by Aldoc expression (gray and white) Scale bar in B applies to all panels in A and B.
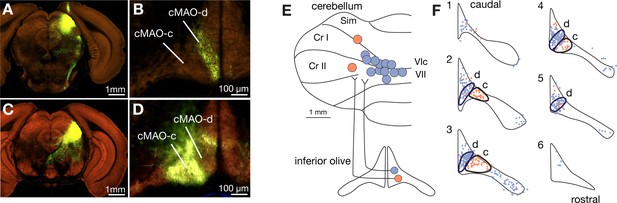
Distinct input and output projections of the caudal medial accessory olive subnuclei ‘d’ and c.
(A–D) Distinct input from the superior colliculus (SC) to subnuclei d and c of medial accessory olive are shown with anterograde tracing experiments from the Allen Connectivity Atlas. Labeled axons from the medial SC (A) were found at the most medial part of the caudal medial accessory olive (cMAO), which we designated as cMAO-d (B). Data were obtained from Experiment #128001349, section 49 for injection site at the SC (A) and section 15 for the labeling in the IO (B). Labeled axons from the lateral SC (C) are found at the cMAO-c (D). Data were obtained from Experiment #146078721, section 54 for injection site at the SC and section 11 for the labeling in the IO. (E) Schematic illustration of experiments to identify projection targets of the subnuclei d and c of the cMAO. Retrograde tracers were injected to various loci in the cerebellar cortex (n = 14 injections to n = 2 males and n = 4 females). Each circle indicates one tracer injection case. Retrobead, BDA, and AAV1.Cre, which retrogradely infects IO neurons, were used as retrograde tracer. Injection cases are color coded on the basis of downstream fastigial areas revealed in Figure 7, in which the lobules VIc/VII and medial Crus I (blue) were upstream of the cDLP (F2 region) and the medial Simplex lobule (Sim)/Crus II (orange) were upstream of the rDLP (F1rDLP region). (F) Mapping of retrogradely labeled IO neurons from the injection sites shown in (E). Individual labeled IO neurons are mapped onto serial coronal drawings of the inferior olive, in which 1 and 6 correspond to the most caudal and rostral levels used in this analysis, respectively. Circumscribed are two clusters of IO neurons; blue dots are those labeled by injections to the lobules VIc/VII and medial Crus I, while orange dots are those labeled by injections to the medial Sim/Crus II. The two clusters were mostly not overlapped. Projection to the medial Sim/Crus II is hallmark of the cMAO-c subnucleus (orange cluster). We termed the other IO neuronal cluster (blue cluster) as "cMAO-d " subnucleus that are medial to the cMAO-c and project to the lobules VIc/VII and medial Crus I.
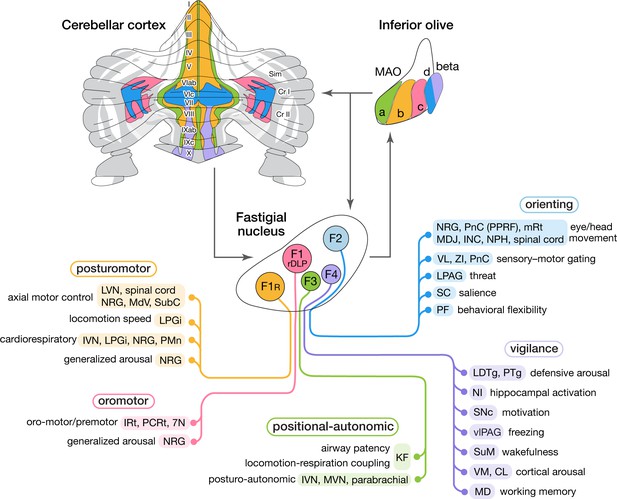
Modular circuit connections of excitatory fastigial projection neurons provide circuit substrates for coordinating five broad organismal functions.
Schematics summarize cerebellar modular circuit connections that link distinct types of fastigial nucleus neurons with specific neurons in the inferior olive, cerebellar cortex, and downstream brain regions. Projection targets of each FN cell type are indicated in shaded colors. Specific functions associated with each collection of projection targets are indicated at the left; proposed broad organismal functions of each module are encircled above. To show the distribution of Purkinje cells associated with each module, a flatmap of the mouse cerebellar cortex with vermis lobules indicated numerically and Aldoc/zebrin stripes indicated in grey, with medial-lateral width expanded 5x for clarity, was redrawn from Fujita et al., 2014. Inferior olive subnuclei of the caudal MAO are denoted a, b, c, d and beta. Circles around each fastigial nucleus cell type indicate relative size and parasagittal position. Abbreviations, 7N, facial nucleus; CL, centrolateral thalamic nucleus; Cr I, Crus I; Cr II, Crus II; INC, interstitial nucleus of Cajal; IRt, intermediate reticular nucleus; IVN, inferior vestibular nucleus; KF, Kölliker-Fuse nucleus; LDTg, laterodorsal tegmental nucleus; LPAG, lateral periaqueductal gray; LPGi, lateral paragigantocellular nucleus; LVN, lateral vestibular nucleus; MAO, medial accessory olive; MD, mediodorsal thalamic nucleus; MDJ, mesodiencephalic junction; MdV, medullary reticular nucleus, ventral; mRt, mesencephalic reticular formation; MVN, medial vestibular nucleus; NPH, nucleus prepositus hypoglossi; NI, nucleus incertus; NRG, nucleus reticularis gigantocellularis; PCRt, parvocellular reticular nucleus; PF, parafascicular thalamic nucleus; PMn, paramedian reticular nucleus; PnC, pontine reticular nucleus, caudal; PPRF, paramedian pontine reticular formation; PTg, pedunculotegmental nucleus; Sim, simplex lobule; SC, superior colliculus; SNc, substantia nigra, pars compacta; SubC, subcoeruleus nucleus; SuM, supramammillary region; VL, ventrolateral thalamic nucleus; vlPAG, ventrolateral periaqueductal gray; VM, ventromedial thalamic nucleus; ZI, zona incerta.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6J | Jackson Laboratory | stock 000664 | |
| Genetic reagent (Mus musculus) | YFP-16; Tg(Thy1-YFP)16Jrs | Dr. Joshua R Sanes | MGI:3505585 | PMID:11086982 |
| Genetic reagent (Mus musculus) | GlyT2-EGFP; Tg(Slc6a5-EGFP)1Uze | Dr. Hanns Ulrich Zeilhofer | MGI:3835459 | PMID:15611994 |
| Genetic reagent (Mus musculus) | Gad2-nls-mCherry; STOCK Gad2tm2(cre)Zjh/J ; Gad22A-mCherry | Jackson Laboratory | stock 023140 | PMID:25913859 |
| Genetic reagent (Mus musculus) | Lhx6-EGFP; Tg(Lhx6-EGFP)BP221Gsat | MMRRC | stock 000246 | PMID:14586460 |
| Genetic reagent (Mus musculus) | L7Cre; B6.129-Tg(Pcp2-cre)2Mpin/J | Jackson Laboratory | stock 004146 | PMID:11105049 |
| Genetic reagent (Mus musculus) | ChatCre; B6N.129S6(B6)-Chattm2(cre)Lowl/J | Jackson Laboratory | stock 018957 | PMID:21284986 |
| Genetic reagent (Mus musculus) | Gad2Cre; B6N.Cg-Gad2tm2(cre)Zjh/J | Jackson Laboratory | stock 019022 | |
| Genetic reagent (Mus musculus) | VgluT2-Cre; STOCK Slc17a6tm2(cre)Lowl/J | Jackson Laboratory | stock 016963 | PMID:21745644 |
| Genetic reagent (Mus musculus) | Ai3; B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J | Jackson Laboratory | stock 007903 | PMID:22446880 |
| Genetic reagent (Mus musculus) | Ai14; B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratory | stock 007908 | PMID:22446880 |
| Genetic reagent (Mus musculus) | Ai32; B6.Cg-Gt(ROSA)26Sortm32(CAG-COP4*H134R/EYFP)Hze/J | Jackson Laboratory | stock 024109 | PMID:22446880 |
| Genetic reagent (Mus musculus) | Sun1-sfGFP; B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J | Jackson Laboratory | stock 021039 | PMID:26087164 |
| Antibody | anti-alpha synuclein (SNCA), mouse monoclonal | BD Bioscience | Cat# 610786, RRID:AB_398107 | dilution 1:100 |
| Antibody | anti-osteopontin (SPP1), goat polyclonal | R and D Systems | Cat# AF808; RRID:AB_2194992 | dilution 1:300 |
| Antibody | anti-calretinin (CALB2), rabbit polyclonal | Millipore | Cat# AB5054; RRID:AB_2068506 | dilution 1:500 |
| Antibody | anti-SMI32 (NEFH), mouse monoclonal | Covance | Cat# SMI-32R-100; RRID:AB_509997 | dilution 1:1000 |
| Antibody | anti-tyrosine hydroxylase (TH), rabbit polyclonal | Millipore | Cat# AB152; RRID:AB_390204 | dilution 1:1000 |
| Antibody | anti-Chat, goat polyclonal | MilliporeSigma | Cat# AB144P; RRID:AB_2079751 | dilution 1:500 |
| Antibody | anti-Aldoc, rabbit polyclonal | Dr. Izumi Sugihara; PMID:15470143 | RRID:AB_2313920 | dilution 1:8000 of14.66 mg/mL stock |
| Antibody | anti-PLCb4, rabbit polyclonal | Santa Cruz Biotechnology | Cat# sc-20760 | dilution 1:100 |
| Antibody | anti-orexin, rabbit polyclonal | Peninsula Laboratories | Cat# T4074; RRID:AB_2315020 | dilution 1:500 |
| Antibody | anti-GFP, rabbit polyclonal | Millipore | Cat# AB3080; RRID:AB_91337 | dilution 1:1000 |
| Antibody | anti-GFP, chicken polyclonal | Aves Labs | Cat# GFP-1020; RRID:AB_2307313 | dilution 1:1000 |
| Antibody | anti-RFP, rabbit polyclonal | MBL International | Cat# PM005; RRID:AB_591279 | dilution 1:1000 |
| Recombinant DNA reagent | AAV1.hSyn.Cre.WPRE.hGH | Penn Vector Core | Lot. CS1087 | titer 4.37 × 1013 GC/ml |
| Recombinant DNA reagent | AAV1.hSyn.eGFP.WPRE.bGH | Penn Vector Core | Lot.CS0500-3CS | titer 2.24 × 1013 GC/ml |
| Recombinant DNA reagent | AAV1.hSyn.TurboRFP.WPRE.rBG | Penn Vector Core | Lot. V3731TI-R | titer 2.15 × 1013 GC/ml |
| Recombinant DNA reagent | AAV2retro.CAG.Cre | UNC Vector Core | Lot. AV7703C | titer 5.3 × 1012 GC/ml |
| Recombinant DNA reagent | AAV9.CAG.Flex.eGFP.WPRE.bGH | Penn Vector Core | Lot. CS0374 | titer 2.28 × 1013 GC/ml |
| Recombinant DNA reagent | AAV9.CAG.Flex.tdTomato.WPRE.bGH | Penn Vector Core | Lot. CS0634 | titer 1.25 × 1013 GC/ml |
| Recombinant DNA reagent | AAV9.CAG.hChR2(H134R)-mCherry.WPRE.SV40 | Penn Vector Core | Lot. CS0753-3CS | titer 1.71 × 1013 GC/ml |
| Recombinant DNA reagent | AAV9.hSyn.eGFP.WPRE.bGH | Penn Vector Core | Lot. CS0354 | titer 8.88 × 1013 GC/ml |
| Recombinant DNA reagent | AAV9.hSyn.TurboRFP.WPRE.rBG | Penn Vector Core | Lot. V4861MI-R | titer 6.64 × 1013 GC/ml |
| Recombinant DNA reagent | RVdG-mC; glycoprotein deleted recombinant rabies virus carrying mCherry gene | GT3 core at the Salk Institute | titer 9.6 × 109 TU/ml | |
| Chemical compound, drug | dextran (3000 MW), conjugated with biotin | Thermo Fisher Scientific | Cat# D7135 | |
| Chemical compound, drug | dextran (3000 MW), conjugated with Texas Red | Thermo Fisher Scientific | Cat# D3328 | |
| Chemical compound, drug | red retrobeads IX | Lumafluor | ||
| Chemical compound, drug | green retrobeads IX | Lumafluor | ||
| Chemical compound, drug | FastBlue | Polysciences, Inc | Cat# 17740–1 |





