Miniaturized 3D bone marrow tissue model to assess response to Thrombopoietin-receptor agonists in patients
Figures
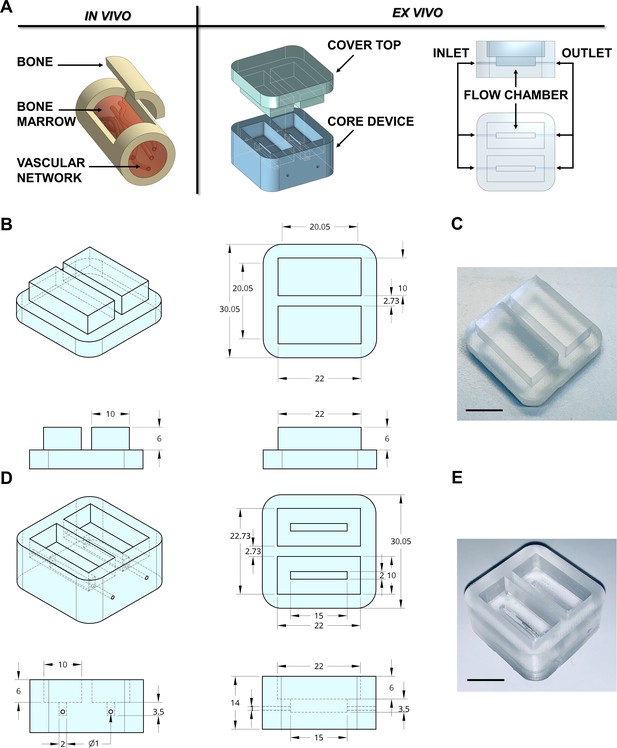
Design of the bone marrow mimicking device.
(A) To mimic the vascularized bone marrow tissue structure ex vivo a double-flow chamber device was designed in two parts. The core contains two separates flow channels dedicated to the perfusion having inlet and outlet ports for connection to a perfusion system. (B,C) The dimension of the polydimethylsiloxane (PDMS) mold cover top and (D,E) the core device is expressed in millimeters. Alternative models of the device are shown in Figure 1—figure supplement 1. The 3D-printed negative mold of the chamber is shown in Figure 1—figure supplement 2.

Various models of the silk bone marrow device.
The devices can be produced from one to four perfusion channels and molded with PDMS (scale bars = 10 mm).
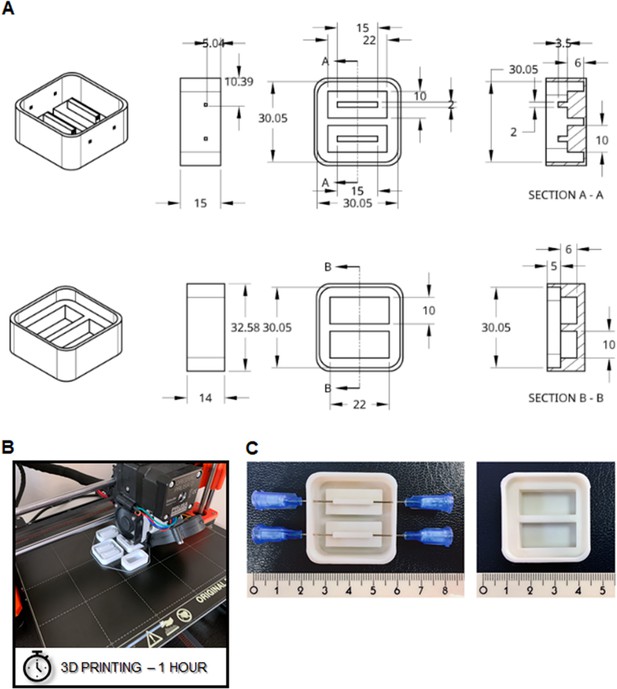
3D printing of the negative mold.
(A) Drawing of negative molds for the core device (upper panel) and the cover tap (lower panel) are disposed in isometric view, right view, top view, and transversal section view, respectively. Dimensions are in millimeters. (B) The production is operated by an FDM 3D printer using PLA and (C) needles are finally mounted to connect the future inlet and outlet for perfusion of the chamber.
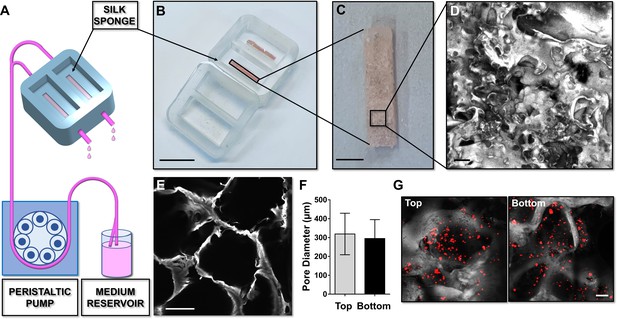
Silk sponge bone marrow perfusion system.
(A–C) A peristaltic pump drives perfusion of the cell culture medium from a reservoir to the device equipped with a silk fibroin sponge prepared directly inside the chamber by dispensing an aqueous silk solution mixed with salt particles (scale bar B = 1.5 cm; scale bar C = 2 mm). After leaching out the salt, the resulting porous silk sponge can be sterilized. (D,E) Confocal microscopy reconstruction of the silk sponge showed the presence of an interconnected alveolar network (scale bar D = 200 µm; scale bar E = 150 µm). (F) The analysis of pore diameters measured on the top and bottom of the scaffold demonstrated no significant differences throughout the scaffold. Results are presented as mean ± SD (n = 150 pore/condition, p=NS). (G) Confocal microscopy analysis of CFSE+ cells cultured within the silk scaffold (red = CFSE; gray = silk; scale bar = 50 µm). The full data set is provided in Figure 2—source data 1.
-
Figure 2—source data 1
Analysis of the pore diameter of the silk scaffolds.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig2-data1-v1.xlsx
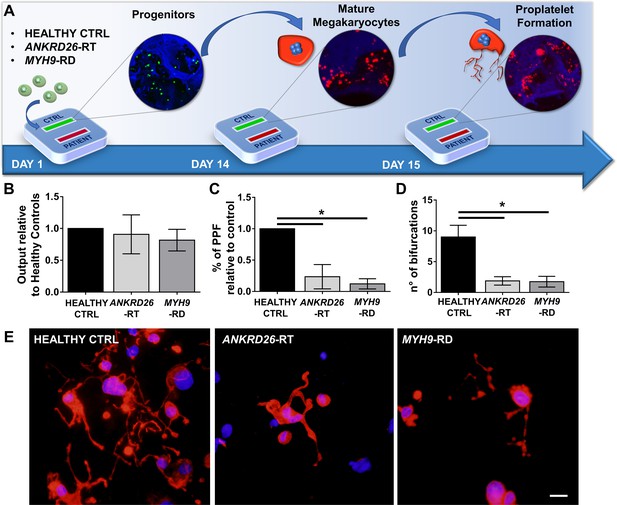
Modeling physiological and pathological megakaryopoiesis.
(A) Megakaryocytes were differentiated from healthy controls and patients affected by MYH9-RD and ANKRD26-RT patients and cultured into the bone marrow device in presence of 10 ng/mL TPO. (B) Output of CD41+CD42b+ megakaryocyte at the end of differentiation relative to healthy controls (n = 12 Healthy Controls, n = 12 MYH9-RD; n = 12 ANKRD26-RT) (C) Percentage of proplatelet formation relative to healthy controls (n = 12 Healthy Controls, n = 12 MYH9-RD; n = 12 ANKRD26-RT; *p<0.01). (D) The number of proplatelet bifurcation per single megakaryocytes in healthy controls and patients (n = 12 Healthy Controls, n = 12 MYH9-RD; n = 12 ANKRD26-RT; *p<0.01). (E) Representative immunofluorescence staining of proplatelet structure (red=β1-tubulin; blue = nuclei; scale bar = 20 µm). All results are presented as mean ± SD. Data from the treatment of healthy controls in the presence of TPO and TPO +EPAG are shown in Figure 3—figure supplement 1. The full data set is provided in Figure 3—source data 1.
-
Figure 3—source data 1
Analysis of megakaryocyte differentiation and proplatelet formation in healthy controls and patients.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig3-data1-v1.xlsx
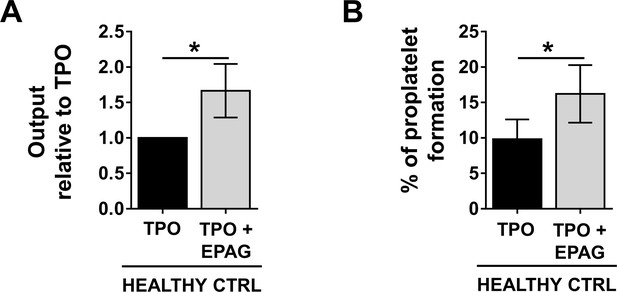
Eltrombopag is effective in promoting thrombopoiesis from healthy controls.
(A) Output of CD41+CD42b+ healthy control megakaryocytes at the end of differentiation cultured in the presence of TPO +EPAG, with respect to TPO alone (n = 6, *p<0.01). (B) The percentage of proplatelet forming-megakaryocytes was calculated as the number of cells displaying long filamentous pseudopods with respect to the total number of round megakaryocytes per analyzed field (n = 6; *p<0.01). All results are presented as mean ± SD. The full data set is provided in Figure 3—source data 1.
-
Figure 3—figure supplement 1—source data 1
Analysis of megakaryocyte output and proplatelet formation in healthy controls in the presence of EPAG.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig3-figsupp1-data1-v1.xlsx
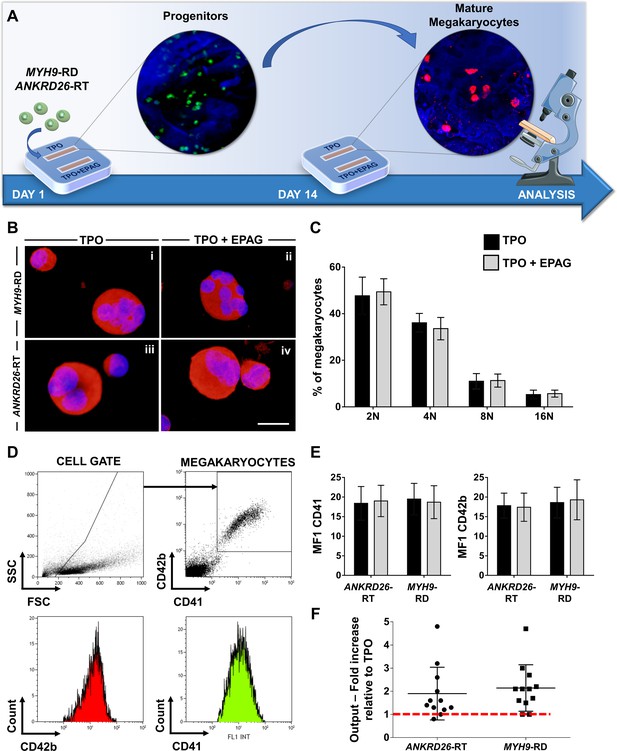
Eltrombopag promotes megakaryocyte differentiation ex vivo.
(A) Megakaryocytes were differentiated from peripheral blood progenitors of patients affected by MYH9-RD or ANKRD26-RT and cultured in the silk bone marrow tissue device in the presence of 10 ng/mL TPO supplemented or not with 500 ng/mL Eltrombopag (EPAG) and analyzed. The figure of the microscope was adapted from Servier Medical Art licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com). (B) Representative immunofluorescence staining of CD61 (red = CD61; blue = nuclei; scale bar = 25 µm) and (C) analysis of ploidy levels at the end of the culture (TPO: n = 3 MYH9-RD; n = 3 ANKRD26-RT; TPO +EPAG: n = 3 MYH9-RD; n = 3 ANKRD26-RT; p=NS). (D) Representative flow cytometry analysis of CD41+CD42b+ megakaryocytes at the end of the culture and (E) statistical analysis of mean fluorescence intensity (MFI) of the markers (TPO: n = 12 MYH9-RD; n = 12 ANKRD26-RT; TPO +EPAG: n = 12 MYH9-RD; n = 12 ANKRD26-RT; p=NS). (F) Output was calculated as the fold increase in the percentage of CD41+CD42b+ cells in presence of TPO +EPAG with respect to the percentage of double-positive cells in presence of TPO alone (ANKRD26-RT: n = 12, p<0.05; MYH9-RD: n = 12, p<0.01). All results are presented as mean ± SD. The full data set is provided in Figure 4—source data 1.
-
Figure 4—source data 1
Analysis of megakaryocyte differentiation.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig4-data1-v1.xlsx
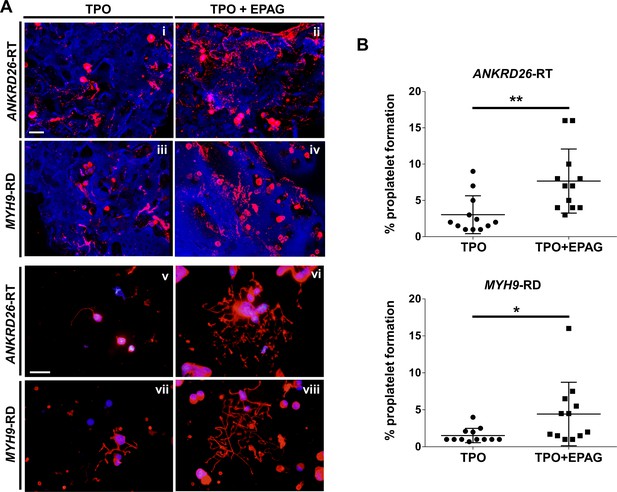
Eltrombopag sustains increased proplatelet formation ex vivo.
(A) Confocal microscopy analysis of 3D megakaryocyte culture imaged at the end of differentiation. Megakaryocytes were elongating proplatelet shafts, which assembled nascent platelets at their terminal ends, within the hollow space of silk pores (red = CD61, blue = silk) (scale bars = 50 µm). (Av-viii) Analysis of proplatelet structure was performed by immunofluorescence staining of the megakaryocyte-specific cytoskeleton component β1-tubulin (red=β1-tubulin; blue = nuclei; scale bar = 25 µm). In both diseases, the representative pictures show increased elongation and branching of proplatelet shafts in presence of TPO +EPAG with respect to TPO alone. (B) The percentage of proplatelet forming megakaryocytes was calculated as the number of cells displaying long filamentous pseudopods with respect to the total number of round megakaryocytes per analyzed field (TPO: n = 12 MYH9-RD; n = 12 ANKRD26-RT; TPO +EPAG: n = 12 MYH9-RD; n = 12 ANKRD26-RT; **p<0.01; *p<0.05). All results are presented as mean ± SD. The full data set is provided in Figure 5—source data 1.
-
Figure 5—source data 1
Analysis of proplatelet formation.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig5-data1-v1.xlsx
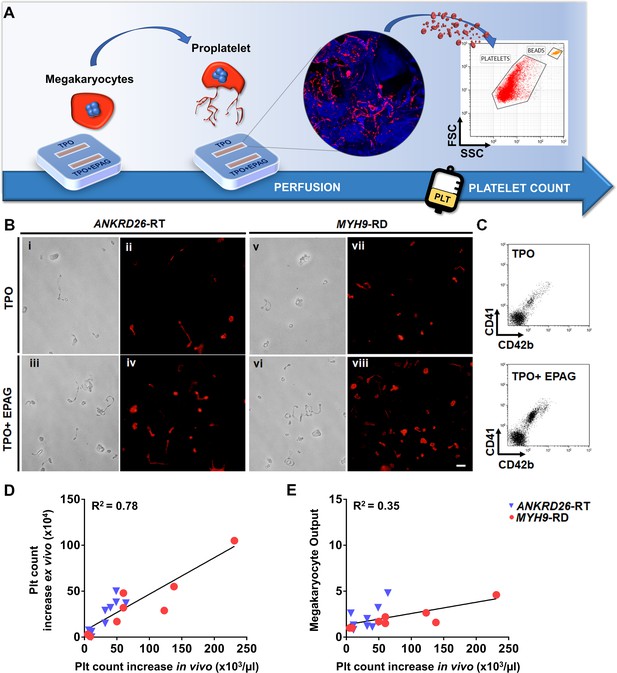
Ex vivo platelet count for predicting response to treatments.
(A) The flow chamber was perfused with culture media and released platelets collected into gas-permeable bags before counting by flow cytometry. (B) Light microscopy and immunofluorescent analysis of the collected medium demonstrated the presence of large pre-platelets, dumbbells, and little discoid platelets having the microtubule coil typically present in resting platelets (red=β1-tubulin, scale bars = 10 µm). (C) Representative flow cytometry analysis of expression of CD41 and CD42b surface markers. (D) Analysis of the correlation between the increase of platelet count analyzed ex vivo and the increase of platelet count observed in vivo from the same patients. For the ex vivo analysis, platelet count was calculated by flow cytometry with counting beads (n = 8 MYH9-RD; n = 9 ANKRD26-RT). (E) Analysis of the correlation between ex vivo megakaryocyte output and the increase of platelet count observed in vivo from the same patients. (n = 8 MYH9-RD; n = 9 ANKRD26-RT). The full data set is provided in Figure 6—source data 1.
-
Figure 6—source data 1
Analysis of platelet count and megakaryocyte output ex vivo, and correlation with platelet count in vivo.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig6-data1-v1.xlsx
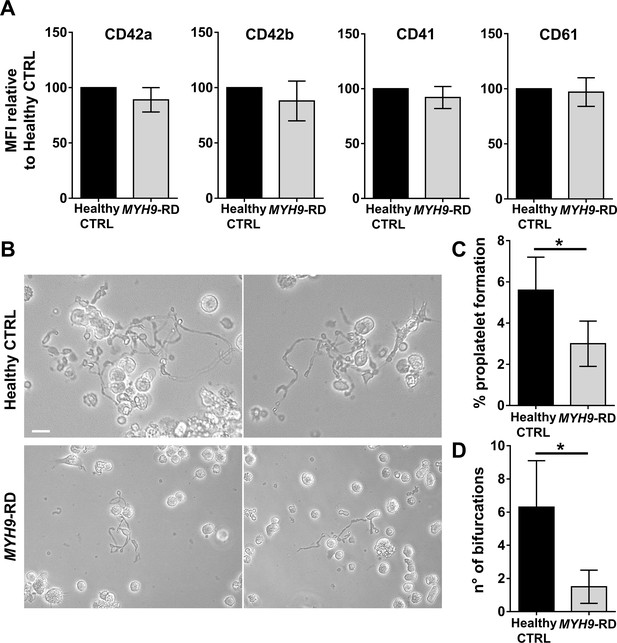
Assessment of iPSC megakaryocyte differentiation.
(A) iPSC clones were cultured for 18 days and analyzed by flow cytometry to assess megakaryocytes differentiation. Histograms show the mean fluorescence intensity (MFI) of MYH9-RD clones for CD42a, CD42b, CD41, and CD61 markers, relative to healthy controls (n = 3 Healthy Controls, n = 3 MYH9-RD; p=NS). (B) Representative images of proplatelet forming-megakaryocytes at day 19 of culture from different iPSC clones. (C) Percentage of proplatelet formation from the different genotypes (n = 6 Healthy Control; n = 6 MYH9-RD (each clone repeated two times); *p<0.05). (D) The number of bifurcation per single megakaryocyte from the different genotypes (*p<0.01). All results are presented as mean ± SD. The full data set is provided in Figure 7—source data 1. Assessment of iPSC clone pluripotency is shown in Figure 7—figure supplement 1. Morphological and genomic characterization of iPSC clones is shown in Figure 7—figure supplement 2. iPSCs haematopoietic differentiation is shown in Figure 7—figure supplement 3. Embryoid Bodies and trilineage differentiation of iPSC clones is shown in Figure 7—figure supplement 4.
-
Figure 7—source data 1
Analysis of iPSC differentiation and proplatelet formation.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig7-data1-v1.xlsx
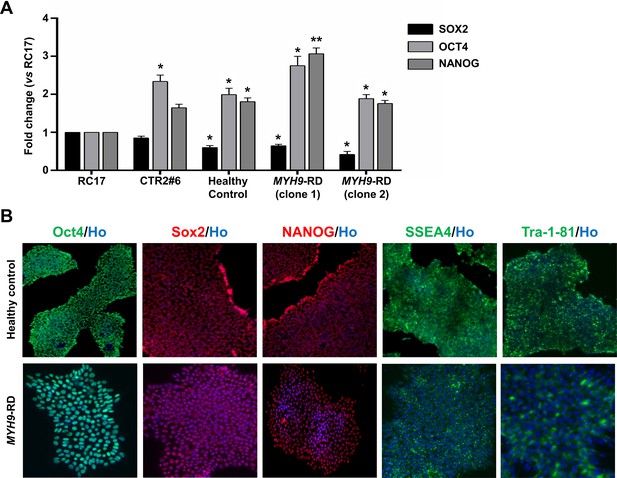
Characterization of iPSCs.
(A) Quantitative RT-PCR analysis of SOX2, NANOG, and OCT4 mRNA expression levels in healthy control and MYH9-RD clones. Expression levels were normalized relative to hESCs RC17. A control iPSC cell line (CTR2#6) was used as internal control. Results are presented as mean ± SEM (n = 2/clone; *p<0.05; **p<0.01). (B) Representative immunofluorescence staining of pluripotency markers OCT4, SOX2, NANOG, SSEA-4, and Tra-1–81.
-
Figure 7—figure supplement 1—source data 1
Analysis of gene expression in iPSCs.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig7-figsupp1-data1-v1.xlsx
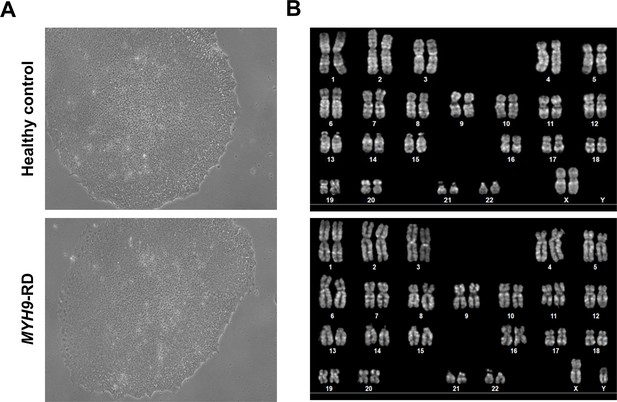
Morphological and genomic characterization of iPSC clones.
(A) Representative phase-contrast images of healthy controls and mutated clones. Magnification ×10. (B) Representative karyotype analysis showed a normal karyotype (healthy controls: upper panel, 46,XX; MYH9-RD bottom panel, 46,XY).
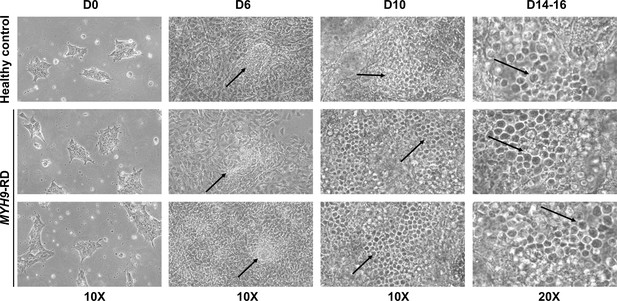
iPSCs hematopoietic differentiation.
Representative phase-contrast microscope images of iPSC colonies at day 0, hematopoietic progenitor.
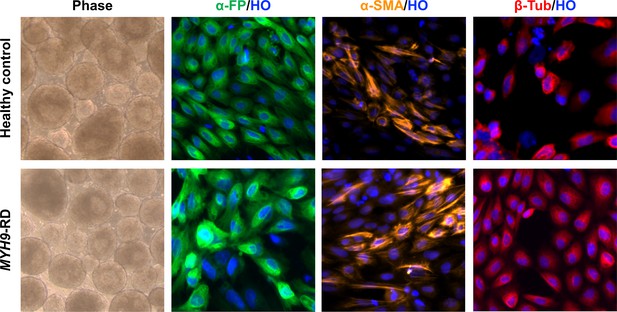
Embryoid Bodies and trilineage differentiation of iPSC clones.
Phase: day 4 embryoid body culture (10 x). After 14 days, differentiated cultures exhibited the presence of cells immune-positive for endodermal (a–FP), mesodermal (a-SMA), and ectodermal (bIII-Tubulin), germ-layer markers. Magnification: ×10.
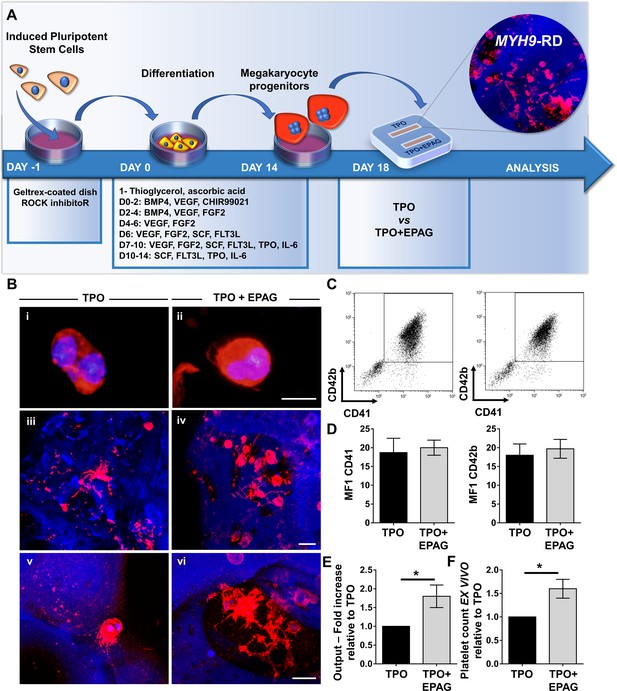
Validation of the system with iPSC mutated clones.
(A) Megakaryocytes were differentiated from iPSCs of patients affected by MYH9-RD and cultured for 14 days in a petri dish before passing into the bone marrow device in presence of 10 ng/mL TPO supplemented or not with 500 ng/mL EPAG. (B) Representative immunofluorescence staining of CD61 megakaryocytes (i,ii) and confocal microscopy analysis (iii-vi) of 3D megakaryocyte culture imaged at the end of differentiation. Megakaryocytes are elongating proplatelet shafts, which assemble nascent platelets at their terminal ends, within the hollow space of silk pores (red = CD61, blue = silk, scale bars = 50 µm). (C) Representative flow cytometry analysis of CD41+CD42b+ megakaryocytes at the end of the culture and (D) statistical analysis of mean fluorescence intensity (MFI) of the markers (n = 3 TPO, n = 3 TPO +EPAG; p=NS). (E) Output was calculated as the number of CD41+CD42b+ cells in presence of TPO +EPAG with respect to the percentage of double-positive cells in presence of TPO alone (n = 3 TPO, n = 3 TPO +EPAG; *p<0.05). (F) Platelet number was calculated by counting beads after perfusing the chamber. The fold increase was calculated as the number of ex vivo platelet count in the presence of TPO +EPAG with respect to TPO alone (n = 3 TPO, n = 3 TPO +EPAG; *p<0.05). All results are presented as mean ± SD. The full data set is provided in Figure 8—source data 1.
-
Figure 8—source data 1
Analysis of iPSC differentiation and platelet production in the presence of EPAG.
- https://cdn.elifesciences.org/articles/58775/elife-58775-fig8-data1-v1.xlsx
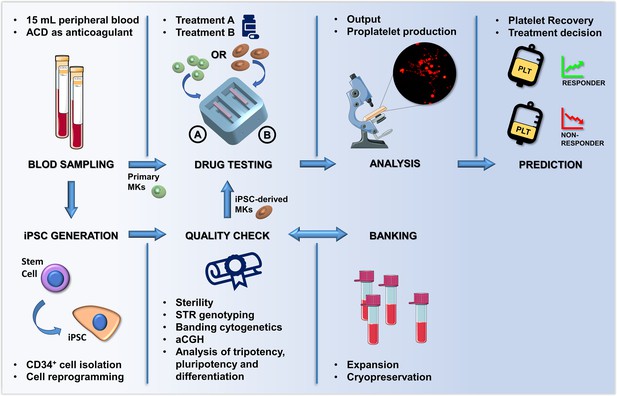
Summary of the proposed workflow.
After sampling, hematopoietic stem and progenitors cell from patients can be differentiated into primary megakaryocytes (MKs) or transformed into induced Pluripotent Stem Cells (iPSCs). iPSC are subjected to quality check, expansion and banking. Megakaryocytic progenitors differentiated either from primary stem cells or iPSCs are seeded within a 3D bone marrow tissue device, cultured in the presence of the tested drug, and analyzed. After perfusion, platelets are collected and counted in order to assess patient-specific response. The figure of the microscope and tubes was adapted from Servier Medical Art licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com).
Tables
Main features of the study population.
| ANKRD26-RT | MYH9-RD | |
|---|---|---|
| Total samples, no. | 12 | 12 |
| M/F | 9/3 | 5/7 |
| Age - mean [range], years | 46 [22-67] | 48 [26-59] |
| Platelet count - mean [range] x109/L | 32 [9-75] | 29 [5-69] |
Main features of the study population treated with Eltrombopag in vivo and ex vivo.
| ANKRD26-RT | MYH9-RD | |
|---|---|---|
| Patients treated with Eltrombopag in vivo and ex vivo, no. | 6† | 7* |
| M/F | 6/3 | 2/6 |
| Age - mean [range], years | 47 [22-67] | 45 [31-59] |
| Platelet count at baseline IN VIVO mean [range], x109/L | 36 [12-75] | 24 [5-69] |
| Increase of platelet count after Eltrombopag treatment IN VIVO‡ - mean [range], x109/L | 34 [7-64] | 88 [5-231] |
| Platelet count EX VIVO – TPO mean [range], x104 | 8.3 [6-13] | 7.8 [5-12] |
| Increase of platelet count EX VIVO§ – TPO + EPAG mean [range], x104 | 24 [0–50] | 36 [1-105] |
-
* of whom one patient repeated two times ex vivo.
† of whom three patients repeated two times ex vivo.
-
‡ Increase of platelet count with Eltrombopag with respect to baseline.
§ Increase of platelet count with Eltrombopag with respect to the untreated counterpart (TPO only).
Comparison of silk-bone marrow models for megakaryocyte culture.
| Di Buduo et al., 2015 | Di Buduo et al., 2017 | Current Manuscript | |
|---|---|---|---|
| Size of the cell-seeding well | 15 × 20 × 5 mm (1500 mm3) | 3.5 × 20 × 5 mm (350 mm3) | 2 × 15 × 3.5 mm (105 mm3) |
| No. of chambers that can be perfused in parallel | Max. 2 | Max. 1 | >2 (up to at least 4) |
| Source of blood | Human Umbilical Cord Blood | Human Umbilical Cord Blood | Human Peripheral Blood |
| Type of hemopoietic stem and progenitor cells | CD34+ | CD34+ | - CD45+ - CD34+-derived iPSCs |
| Type of cells seeded | CD34+-derived megakaryocytes | CD34+-derived megakaryocytes | - CD45+-derived megakaryocyte progenitors - iPSC-derived megakaryocyte progenitors |
| No. of cells seeded | 2.5 × 105 | 4 × 105 | 5 × 104 |
| Time of the culture | 24 hr | 24 hr | >7 days |
| Major application | Studying mechanisms of thrombopoiesis | Proof of concept for scaling up platelet production | Drug Testing in individual patients |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (human) | ANKRD26 – ankyrin repeat domain 26 | GenBank | THC2, bA145E8.1 | OMIM 188000 |
| Gene (human) | MYH9 – myosin heavy chain 9 | GenBank | BDPLT6, DFNA17, EPSTS, FTNS, MATINS, MHA, NMHC-II-A, NMMHC-IIA, NMMHCA | OMIM 155100 |
| Genetic reagent (human) | STR D21S11, D7S820, CSF1PO, TH01, D13S317, D16S539, vWA, TPOX, D5S818 Amelogenin | Promega GenePrint | Genetic multi-locus human DNA profile Genetic loci | Cell line authentication |
| Cell line (human) | Human embryonic stem cell | ISENET Biobank | Embryonic stem cell line RCe021-A (RC-17) Rosalin cells, RRID:CVCL_L206 | Control embryonic stem cell (human) |
| Cell line (human) | iPSC generated from healthy control | This paper | CTR2#6 | Control iPSC (human) |
| Cell line (human) | iPSC generated from healthy control | This paper | CPN | Control iPSC (human) |
| Cell line (human) | iPSC generated from MYH9-RD patient | This paper | MH | Patient iPSC (human) |
| Transfected construct (human) | Sindai virus OCT4, KLF4, Sox2, cMyc | Life Technologies | CytoTune-iPS Sendai Reprogramming | Generation of iPSCs (human) |
| Biological sample (human) | Peripheral Blood | - | - | - |
| Commercial assay or kit | CytoTune-iPS 2.0 Sendai Reprogramming Kit | Invitrogen/Thermo Fisher Scientific | #A16517 | Generation of iPSCs (human) |
| Commercial assay or kit | Venor GeM | Minerva Biolabs | #11–1250 | Mycoplasma detection |
| Commercial assay or kit | Qiagen DNA Mini Kit | Qiagen | #A31881 | DNA extraction |
| Commercial assay or kit | TruCount | Becton - Dickinson | #340334 | FACS Counting Beads |
| Commercial assay or kit | Slide-A-Lyzer Dyalisis Cassettes, 3.5K MWCO, 12 mL | Thermo Scientific | #66110 | Dyalisis |
| Commercial assay or kit | MiniMACS Starting Kit | Miltenyi Biotec | # 130-090-312 | Cell sorting |
| Antibody | Anti-Human CD61 (Mouse Monoclonal) | Beckman Coulter | #IM0540, RRID:AB_2889176 | 1:100 |
| Antibody | Anti - CD45 Microbeads (Mouse Monoclonal) | Miltenyi Biotec | #130-045-801, RRID:AB_2783001 | 20 μl / 107 cells |
| Antibody | Anti - CD61 Microbeads (Mouse Monoclonal) | Miltenyi Biotec | #130-051-101 RRID:AB_2889174 | 20 μl / 107 cells |
| Antibody | Alexa Fluor 594 (Goat Anti Mouse) | ThermoFisher Scientific | # A11005, RRID:AB_2534073 | 1:500 |
| Antibody | FITC anti-human CD41 Antibody (Mouse Monoclonal) | BioLegend | #303703, RRID:AB_314373 | 5 μl Megakaryocytic marker |
| Antibody | PE anti-human CD42b Antibody (Mouse Monoclonal) | BioLegend | #303905, RRID:AB_314385 | 5 μl Megakaryocytic marker |
| Antibody | a-FP (Mouse Monoclonal) | R and D system | #MAB1369, RRID:AB_2258005 | 1:50 Trilineage differentiation |
| Antibody | a-SMA (Mouse Monoclonal) | Millipore Sigma | #A2547, RRID:AB_476701 | 1:200 Trilineage differentiation |
| Antibody | bIII-Tubulin (Mouse Monoclonal) | Millipore Sigma | #MAB1637, RRID:AB_2210524 | 1:100 Trilineage differentiation |
| Antibody | SOX2 (Rabbit Monoclonal) | Abcam, | #Ab97959, RRID:AB_2341193 | 1:300 Pluripotency markers |
| Antibody | Nanog (Mouse Monoclonal) | Millipore | #MABD24, RRID:AB_11203826 | 1:500 Pluripotency markers |
| Antibody | SSEA4 (Mouse Monoclonal) | Millipore | #MAB4304, RRID:AB_177629 | 1:50 Pluripotency markers |
| Antibody | Tra1-81 (Mouse Monoclonal) | Millipore | #MAB4381C3, RRID:AB_2889175 | 1:50 Pluripotency markers |
| Sequence-based reagent | GenePrint 10 System | Promega | #B9510 | STR genotyping |
| Peptide, recombinant protein | Recombinant Human Thrombopoietin (TPO) | Peprotech | #300–18 | Cytokine |
| Peptide, recombinant protein | Recombinant Human interleukin-11 (IL-11) | Peprotech | #200–11 | Cytokine |
| Peptide, recombinant protein | Recombinant Human interleukin-6 (IL-6) | Peprotech | #200–6 | Cytokine |
| Peptide, recombinant protein | Recombinant Human Fibroblast Growth Factor (FGF) | Peprotech | #100-18B | Cytokine |
| Peptide, recombinant protein | Recombinant Fms-related tyrosine kinase three ligand (Flt3L) | Peprotech | #300–19 | Cytokine |
| Peptide, recombinant protein | Recombinant Human Stem Cell Factor (SCF) | Peprotech | #300–07 | Cytokine |
| Peptide, recombinant protein | Recombinant Human VEGF165 | Peprotech | #100–20 | Cytokine |
| Chemical compound, drug | CHIR 99021 | Tocris | #4423 | Pharmacological Inhibitor |
| Chemical compound, drug | Lithium Bromide (LiBr) | Sigma-Aldrich | #213225 | Silk processing |
| Chemical compound, drug | Penicillin – Streptomycin 100X | Euroclone | #EB3001D | Antibiotics |
| Chemical compound, drug | Paraformaldehyde (PFA) | Sigma - Aldrich | #158127 | Fixation |
| Chemical compound, drug | Triton X-100 | Sigma - Aldrich | #X100 | Permeabilization |
| Chemical compound, drug | Eltrombopag | Novartis | N/A | Drug Testing |
| Software algorithm | GeneMapper Software version 4.0 | Applied Biosystem | #4440915, RRID:SCR_014290 | Genotyping Analysis |
| Other | Human Fibronectin | Becton Dickinson | # 354008 | Silk functionalization |
| Other | Poly(lactic acid) | FormFutura | - | Scaffolding |
| Other | Pluronic F-127 | Sigma-Aldrich | #P2443 | 25% |
| Other | PDMS (SYLGARD 184 Silicone Elastomer) | Dow Corning | # 1673921 | Scaffolding |
| Other | Dulbecco’s Phosphate Buffered Saline (PBS) | Euroclone | #ECB4053L | Saline buffer |
| Other | Essential 8 Flex media | Gibco/Thermo Fisher | #A2858501 | Culturing media |
| Other | StemSpan SFEM Medium | Voden | #09650 | Culture medium |
| Other | E8 medium | Gibco/Thermo Fisher Scientific | #A1517001 | Culture medium |
| Other | L-Glutamine 100X | Euroclone | #ECB300D | 1% |
| Other | Hoechst 33258 | Sigma - Aldrich | #861405 | 1:10000 |
| Other | ProLong Gold antifade reagent | Invitrogen | #P36980 | Microscopy |
| Other | CFSE Cell Division Tracker Kit | Biolegend | #423801 | 5 mM |
| Other | Geltrex | Gibco/Thermo Fisher Scientific | #A1413202 | Matrix |
| Other | VTN-N | Gibco/Thermo Fisher Scientific | #A14700 | Matrix |
| Other | KaryoMax Colcemid | Gibco - Sigma | 329749009 | Chromosome/metaphase banding Genomic stability |
| Other | Quinacrine | Sigma/Merck | Q3251 | Chromosome/metaphase banding Genomic stability |
| Other | aCGH | Agilent Technologies | SurePrint G3 Human CGH Microarray 8 × 60K | DNA Analytics software v.5CNVs |
STR-genotype of iPSCs and donor cells.
Numbers in each locus refer to the number of repeats in each allele.
| Cell line | TH01 | D21S11 | D5S818 | D13S317 | D7S820 | D16S539 | CSF1PO | AMEL | vWA | TPOX |
|---|---|---|---|---|---|---|---|---|---|---|
| CPN | 9.3, 9.3 | 28, 33.2 | 12, 12 | 8,11 | 10,11 | 11,11 | 10,12 | X,X | 16,16 | 8,8 |
| MH | 7,7 | 32.2, 33.2 | 12,13 | 11,13 | 9,11 | 12,12 | 11,11 | X,Y | 15,16 | 11,11 |
| MH-1 | 7,7 | 32.2, 33.2 | 12,13 | 11,13 | 9,11 | 12,12 | 11,11 | X,Y | 15,16 | 11,11 |
| MH-11 | 7,7 | 32.2, 33.2 | 12,13 | 11,13 | 9,11 | 12,12 | 11,11 | X,Y | 15,16 | 11,11 |
Molecular karyotype.
aCGH analysis was performed by SurePrint G3 Human CGH Microarray Kit 60K and revealed CNVs on chromosomes 2 and 3 in MH-11 cells. No genomic instability was observed in CPN cells.
| Cell line | Molecular karyotype |
|---|---|
| CPN | arr[hg18] (1–22,X)x2 |
| MH-11 | arr[hg18] 2p16.1 (60,966,923–61,003,123)x6, 3q13.31(117,895,555-118,070,129)x7 |
Primer sequences for qRT-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| SOX2 | GACAGAGCCCATTTTCTCCA | AAATCCTGTCCTCCCATTCC |
| NANOG | TATGCCTGTGATTTGTGGGC | GTTTGCCTTTGGGACTGGTG |
| OCT4 | AGAGGATCACCCTGGGATAT | CGCCGGTTACAGAACCACAC |
| GAPDH | TGTTCGACAGTCAGCCGCAT | TAAAAGCAGCCCTGGTGACC |
| Blood 2015 | Biomaterials 2017 | Current Manuscript | |
|---|---|---|---|
| Size of the cell-seeding well | 15x20x5 mm (1500mm3) | 3.5x20x5 mm (350mm3) | 2x15x3.5 mm (105mm3) |
| No. of chambers that can be perfused in parallel | Max. 2 | Max. 1 | > 2 (up to at least 4) |
| Source of blood | Human Umbilical Cord Blood | Human Umbilical Cord Blood | Human Peripheral Blood |
| Type of haemopoietic stem and progenitor cells | CD34+ | CD34+ |
|
| Type of cells seeded | CD34+-derived megakaryocytes | CD34+-derived megakaryocytes |
|
| No. of cells seeded | 2.5x105 | 4x105 | 5x104 |
| Time of the culture | 24 hours | 24 hours | > 7 days |
| Major application | Studying mechanisms of thrombopoiesis | Producing a high number of platelets | Drug Testing in individual patients |





